Abstract
Overexpression of miR-155 in nasopharygeal carcinoma (NPC) is partly driven by Epstein-Bar virus infection. However the role of miR-155 in NPC oncogenesis is unclear. This study showed that miR-155 inhibitor could inhibit the cell migration in NPC cell lines. ZDHHC2 was identified as a direct target of miR-155 and downregulation of ZDHHC2 prompted cell migration in NPC. Furthermore, reduced ZDHHC2 expression was associated significantly with metastasis and poor survival of NPC patients. Collectively, inhibition of miR-155 suppresses cell migration in NPC through targeting ZDHHC2. The potential of miR-155 and ZDHHC2 as therapeutic targets in NPC should be further investigated.
Keywords: Nasopharyngeal carcinoma, miR-155, ZDHHC2, cell migration
Introduction
Nasopharyngeal carcinoma (NPC) is a tumor arising from the mucosal epithelium covering the nasopharyngeal surface and is one of the most common malignancies in South-China, Southeast-Asia and North Africa [1]. Genetic alterations, Epstein-Barr virus (EBV) infection and other environmental factors were reported to be associated with risk for NPC [2,3]. NPC has a obvious clinical-pathological behavior of loco-regional recurrence and metastasis, which is different from other types of head and neck cancers [4]. However to date the pathogenesis of NPC is not investigated clearly.
MicroRNAs is an abundant class of non-coding RNAs, typically 20-23 nucleotides in length and they are involved in many cellular processes including apoptosis, differentiation, proliferation, and metabolism [5]. In the microRNAs family, microRNA 155 (miR-155) is located on chromosome 21 and transcribed from the B-cell integration cluster. MiR-155 plays important roles in many human tumors, including breast cancer [6-8], leukemia [9], melanoma [10], lymphoma [11-13], cervical cancer [14], hepatocellular carcinoma [15], pancreatic cancer [16], lung cancer [17,18], colon cancer [6], and gastric adenocarcinoma [19]. We have also reported that miR-155 is upregulated in NPC, which is partly driven by EBV encoded LMP1 and LMP2A [20]. However the function of miR-155 in NPC is unclear yet.
Zinc finger, DHHC-type containing 2 (ZDHHC2), also known as reduced expression associated with metastasis protein (REAM), is one member of DHHC protein family of protein acyltransferases (PATs). It is located in chromosome 8p21.3-22 [21], where frequent loss of heterozygosity has been detected in various types of metastatic cancers, including colorectal cancer [22], hepatocellular carcinoma [22], prostate cancer [23], non-small cell lung cancer [22], urinary bladder cancer [24], breast cancer [25]. Reduced of ZDHHC2 was found to be associated with lymph node metastasis and poor prognosis in gastric adenocarcinoma [26], and the mRNA level of ZDHHC2 expression was significantly reduced in primary and metastatic foci of advanced colorectal cancer [21]. However the expression pattern of ZDHHC2 has not been investigated in NPC yet. In view of the proposed roles of miR-155 and ZDHHC2 in cancer, we aimed to investigate the function of miR-155 and ZDHHC2 and their potential relationship in NPC.
Materials and methods
Patients and tissue samples
The study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center (Guangzhou 510060, China). All samples used in this study were anonymous and collected from patients for routine pathology use. The patient records/information was de-identified as well as anonymized, prior to analysis. No informed consent (written or verbal) was obtained for use of retrospective tissue samples from the patients in this study. In this retrospective study, archival formalin-fixed, paraffin-embed-ded (FFPE) tissue specimens from 124 primary NPC patients (29 females and 95 males; aged from 21 to 77 years; median, 48 years) who underwent radical radiotherapy with or without chemotherapy from 1999 and 2007 were obtained from the Sun Yat-sen University Cancer Center (Guangzhou, China, 510060). The disease stages of all patients were classified or reclassified according to the China 1992 NPC staging system [27]. Of the 124 primary NPC patients, 4 were classified as stage I, 26 as stage II, 64 as stage III, and 28 as stage IV, and 2 cases without stage information available.
Cell lines
Human NPC cell lines CNE1 (EBV negative, from Cancer Center, Sun Yat-sen University, China), TW03 (EBV negative, the generous gift of Prof. Chin-Tarng Lin, National Taiwan University Hospital) [28] were cultured in 1640 (Gibco USA) containing 10% fetal calf serum (FCS), and Human Embryonic Kidney 293 T cells (from American Tissue Culture Collection, ATCC, Mana-ssas, VA) were cultured in DMEM (Gibco USA) containing 10% fetal calf serum (FCS). All the cell lines were grown in a humidified incubator at 37°C with 5% CO2.
