Abstract
Background: MicroRNAs (miRNAs) are a class of small non-coding RNAs that have been suggested to play an essential role in tumorigenesis. miR-206 functions as a tumor suppressor in several cancers. However, its role in non small cell lung cancer (NSCLC) remains unclear. Methods: Expression levels of miR-206 in NSCLC tissues and cell lines were determined by quantitative real-time PCR (qRT-PCR). Then, we investigated the role of miR-206 on NSCLC cell proliferation, migration and invasion. Furthermore, luciferase reporter assay was performed to confirm the target gene of miR-206 and the results were validated in NSCLC cells. Results: In the present study, our results showed that miR-206 was decreased in NSCLC tissues compared with adjacent non-tumor tissues. Forced overexpression of miR-206 significantly inhibited cell proliferation, migration and invasion of NSCLC cells. SOX9 was found to be a target of miR-206. Furthermore, down-regulation of SOX9 by shRNA performed similar effects with overexpression of miR-206. Conclusions: Our study suggested that miR-206 acts as tumor suppressor in NSCLC partially via targeting SOX9.
Keywords: Non small cell lung cancer, miR-206, SOX9, proliferation, invasion
Introduction
Lung cancer is the leading cause of cancer-related deaths around the world and the majority of lung cancer cases are non small cell lung cancer (NSCLC), which accounts for approximately 80% of all lung cancer cases [1]. Despite the improvements in therapeutic modalities, the 5-year survival rate of NSCLC patients is still around 15% [2]. Thus, it is necessary to explore the molecular mechanisms and to facilitate the treatment of NSCLC.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules, 19-25 nucleotides in length that regulate gene expression post-transcriptionally by targeting mRNAs [3]. MiRNAs bind to the 3’-untranslated region (3’UTR) of target mRNAs leading to translational repression or degradation of mRNA [4]. Accumulating evidence shows that miRNAs play critical roles in the regulation of cancer initiation and progression [5]. MiRNAs might serve as new therapeutic strategies for cancers, as they can act as tumor suppressors or as oncogenes dependent on their target mRNAs [6]. For example, Zhao et al showed expression of the miR-497 was significantly decreased in renal cancer and associated with tumor stage, histological grade and lymph node metastases [7]. Sun et al revealed that miR-646 was significantly decreased in osteosarcoma and inhibited cell proliferation, migration and invasion by targeting FGF2 [8]. Yang et al suggested that miR-221/222 were significantly upregulated in human glioma and promote glioma cell invasion and angiogenesis by targeting TIMP2 [9]. However, the role of miR-206 in NSCLC remains largely unknown.
In the present study, our results showed that miR-206 was substantially decreased in NSCLC tissues and cell lines, and ectopic overexpression of miR-206 significantly suppressed the proliferation, migration and invasion of NSCLC cells. SOX9 was found to be a target of miR-206, and down-regulation of SOX9 by shRNA performed similar effects with overexpression of miR-206 in NSCLC cells.
Materials and methods
Tissue samples and cell lines
Paired NSCLC and adjacent non-tumor tissues were obtained from 28 patients with informed consent at Huaihe Hospital of Henan University. Tissues were immediately snap frozen and stored at -80°C. Three NSCLC cell lines (A549, H157 and H520) and a normal lung bronchus epithelial cell line BEAS-2B were obtained from American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Invitrogen), and incubated in 5% CO2 humid atmosphere at 37°C.
Transfection
miR-206 mimics and miR-206 negative control (miR-NC) were purchased from RiboBio (Guangzhou). siRNA/SOX9 (siR-SOX9) and its negative control oligonucleotide (siR-NC) were obtained from Applied Biosystems. The transfection were performed using Lipofectamine 2000 (Invitrogen) according to the instructions provided by the manufacturer. The transected cells were resuspended and cultured in regular culture medium for 24 h before analysis.
Construction of plasmid vectors
Previously, the pcDNA/SOX9 vector with the SOX9 coding region was successfully constructed and conserved by our lab. To construct a luciferase reporter vector, the SOX9 3’UTR fragment containing putative binding sites for miR-206 was amplified by PCR using the following primers: sense 5’-TGGAGACTTCTGAACGAGAG-3’ and reverse 5’-CTTGAAGATGGCGTTGGG-3’, and inserted into downstream of the luciferase gene in the pLuc luciferase vector (Ambion) and named SOX9 3’UTR-Wild. Site-directed mutagenesis of the miR-206 target-site in the SOX9-3’UTR was performed using the Quick-change mutagenesis kit (Stratagene) and named SOX9 3’UTR-Mut according to the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR
Total RNA isolated from cells or tissues using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was obtained using M-MLV (Promega). The relative level of miR-206 was detected by using SYBR®Premix Ex TaqTM Kits (TaKaRa) on an ABI 7900 thermocycler (Applied Biosystems). The primers were purchased from GeneCopoeia (Carlsbad). The relative expression level was calculated by normalization with the signal for U6 expression using 2-ΔΔCt method.