MiRNA and siRNA transfections
Before transfection, 2×105 cells per well were plated into 6-well plates and grown for one day in antibiotic-free medium containing 10% FCS. When the cell confluence was reached to 40% to 60%, cells were transfected with 100nM miR-155 Anti-miRTM miRNA Inhibitor (Cat#: AM12601, Ambion, USA), or Anti-miRTM miRNA Inhibitors-Negative Control #1 (Cat#: AM17010, Ambion, USA), or 100 nM four individual ZDHHC2 specific siRNAs (Please see Figure S1 for detailed siRNA sequence) and a negative control siRNA by using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Transfected cells were grown at 37°C for 6 h, followed by incubation with complete medium. For q-PCR and Western blot analysis, cells were harvested for RNA and protein respectively 48 h post-transfection.
Luciferase reporter assays
The whole 3’UTR sequences of ZDHHC2 were cloned to downstream from the luciferase reading frame in the plasmid pmiR-Glo-report-Vector (Genepharma, China). Mutations were made in these two putative miR-155 target sites (433-439 and 2334-2340). All inserts were sequenced in their entirety to verify polymerase fidelity. Luciferase reporter assays were performed by transiently transfecting HEK 293T cells respectively with 200 ng of with pmiR-Glo-ZDHHC2-3’UTR-wild, pmiR-Glo-ZDHHC2-3’UTR-433mut, or pmiR-Glo-ZDHHC2-3’UTR-2334mut, together with 30 nM miR-155 Pre-miRTM miRNA Precursor Molecules (Cat#: PM12601, Ambion, USA), or Pre-miRTM miRNA Precursor Molecules-Negative Control #1 (Cat#: AM17110, Ambion, USA) and 200 ng of pCMV-Renilla (internal control) using Lipofectamine 2000 (Invitrogen) respectively. Firefly and Renilla luciferase activities were measured consecutively by using Dual Luciferase Assay (Cat#: E1910, Promega, USA) 48 hr after transfection. Firefly luciferase values have been normalized to Renilla, and the ratio of firefly/renilla was presented.
PCR assays
For miRNAs quantitive realtime PCR (qPCR) assay, total RNA from cell lines was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, then was treated with RNase free DNase I (Cat#: 04716728001, Roche). The miR-155 quantitive realtime PCR assay was performed by TaqMan MicroRNA Assays (Cat#: 4373124, Applied Biosystems, USA) and RNU6B (Cat#: 4373381, Applied Biosystems, USA) was used as internal control. For mRNA qPCR assay, total RNA was extracted from cell lines using TRIzol reagent (Invitrogen). After reverse transcription of the total RNA, the first-strand cDNA was then used as template for detection of ZDHHC2, expression by quantitative real time PCR (qPCR) with the SYBR Green I chemistry (Power SYBR Green PCR Master Mix, CAT#: 4367659, ABI Inc., USA). GAPDH was used as internal control. The primers were ZDHHC2 (Forward: TCT TAG GCG AGC AGC CAA GGA T and Reverse: CAG TGA TGG CAG CGA TCT GGT T); GAPDH (Forward: AGC CAC ATC GCT CAG ACA C and Reverse: GCC CAA TAC GAC CAA ATC C). The relative expression level was determined as 2-ΔΔCt. Data are presented as the expression level relative to the calibrator (control sample), with the standard error of the mean of triplicate measures for each test sample.
Western-blot assays
Cells were harvested and lysed with RIPA buffer (Upstate, USA) at 48 h post-transfection. Equal amounts of denatured protein sample were separated by SDS-PAGE and were then transferred electrophoretically to PVDF membranes (Pall, USA) for immunoblot analysis. Antibodies used for immunoblot analysis were against ZDHHC2 (1:1000 dilution, Cat#: AP 5592a, Abgent, CA) and an anti-GAPDH antibody (1:5,000 dilution, sc-32233, Santa Cruz, USA) was used as loading control. All protein bands were detected using an enhanced chemiluminescent (ECL) Western blot Kit (Cell Signaling Technology, USA).
Transwell assay
For the migration assay, 5×104 cells were plated in the top chamber with a non-coated membrane (24-well insert; 8-μm pore size; BD Biosciences, USA). The cells were plated in medium without serum, and medium supplemented with 10% serum was used as a chemoattractant in the lower chamber. The cells were incubated for 24 h. Cells that didn’t migrate through the pores, were removed by a cotton swab. Filters were fixed with 90% ethanol, stained with 0.1% crystal violet, and photographed. Cell numbers were counted under microscope.
Wound-healing assay
The cultured cells were transfected with 100 nM miR-155 inhibitor or negative control. At 48 h post-transfection, the cells were allowed to reach confluence before dragging a 200 ul sterile pipette tip through the mono-layer. The cells were then washed and allowed to migrate for additional 24 h. At 0, 6, 12 and 24 h post-wounding, images were captured. The experiment was performed in triplicate.
Methylthiazolyl blue tetrazolium (MTT) assay
Cell growth was determined using MTT (Sigma-Aldrich, USA) spectrophotometric dye assay. At 24 h post-transfection with miR-155 inhibitor or negative control, cells were seeded into 96-well plates (4×103 cells/well), when the cells were adhered (about culturing for 6 h), DDP was added to each well according to the concentration gradient (0 nM, 12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM, 400 nM, 800 nM) and each concentration gradient was repeated thrice. After culturing for 48 h, MTT (10 μL per well) was added. The mixtures were further cultured for 4 h and the liquids were discarded. To each well 100 μL dimethylsulfoxide (DMSO) was added and the plate was shaken for 10 min. The absorbance at 570 nm was measured using an automatic ELISA reader. The inhibition rate (%) = (1-A~experimental group/A~control group) ×100.
Immunohistochemistry
In our study, primary antibodies against ZDHHC2 (1:200 dilution, Cat#: AP 5592a, Abgent, CA) were used. Briefly, tissue sections were de-waxed, incubated with hydrogen peroxide for 10 minutes, incubated in retrieval buffer solution for antigen recovery, blocked with normal serum for 10 minutes and incubated with a primary antibody for 60 minutes, followed by detection using a Catalyzed Signal Amplification Kit (DAKO, USA); signal was visualized using diaminobenzidine. Non-immune rabbit serum was substituted for the primary antibody as a negative control. The immunohistochemistry results were evaluated and scored by a senior pathologist without knowledge of the clinicopathological outcomes of the patients. A semiquantitative estimation was made by using a composite score obtained by adding the values of the staining intensity and the relative abundance of positive cells. The intensity was graded as 0 (no staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). The abundance of the positive cells was graded from 0 to 3 (0.5% positive cells; 1. 5-25%; 2. 26-50%; 3. 51-75%; 4. >75%). A composite score greater than the median value was considered as high expression, and composite scores less than or equal to the median value were considered as low expression.
Statistical analysis
All the data in this study was analyzed by SPSS 13.0 software. The association between ZDHHC2 and clinical-pathological parameters were assessed by Chi-Square test. Kaplan-Meier analysis and log-rank tests were used to compare the difference in survival curves. It was considered as significant differences when P<0.05.
Results
Inhibition of miR-155 suppresses cell migration in nasopharyngeal carcinoma
To investigate the function of miR-155 in NPC, miR-155 inhibitor (Ambion, USA) and miRNA inhibitor control (Ambion, USA) were transiently transfected to two EBV negative NPC cell lines CNE1 and TW03 respectively, and then both transwell assay and wound healing assay were performed to determine if miR-155 could influence the cell migration ability in NPC cells. miR-155 was reduced by 81%, and 66% in CNE1 and TW03 cells respectively by miR-155 inhibitor by qPCR dectection at 48 hours posttransfection (Figure 1A). The CNE1 and TW03 cells transfected with miR-155 inhibitor (Ambion, USA) and miRNA inhibitor control (Ambion, USA) were used for transwell migration assays at 48 hours posttransfection. The average number of migrated CNE1 cells transfected with miR-155 inhibitor was 201±9, which was sigfinicantly less than that (388±10) of CNE1 cells transfected with inhibitor control (P<0.001 ) (Figure 1B and 1C). The average number of migrated cells was 69±4 and 112±7, repectively, in miR-155 inhibitor and inhibitor control treated TW03 cells. There was statistically difference (P<0.001) (Figure 1B and 1C). Notably, wound healing assay futher demonstrated that miR-155 inhibitor could reduce the cell migration ability in NPC CNE1 and TW03 cells (Figure 1D). Moreover MTT assay indicated that miR-155 could also inhibit cell proliferation in NPC CNE1 and TW03 cells (Figure S2).
Figure 1.

miR-155 inhibitor suppresses cell migration in nasopharyngeal carcinoma CNE1 and TW03 cells. A. The relative miR-155 expression in CNE1 and TW03 cells transfected with miR-155 inhibitor (100 nM) and inhibitor negative control (100 nM) by qPCR detection, and U6 was used as a loading control. B and C. The migrational ability of CNE1 cells and TW03 cells was analyzed by transwell migration assays at 48 h post-transfection with a miR-155 inhibitor (100 nM) and a negative control (100 nM). D. The migrational ability of CNE1 cells and TW03 cells was analyzed by wound healing assays at 48 h post-transfection with a miR-155 inhibitor (100 nM) and a negative control (100 nM).
ZDHHC2 is the direct target of miR-155
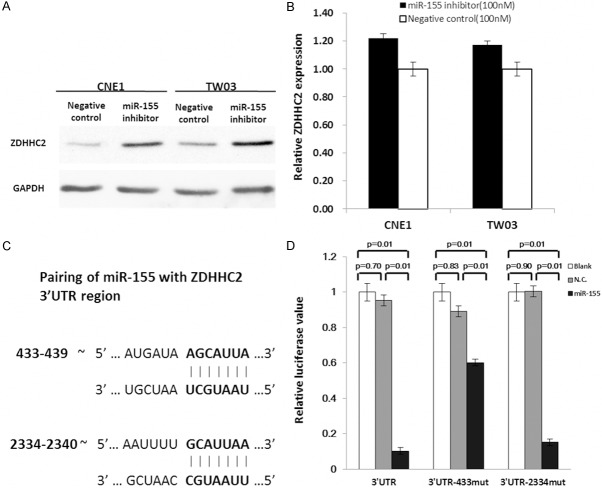
We found that ZDHHC2 protein was increased in miR-155 inhibitor transfected CNE1 and TW03 cells, compared with inhibitor control transfected cells (Figure 2A), but ZDHHC2 mRNA was not changed significantly (Figure 2B). Analyzing the sequence of ZDHHC2 3’UTR region by TargetScan [29], we found two potential miR-155 target sites in ZDHHC2 3’UTR region (base sequence 433-439 and 2334-2340) (Figure 2C).