Cell proliferation assay
Transfected cells were seeded into 96-well plates with a density of 4000 cells/well, and cultured for different time. 10 μL of MTT was added into each well, and incubated for 4 h. Then the supernatant was discarded, and 200 μL of DMSO was added to each well. Optical density (OD) was detected at the wavelength of 490 nm. Data were derived from three independent experiments.
Migration and invasion assays
For migration, 5×104 transfected cells were seeded into the upper chamber (Millipore). For invasion, 1×105 transfected cells were seeded into the upper chamber precoated with Matrigel (Sigma). DMEM containing 10% FBS was used as a chemoattractant, and added to the lower chamber. After 24 h incubation, membranes were stained with 0.5% crystal violet for 20 min. Invaded cells on the lower membrane were counted under a microscope (Olympus).
Luciferase assay
The constructs were sequenced and named pLuc-SOX9-Wt or pLuc-SOX9-Mut. For reporter assays, A549 cells were cultured in 24-well plates and each transfected with 100 ng of pLuc-SOX9-Wt or pLuc-SOX9-Mut and 50 nM of miR-206 mimics using Lipofectamine 2000. 48 h after transfection, cells were harvested and assayed with Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer’s instructions.
Western blotting
Total protein from cells were lysed by RIPA buffer. Equal amount of proteins were separated by 10% SDS-PAGE, and transferred to PVDF membrane (Millipore), and incubated with primary antibodies against SOX9 (Cell Signaling) and β-actin (Abcam), followed by HRP labeled secondary antibodies for 1 h at room temperature. Blots were visualized the ECL Detection Reagents (Amersham Biosciences).
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software (IBM). All data from three independent experiments were expressed as mean ± SD. Statistical differences were determined by ANOVA or Student t test. P < 0.05 was considered statistically significant.
Results
miR-206 was downregulated in NSCLC tissues and cell lines
Expression levels of miR-206 were detected from NSCLC tissues and cell lines was examined by qRT-PCR as previously described. As shown in Figure 1A, the relative level of miR-206 expression was significantly lower in NSCLC tissues than in corresponding non-tumor tissues. Furthermore, miR-206 was also decreased in three NSCLC cell lines compared with the normal lung bronchus epithelial cell line BEAS-2B (Figure 1B). Thus, the data indicated that downregulation of miR-206 might play important roles in lung cancer tumorigenesis.
Figure 1.
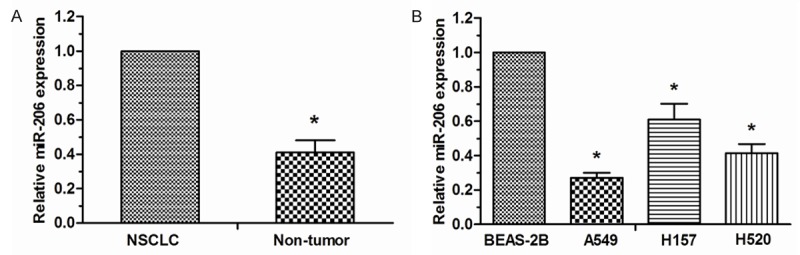
Decreased miR-206 expression in NSCLC tissues and cell lines. A. qRT-PCR analysis of miR-206 expression in NSCLC tissues and adjacent on-tumor tissues from 28 patients. B. qRT-PCR analysis of miR-206 expression in NSCLC cell lines (A549, H157, and H520) and lung bronchus epithelial cell line BEAS-2B. *P < 0.05.
miR-206 suppressed NSCLC cell proliferation, migration and invasion
To investigate the effect of miR-206 expression on cell proliferation, migration and invasion of NSCLC cells, miR-206 mimics was transiently transfected into A549 cells. 48 h after transfection, the level of miR-206 expression was determined by qRT-PCR (Figure 2A). MTT assay showed that overexpression of miR-206 significantly suppressed cell proliferation of A549 cells compared with cell transfected with miR-NC (Figure 2B). Furthermore, we investigated the effect of miR-206 on NSCLC cell migration and invasion. transwell migration assay revealed that upregulation of miR-206 dramatically inhibited tumor cell migration in A549 cells compared with miR-NC group (Figure 2C). Similarly, transwell invasion assay demonstrated that miR-206 markedly decreased the invasive capacity of A549 cells (Figure 2D).