Figure 2.
ZDHHC2 is the direct target of miR-155. A. The expression of ZDHHC2 was analyzed by Western-blot at 48 h post-transfection in CNE1 cells and TW03 cells. The miR-155 inhibitor could upregulate ZDHHC2 expression in protein level. B. In q-PCR detection, the ZDHHC2 mRNA was not changed significantly after transfection. GAPDH was used as a loading control. C. Pairing of miR-155 with ZDHHC2 3’UTR region. D. Overexpression of miR-155 by the miR-155 mimic resulted in a significant decrease in luciferase signals of pmiR-Glo-ZDHHC2-3’UTR, pmiR-Glo-ZDHHC2-3’UTR–433-mut and pmiR-Glo-ZDHHC2-3’UTR-2334-mut transfected HEK293 cells.
To test whether ZDHHC2 responds to miR-155 through direct 3’UTR interactions, we cloned the wild type of 3’UTR of ZDHHC2 (3’UTR), and two mutated potential miR-155 target sites in ZDHHC2 3’UTR region (3’UTR-433mut and 3’UTR-2334mut) respectively into a reporter plasmid downstream of luciferase. The luciferase reporter assays were performed by transiently transfecting HEK 293T cells respectively, with pmiR-Glo-ZDHHC2-3’UTR, pmiR-Glo-ZDHHC2-3’UTR-433mut, or pmiR-Glo-ZDHHC2-3’UTR-2334 mut, together with miR155 mimic (Ambion, USA), mimic control (negative control), or blank and pCMV-Renilla (internal control). After 48 hr transfection, a dual-luciferase reporter assay system ( Promega, USA) was used to detect luciferase expression. MiR155 mimic could downregulate the ratio of luciferase significantly in pmiR-Glo-ZDHHC2-3’UTR group (P=0.01), pmiR-Glo-ZDHHC2-3’UTR-433mut group (P=0.01) and pmiR-Glo-ZDHHC2-3’UTR-2334 mut group (P=0.01), compared to negative control siRNA transfected groups and blank groups (Figure 2D). Taken together, these data confirmed that miR-155 could directly target ZDHHC2-3’UTR region throught the sites of ZDHHC2-3’UTR 433~439 and 2334~2340 base sequence.
Inhibition of ZDHHC2 promotes cell migration in nasopharyngeal carcinoma
To investigate the function of ZDHHC2 in NPC, four ZDHHC2 sepcific siRNAs (ZDHHC2-homo-824, ZDHHC2-homo-1101, ZDHHC2-homo-1175 and ZDHHC2-homo-1295) and negative control were transiently transfected to two EBV negative NPC cell lines CNE1 and TW03, and then both transwell assay and wound healing assay were performed to determine if ZDHHC2 could influence the cell migration ability in NPC cells. Besides ZDHHC2-homo-1295, the other three ZDHHC2 siRNAs (ZDHHC2-homo-824, ZDHHC2-homo-1101 and ZDHHC2-homo-1175) could knock down both ZDHHC2 mRNA and ZDHHC2 protein significantly in NPC CNE1 and TW03 cells (Figure 3A and 3B). The average number of migrated CNE1 cells transfected with ZDHHC2-homo-824, ZDHHC2-homo-1101 and ZDHHC2-homo-1175 was 464±7, 350±6, 295±7, which was sigfinicantly more than that of CNE1 cells transfected with ZDHHC2-homo-1295 and negative control (264±4 and 237±5) (P=0.015) (Figure 4A and 4B). Similar results was found in NPC TW03 cells (Figure 4A and 4B).
Figure 3.

The inhibition effect of four ZDHHC2 siRNAs in nasopharyngeal carcinoma CNE1 and TW03 cells. A. The relative ZDHHC2 expression were analyzed by q-PCR detection in CNE1 and TWO3 cells at 48 h post-transfection with a negative control, ZDHHC2-homo-824, ZDHHC2-homo-1101, ZDHHC2-homo-1175 and ZDHHC2-homo-1295 respectively. GAPDH was used as a loading control. B. The expression of ZDHHC2 was analyzed by Western-blot at 48 h post-transfection in CNE1 cells and TWO3 cells, and GAPDH was used as a loading control.
Figure 4.

ZDHHC2 inhibits cell migration in nasopharyngeal carcinoma CNE1 and TW03 cells. A. The migrational ability of CNE1 cells and TWO3 cells transfected with ZDHHC2-homo-824, ZDHHC2-homo-1101, ZDHHC2-1175, ZDHHC2-homo-1295 and negative control, was analyzed by Transwell Migration Assays. B. ZDHHC2-homo-824, ZDHHC2-homo-1101, ZDHHC2-1175 could increase the cells migrational ability of CNE1 and TW03, comparing to the negative control group.
Reduced expression of ZDHHC2 correlates with metastasis and poor prognosis of nasopharyngeal carcinoma
To investigate the association between ZDHHC2 expression and clinicopathological parameters of NPC patients, paraffin-embedded tissues section (n=124) with histopathologically confirmed NPC were examined using immunohistochemistry. The ZDHHC2 was observed predominantly in the cytoplasm of NPC tumor cells, and low expression of ZDHHC2 was observed in 50% (62/124) of NPC cancer patients (Figure 5A). Interestingly, reduced ZDHHC2 expression was associated significantly with metastasis (P=0.011) and T stage (P=0.035) (Table 1). No significant association was seen between ZDHHC2 expression and age, gender, N stage, TNM stage and recurrence (Table 1).
Figure 5.

Low expression of ZDHHC2 predicts poor prognosis of nasopharyngeal carcinoma patients. A. Two representative images show high and low expression of ZDHHC2 in NPC tumor cells. B. Kaplan-Meier curves for disease-free survival (DFS) of the 124 NPC patients; C. Kaplan-Meier curves for DFS in NPC patients with low level and high level ZDHHC2 expression; D. Kaplan-Meier curves for overall survival (OS) of the 124 NPC patients; E. Kaplan-Meier curves for OS in NPC patients with low level and high level ZDHHC2 expression.
Table 1.
Correation between ZDHHC2 expression and clinical-pathological parameters of NPC
| Parameters | Cases (n) | Missing (n) | ZDHHC2 Expression | Sig. | |
|---|---|---|---|---|---|
|
| |||||
| L.E. (n) | H.E. (n) | ||||
| Gender | 0 | ||||
| Female | 29 | 16 | 13 | 0.138 | |
| Male | 95 | 46 | 49 | ||
| Age | 0 | ||||
| ≤48 y | 64 | 34 | 30 | 0.111 | |
| >48 y | 60 | 28 | 32 | ||
| T stage | 4 | ||||
| T1+T2 | 50 | 30 | 20 | 0.035* | |
| T3+T4 | 70 | 31 | 39 | ||
| N stage | 4 | ||||
| N0 | 29 | 14 | 15 | 0.161 | |
| N1+N2+N3 | 91 | 47 | 44 | ||
| TNM stage | 2 | ||||
| I+II | 30 | 18 | 12 | 0.086 | |
| III+IV | 92 | 44 | 48 | ||
| Recurrence | 0 | ||||
| No | 85 | 39 | 46 | 0.062 | |
| Yes | 39 | 23 | 16 | ||
| Metastasis | 0 | ||||
| No | 95 | 42 | 53 | 0.011* | |
| Yes | 29 | 20 | 9 | ||
ps: L.E.: Low Expression; H.E.: High Expression;
P value < 0.05.
To investigate the prognostic value of ZDHHC2 expression in NPC patients, disease free survival (DFS) and overall survival (OS) analysis were performed in these 124 NPC cases, and the five-year DFS rate was 65.7% for these patients (Figure 5B). The five-year DFS rate was 55.4% for patients with low ZDHHC2 expression (n=62), and 73.4% for patients with high ZDHHC2 expression (n=62), which was a significant difference (P=0.031, Figure 5C). The five-year OS rate was 77.9% for these 124 NPC patients (Figure 5D). the patients with the low levels of ZDHHC2 expression (n=62) had a poorer prognosis than the patients with high levels of ZDHHC2 expression (n=62) (P=0.011, Figure 5E). Univariate and multivariate analyses were performed to compare the impact of ZDHHC2 expression and other clinicopathological parameters on prognosis. Univariate analyses showed that 3 factors, including ZDHHC2 expression (P=0.014), metastasis status (P=0.001) and age (P=0.001) were prognostic predictors of OS in NPC patients. Multivariate analyses indicated the relative risk of death in patients with high ZDHHC2 expression tumors was lower than that of patients with low ZDHHC2 expression tumors (HR=0.213, 95% CI=0.095-0.480). ZDHHC2 expression had a significant, independent predictive value for survival of NPC patients (P=0.001). Moreover metastasis status (P=0.001) and age (P=0.001) were also independent prognosis predictors for NPC patients (Table 2).
Table 2.