Figure 2.
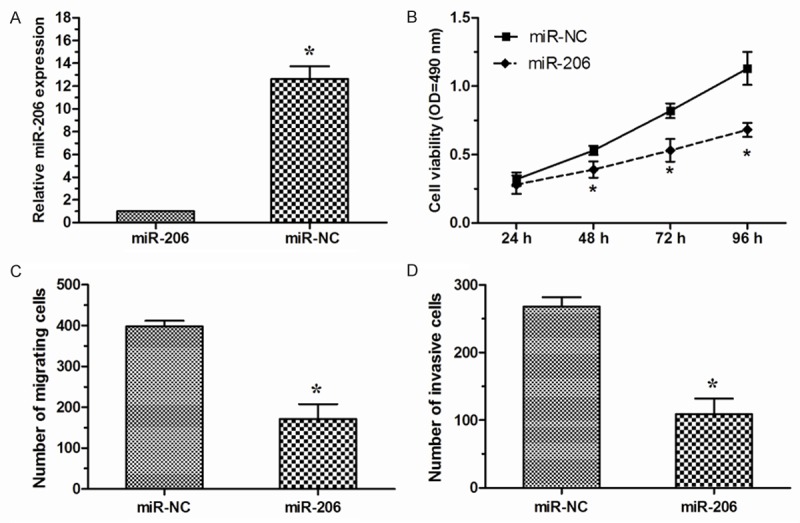
miR-206 inhibited NSCLC cell proliferation, migration and invasion in vitro. A. A549 cells were transfected with miR-206 mimics or miR-NC. The expression level of miR-206 was detected by qRT-PCR. B. MTT assay was performed to investigate the effect of miR-206 on the proliferation of A549 cells. C. Transwell migration assay was performed in A549 cells transiently transfected with miR-206 mimics or miR-NC. D. Transwell invasion assay showing the invasion ability of A549 cells transiently transfected with miR-206 mimics or miR-NC. *P < 0.05.
SOX9 was a target of miR-206 in NSCLC cells
To identify the target of miR-206 in NSCLC, TargetScan 6.2 was used to screen the target gene of miR-206. SOX9 was predicted to be a target of miR-206 (Figure 3A). Luciferase reporter assay showed that miR-206 significantly decreased the luciferase activity of the Wt 3’UTR but not the Mut 3’-UTR of SOX9 in A549 cells (Figure 3B). In addition, overexpression of miR-206 significantly inhibited SOX9 expression (Figure 3C).
Figure 3.
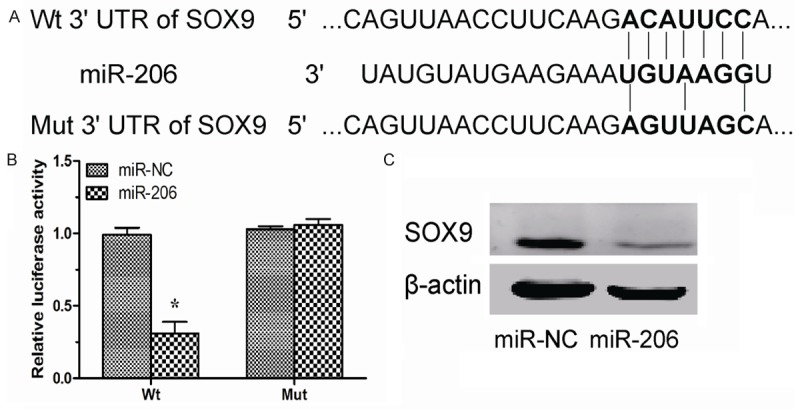
SOX9 was a target of miR-206 in NSCLC cells. A. The potential miR-206 binding sequence of SOX9 3’UTR and the mutant. B. A549 cells were co-transfected with miR-206 mimics or miR-NC with Wt or Mut SOX9 3’UTR. Luciferase activity was assayed. C. Protein level in A549 cells transfected with miR-206 mimics or miR-NC was detected by Western blotting. β-actin was used as a control. *P < 0.05.
Decreased expression of SOX9 showed similar effect with miR-206 overexpression
To study the effect of SOX9 on NSCLC cell growth, A549 cells were transfected with siR-SOX9 or siR-NC. Inhibition of SOX9 by shRNA significantly suppressed the proliferation, migration and invasion of A549 cells (Figure 4A-C). The effect of siR-SOX9 was confirmed by Western blotting (Figure 4D).
Figure 4.