Univariate and multivariate analysis of various prognostic parameters for OS in NPC
| Variables | Univariate analysis | Mutivariate analysis | ||
|---|---|---|---|---|
|
| ||||
| HR (95% CI) | P | HR (95% CI) | P | |
| Gender (F/M) | 1.055 (0.515, 2.163) | 0.884 | 0.650 (0.283, 1.495) | 0.311 |
| Age (>48 y/≤48 y) | 3.890 (1.905, 7.944) | 0.001 | 7.474 (3.189, 17.514) | 0.001 |
| T stage (T3+T4/T1+T2) | 0.915 (0.479, 1.749) | 0.789 | 1.561 (0.612, 3.985) | 0.352 |
| N stage (N1+N2+N3/N0) | 1.070 (0.506, 2.263) | 0.858 | 0.825 (0.357, 1.906) | 0.653 |
| TNM stage (III+IV/I+II) | 1.211 (0.575, 2.550) | 0.614 | 1.155 (0.380, 3.504) | 0.780 |
| Recurrence (Yes/No) | 1.561 (0.830, 2.937) | 0.167 | 0.777 (0.371, 1.630) | 0.505 |
| Metastasis (Yes/No) | 6.036 (3.225, 11.295) | 0.001 | 11.638 (4.995, 27.117) | 0.001 |
| ZDHHC2 (H.E./L.E.) | 0.441 (0.231, 0.845) | 0.014 | 0.213 (0.095, 0.480) | 0.001 |
ps: L.E.: Low Expression; H.E.: High Expression.
Discussion
MiR-155 is considered as a typical multifunctional microRNA, which is involved in many other different biological processes including inflammation, haematopoiesis and immunity. MiR-155 also plays a pivotal role in many human cancers, and many studies have reported on the role of miR-155 in cancer development either as an oncomiR or as an oncosuppressor-miR [30]. Expression of miR-155 was upregulated in many human cancers with high proliferative activity and decreased apoptotic capability, including breast cancer [8,31], B-cell lymphoma [12], hepatocellular carcinoma [15], cervical cancer [32], colon cancer [33] and lung cancer [34]. Moreover, upregulation of miR-155 has been also reported to promote the migration and invasion in many human cancers, including colorectal cancer [35], breast cancer [36], pancreatic cancer [37] and hepatocellular carcinoma [38]. However on the other hand, miR-155 was found to be downregulated and acted as an oncosuppressor-miR in adult Burkitt lymphoma [39], acute myeloid leukaemia [40], gastric cancer [41,42], ovarian cancer [43] and melanoma [44]. In the present study, we report that miR-155 inhibitor could suppress cell migration as well as cell proliferation in NPC CNE1 and TW03 cells in vitro. These data proposed that miR-155 acts as an oncomiR in NPC. In our previous study, we have reported that upregulation of miR-155 in NPC is partly driven by EBV encoded LMP1 and LMP2A [20]. Hence as an oncomiR, miR-155 may play an important role in oncogenesis of EBV encoded LMP1 and LMP2A in NPC.
MiRNAs could regulate about 30% of all protein-encoding genes of the human genome [45], and each miRNA could have many direct target transcripts while each transcript could be regulated by one or more miRNAs [46]. At present, several direct targets of miR-155 have been identified in may human cancers. Loss of C/EBPbeta (CCAAT/enhancer binding protein, beta) mediated by miR-155 shifts the TGF-beta response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer [36], and miR-155 could promote the proliferation of breast cancer cells through down-regulation of SOCS1 (Suppressor of cytokine signaling 1) [8] and FOXO3a (forkhead Forkhead box O3) [31]. MiR-155 affected pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1 [37]. MiR-155 has also been reported to be involved in the development of lymphoma by targeting SMAD5 (SMAD family member 5) [47] and SHIP1 (inositol polyphosphate-5-phosphatase) [48]. In our study, we found that miR-155 inhibitor could increase endogenous ZDHHC2 protein expression in NPC CNE1 and TW03 cells, and luciferase reporter assay was performed to identify ZDHHC2 as direct targets of miR-155. This is the first report that ZDHHC2 is a direct target of miR-155. Moreover knockdown of ZDHHC2 could promote cell migiration in NPC CNE1 and TW03 cells, but didn’t affect the cell proliferation in NPC (data was not shown). These data suggested that miR-155 regulated cell migiration in NPC through targeting ZDDHC2 and besides ZDHHC2, there should be some other targets of miR-155 which has not been identified in NPC.
ZDHHC2 originally named as reduced expression associated with metastasis protein (REAM), has been proposed as a putative tumor/metastasis suppressor gene. The mRNA level of ZDHHC2 expression was significantly reduced in primary and metastatic foci of advanced colorectal cancer [21] and reduced of ZDHHC2 was found to be associated with metastasis and poor prognosis in gastric adenocarcinoma [26] and hepatocellular carcinoma [49,50]. In the present study, reduction of ZDHHC2 expression was observed in 50.0% (62/124) of NPC patients, and was associated significantly with metastasis (P=0.011) and T stage (P=0.035) of NPC patients. Furthermore, our results indicated that ZDHHC2 expression had a significant, independent predictive value for survival of NPC patients (P=0.001). Hence, our results also proposed that ZDHHC2 was a putative tumor/metastasis suppressor in NPC, which were consistent with previous studies of ZDHHC2. As one member of DHHC protein family of protein acyltransferases (PATs), ZDHHC2 is directly involved in the palmitoyl transfer reaction [51]. CKAP4/p63, a cell surface receptor for antiproliferative factor (APF) [52], has been identied as a substrate of ZDHHC2 related to its tumor/metastasis suppressor function [53]. Moreover tetraspanins CD9 was also identified as the substrate of ZDHHC2 related to its putative tumor/metastasis suppressor [54]. It is possible that hypopalmitoylation of both CKAP4 and CD9 may increase tumor or metastatic behavior.
Taken together, inhibition of miR-155 suppresses cell migration in NPC through targeting ZDHHC2, and downregaultion of ZDHHC2 is associated with metastasis and poor prognosis of NPC patients. Further studies are needed to validate and clarify the mechanism of miR-155 and its emerging targets in NPC, and the potential of miR-155 and ZDHHC2 as therapeutic targets for NPC should be further investigated.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81172565 to YL and No. 81202135 to ZD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
Abbreviations
- NPC
Nasopharyngeal Carcinoma
- ZDHHC2
Zinc finger, DHHC-type containing 2
- EBV
Epstein-Bar virus
Supporting Information
References
- 1.Licitra L, Bernier J, Cvitkovic E, Grandi C, Spinazze S, Bruzzi P, Gatta G, Molinari R. Cancer of the nasopharynx. Crit Rev Oncol Hematol. 2003;45:199–213. doi: 10.1016/s1040-8428(01)00210-4. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Mahanta J. Aetiology of nasopharyngeal carcinoma. A review. Indian J Cancer. 1998;35:47–56. [PubMed] [Google Scholar]
- 3.Shao JY, Li YH, Gao HY, Wu QL, Cui NJ, Zhang L, Cheng G, Hu LF, Ernberg I, Zeng YX. Comparison of plasma Epstein-Barr virus (EBV) DNA levels and serum EBV immunoglobulin A/virus capsid antigen antibody titers in patients with nasopharyngeal carcinoma. Cancer. 2004;100:1162–1170. doi: 10.1002/cncr.20099. [DOI] [PubMed] [Google Scholar]
- 4.Farias TP, Dias FL, Lima RA, Kligerman J, de Sa GM, Barbosa MM, Goncalves FB Jr. Prognostic factors and outcome for nasopharyngeal carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129:794–799. doi: 10.1001/archotol.129.7.794. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 9.Vargova K, Curik N, Burda P, Basova P, Kulvait V, Pospisil V, Savvulidi F, Kokavec J, Necas E, Berkova A, Obrtlikova P, Karban J, Mraz M, Pospisilova S, Mayer J, Trneny M, Zavadil J, Stopka T. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood. 2011;117:3816–3825. doi: 10.1182/blood-2010-05-285064. [DOI] [PubMed] [Google Scholar]
- 10.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 11.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 12.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang Y, Zhao J, McCrae MA, Zhuang H. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118:2431–2442. doi: 10.1002/cncr.26566. [DOI] [PubMed] [Google Scholar]
- 16.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 17.Donnem T, Eklo K, Berg T, Sorbye SW, Lonvik K, Al-Saad S, Al-Shibli K, Andersen S, Stenvold H, Bremnes RM, Busund LT. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J Transl Med. 2011;9:6. doi: 10.1186/1479-5876-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Chen Q, Lai R, Wu X, Liu F, Xu G, Ji Y. Elevated expression of mature miR-21 and miR-155 in cancerous gastric tissues from Chinese patients with gastric cancer. J Biomed Res. 2010;24:187–197. doi: 10.1016/S1674-8301(10)60028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX, Shao JY, Ernberg I. Upregulation of MiR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T, Matsuda H, Arakawa H, Nakamura Y. Isolation of a novel gene on 8p21.3-22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosomes Cancer. 2000;29:9–15. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1001>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara Y, Ohata H, Emi M, Okui K, Koyama K, Tsuchiya E, Nakajima T, Monden M, Mori T, Kurimasa A, et al. A 3-Mb physical map of the chromosome region 8p21.3-p22, including a 600-kb region commonly deleted in human hepatocellular carcinoma, colorectal cancer, and non-small cell lung cancer. Genes Chromosomes Cancer. 1994;10:7–14. doi: 10.1002/gcc.2870100103. [DOI] [PubMed] [Google Scholar]
- 23.Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N, Jacobs SC, Robinson JC, Epstein JI, Walsh PC, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 1993;53:3869–3873. [PubMed] [Google Scholar]
- 24.Knowles MA, Shaw ME, Proctor AJ. Deletion mapping of chromosome 8 in cancers of the urinary bladder using restriction fragment length polymorphisms and microsatellite polymorphisms. Oncogene. 1993;8:1357–1364. [PubMed] [Google Scholar]
- 25.Yaremko ML, Kutza C, Lyzak J, Mick R, Recant WM, Westbrook CA. Loss of heterozygosity from the short arm of chromosome 8 is associated with invasive behavior in breast cancer. Genes Chromosomes Cancer. 1996;16:189–195. doi: 10.1002/(SICI)1098-2264(199607)16:3<189::AID-GCC6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 26.Yan SM, Tang JJ, Huang CY, Xi SY, Huang MY, Liang JZ, Jiang YX, Li YH, Zhou ZW, Ernberg I, Wu QL, Du ZM. Reduced expression of ZDHHC2 is associated with lymph node metastasis and poor prognosis in gastric adenocarcinoma. PLoS One. 2013;8:e56366. doi: 10.1371/journal.pone.0056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min H, Hong M, Ma J, Zhang E, Zheng Q, Zhang J, Zhang F, Su Y, Qiu F. A new staging system for nasopharyngeal carcinoma in China. Int J Radiat Oncol Biol Phys. 1994;30:1037–1042. [PubMed] [Google Scholar]