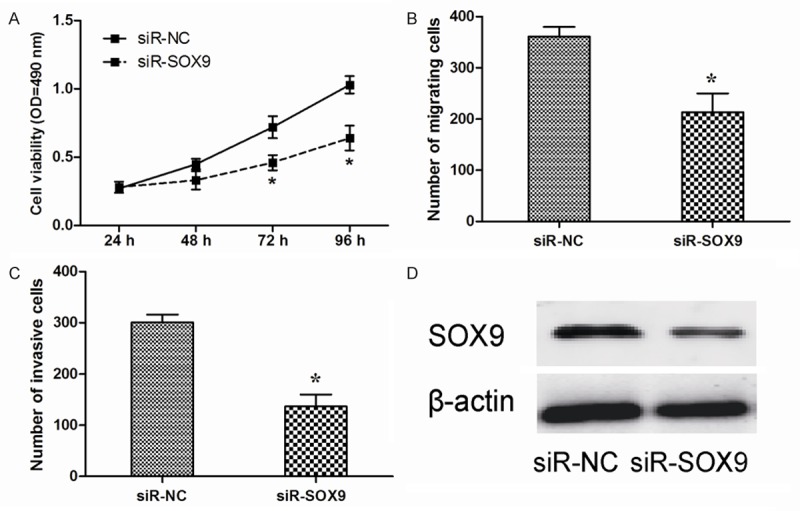
Inhibition of SOX9 showed similar effect with miR-206 overexpression. A. The vitality of A549 cells transfected with siR-SOX9 or siR-NC was detected using the MTT assay. B. Transwell migration assay was used to detect migration ability of A549 cells transfected with siR-SOX9 or siR-NC. C. Transwell invasion assay was used to detect invasion ability of A549 cells transfected with siR-SOX9 or siR-NC. D. Protein level of SOX9 was detected by Western blotting in A549 cells transfected with siR-SOX9 or siR-NC. *P < 0.05.
Discussion
MiRNAs are important regulators in human cancer and their roles as therapeutic targets have been proposed. MiRNA deregulation in NSCLC suggests that miRNAs are involved in the initiation and progression of this disease. In this study, we detected low miR-206 expression and identified SOX9 as a miR-206 target in NSCLC.
miR-206, a member of the muscle-specific miR-1 family of muscle specific microRNAs, is a skeletal muscle specific miRNA involved in muscle development [10]. However, lots of studies revealed that miR-206 is closely related to tumor progression. For example, Wang et al showed that miR-206 was decreased in triple-negative breast cancer (TNBC) and repressed tumor cell migration through direct targeting of CORO1C in TNBC cells [11]. Lin et al found that miR-206 functioned as a tumor suppressor and directly targeted K-Ras in human oral squamous cell carcinoma [12]. Wang et al suggested that miR-206 attenuated tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer [13]. Zhang et al indicated that miR-206 was significantly decreased in gastric cancer and miR-206 inhibited gastric cancer metastasis by negatively regulating expression of PAX3, furthermore, their results revealed that miR-206-PAX3-MET signaling was critical to gastric cancer metastasis [14]. Here, we reported that miR-206 was decreased in NSCLC tissues and cell lines compared to adjacent non-tumor tissues and normal lung bronchus epithelial cell line BEAS-2B, indication a potential tumor suppressive function of miR-206. We demonstrated that miR-206 suppressed cell proliferation, migration and invasion ability of NSCLC cells in vitro, and provided evidence that these effects were partially mediated by suppressing SOX9 expression. Furthermore, our data revealed that SOX9 knockdown performed similar effects of miR-206 overexpression on NSCLC cells. These results supported SOX9 as a direct target gene of miR-206, important in regulating NSCLC progression.
Sry-related high-mobility group (HMG) box 9 (SOX9) is a transcription factor that plays key roles in development, differentiation and lineage commitment in various tissues, including the intestine, liver and pancreas [15-17]. Recent research found that SOX9 is also involved in carcinogenesis in a variety of human cancers. For example, Liu et al suggested that overexpression of SOX9 increased cell migration and invasion and markedly promoted the EMT process. Furthermore, their results revealed that SOX9 stimulated metastasis through activating Wnt/β-catenin signaling [18]. Grimont et al suggested that SOX9 was overexpression in pancreatic ductal adenocarcinoma and regulated ERBB signaling in pancreatic cancer development [19]. Hiraoka et al demonstrated that knockdown of SOX9 suppressed the proliferation and tumorigenicity of glioblastoma cells [20]. In the present study, we found that inhibition of SOX9 remarkably suppressed the proliferation, migration and invasion of NSCLC cells. Our study expanded the function of SOX9 in NSCLC.