- 28.Lin CT, Chan WY, Chen W, Huang HM, Wu HC, Hsu MM, Chuang SM, Wang CC. Characterization of seven newly established nasopharyngeal carcinoma cell lines. Lab Invest. 1993;68:716–727. [PubMed] [Google Scholar]
- 29.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 31.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, Cheng JQ. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Lao G, Liu P, Wu Q, Zhang W, Liu Y, Yang L, Ma C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014;35:11933–8. doi: 10.1007/s13277-014-2479-7. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Yang J, Lv X, Liu K, Gao C, Xing Y, Xi T. miR-155 regulates the proliferation and cell cycle of colorectal carcinoma cells by targeting E2F2. Biotechnol Lett. 2014;36:1743–1752. doi: 10.1007/s10529-014-1540-3. [DOI] [PubMed] [Google Scholar]
- 34.Zang YS, Zhong YF, Fang Z, Li B, An J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012;19:773–778. doi: 10.1038/cgt.2012.60. [DOI] [PubMed] [Google Scholar]
- 35.Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375–1380. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 36.Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, Roswall P, Pietras K, Sund M, Religa P, Fuxe J. MiR-155-mediated loss of C/EBPbeta shifts the TGF-beta response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene. 2013;32:5614–5624. doi: 10.1038/onc.2013.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, Li H, Wu W, Jiang T, Qiu Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol Rep. 2013;30:1223–1230. doi: 10.3892/or.2013.2576. [DOI] [PubMed] [Google Scholar]
- 38.Han ZB, Chen HY, Fan JW, Wu JY, Tang HM, Peng ZH. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol. 2012;138:153–161. doi: 10.1007/s00432-011-1076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 40.Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V, Ma DD. MicroRNA-155 as an inducer of apoptosis and cell differentiation in Acute Myeloid Leukaemia. Mol Cancer. 2014;13:79. doi: 10.1186/1476-4598-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY, Yan M, Qu QL, Zhu ZG, Liu BY. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960–1966. doi: 10.3892/or.2012.1719. [DOI] [PubMed] [Google Scholar]
- 42.Sun S, Sun P, Wang C, Sun T. Downregulation of microRNA-155 accelerates cell growth and invasion by targeting c-myc in human gastric carcinoma cells. Oncol Rep. 2014;32:951–956. doi: 10.3892/or.2014.3288. [DOI] [PubMed] [Google Scholar]
- 43.Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587:1434–1439. doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 44.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, Di Leva G, Ferracin M, Volinia S, Bonmassar E, Croce CM, D’Atri S. Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35:393–400. [PubMed] [Google Scholar]
- 45.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 46.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84:1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 47.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng C, Zhang Z, Wu J, Lv Z, Tang J, Xie H, Zhou L, Zheng S. A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:832712. doi: 10.1155/2014/832712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li SX, Tang GS, Zhou DX, Pan YF, Tan YX, Zhang J, Zhang B, Ding ZW, Liu LJ, Jiang TY, Hu HP, Dong LW, Wang HY. Prognostic significance of cytoskeleton-associated membrane protein 4 and its palmitoyl acyltransferase DHHC2 in hepatocellular carcinoma. Cancer. 2014;120:1520–1531. doi: 10.1002/cncr.28593. [DOI] [PubMed] [Google Scholar]
- 51.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho) physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Conrads TP, Tocci GM, Hood BL, Zhang CO, Guo L, Koch KR, Michejda CJ, Veenstra TD, Keay SK. CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J Biol Chem. 2006;281:37836–37843. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Planey SL, Ceballos C, Stevens SM Jr, Keay SK, Zacharias DA. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol Cell Proteomics. 2008;7:1378–1388. doi: 10.1074/mcp.M800069-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma C, Yang XH, Hemler ME. DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol Biol Cell. 2008;19:3415–3425. doi: 10.1091/mbc.E07-11-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.