In summary, our study showed that miR-206 was downregulated in NSCLC tissues and cell lines, and overexpression of miR-206 significantly inhibited cell proliferation, migration and invasion of NSCLC cells through targeting SOX9. Our data provide new insight into the mechanism of NSCLC, and identified miR-206/SOX9 link provided a potential therapeutic target to treatment of NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Ihde D, Souhami B, Comis R, Gregor A, Hansen H, Johnson B, Murray N, Postmus P, Rocmans P, Saijo N, Stout R, Turrisi A, Wagner H. Small cell lung cancer. Lung Cancer. 1997;17(Suppl 1):S19–21. doi: 10.1016/s0169-5002(97)00039-1. [DOI] [PubMed] [Google Scholar]
- 2.Ridge CA, McErlean AM, Ginsberg MS. Epidemiology of lung cancer. Semin Intervent Radiol. 2013;30:93–98. doi: 10.1055/s-0033-1342949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X, Zhao Z, Xu W, Hou J, Du X. Down-regulation of miR-497 is associated with poor prognosis in renal cancer. Int J Clin Exp Pathol. 2015;8:758. [PMC free article] [PubMed] [Google Scholar]
- 8.Sun XH, Geng XL, Zhang J, Zhang C. miRNA-646 suppresses osteosarcoma cell metastasis by downregulating fibroblast growth factor 2 (FGF2) Tumour Biol. 2015;36:2127–34. doi: 10.1007/s13277-014-2822-z. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Wang W, Zhou C, Xi W, Yuan L, Chen X, Li Y, Yang A, Zhang J, Wang T. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 2015;36:3763–3773. doi: 10.1007/s13277-014-3017-3. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, Williams C. miR-206 inhibits cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer. Mol Oncol. 2014;8:1690–1702. doi: 10.1016/j.molonc.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin F, Yao L, Xiao J, Liu D, Ni Z. Mir-206 functions as a tumor suppressor and directly targets K-ras in human oral squamous cell carcinoma. Onco Targets Ther. 2014;7:1583–91. doi: 10.2147/OTT.S67624. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang XW, Xi XQ, Wu J, Wan YY, Hui HX, Cao XF. microRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol Rep. 2015;33:1402–1410. doi: 10.3892/or.2015.3731. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Xia L, Zhao L, Chen Z, Shang X, Xin J, Liu M, Guo X, Wu K, Pan Y. Activation of PAX3-MET pathways due to miR-206 loss promotes gastric cancer metastasis. Carcinogenesis. 2015;36:390–9. doi: 10.1093/carcin/bgv009. [DOI] [PubMed] [Google Scholar]
- 15.Shi Z, Chiang CI, Mistretta TA, Major A, Mori-Akiyama Y. SOX9 directly regulates IGFBP-4 in the intestinal epithelium. Am J Physiol Gastrointest Liver Physiol. 2013;305:G74–83. doi: 10.1152/ajpgi.00086.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paganelli M, Nyabi O, Sid B, Evraerts J, El Malmi I, Heremans Y, Dolle L, Benton C, Calderon PB, van Grunsven L, Heimberg H, Campard D, Sokal E, Najimi M. Down-regulation of Sox9 expression associates with hepatogenic differentiation of human liver mesenchymal stem/progenitor cells. Stem Cells Dev. 2014;23:1377–1391. doi: 10.1089/scd.2013.0169. [DOI] [PubMed] [Google Scholar]
- 17.Delous M, Yin C, Shin D, Ninov N, Carten JD, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Liu Z, Jiang B, Peng R, Ma Z, Lu J. SOX9 Overexpression Promotes Glioma Metastasis via Wnt/β-Catenin Signaling. Cell Biochem Biophys. 2015:1–8. doi: 10.1007/s12013-015-0647-z. [DOI] [PubMed] [Google Scholar]
- 19.Grimont A, Pinho AV, Cowley MJ, Augereau C, Mawson A, Giry-Laterriere M, Van den Steen G, Waddell N, Pajic M, Sempoux C, Wu J, Grimmond SM, Biankin AV, Lemaigre FP, Rooman I, Jacquemin P. SOX9 regulates ERBB signalling in pancreatic cancer development. Gut. 2014 doi: 10.1136/gutjnl-2014-307075. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Kaneko R, Nasu-Nishimura Y, Koyama-Nasu R, Kawasaki Y, Akiyama T. SOX9-mediated upregulation of LGR5 is important for glioblastoma tumorigenicity. Biochem Biophys Res Commun. 2015;460:216–21. doi: 10.1016/j.bbrc.2015.03.012. [DOI] [PubMed] [Google Scholar]


