Abstract
Previous studies have identified 8q24 as an important region to prostate cancer (PCa) susceptibility. The aim of this study was to investigate the role of six genetic variants on 8q24 (rs1447295, A; rs6983267, G; rs6983561, C; rs7837688, T; rs10090154, T and rs16901979, A) on PCa risk in Chinese population. Online electronic databases were searched to retrieve related articles concerning the association between 8q24 variants and PCa risk in men of Chinese population published between 2000 and 2014. Odds ratio (ORs) with its 95% correspondence interval (CI) were employed to assess the strength of association. Total eleven case-control studies were screened out, including 2624 PCa patients and 2438 healthy controls. Our results showed that three risk alleles of rs1447295 A (OR=1.35, 95% CI=1.19-1.53, P<0.00001), rs6983561 C (C vs. A: OR=1.41, 95% CI=1.21-1.63, P<0.00001) and rs10090154 T (T vs. C: OR=1.48, 95% CI=1.22-1.80, P<0.00001) on8q24 were significantly associated with PCa risk in Chinese population. Furthermore, genotypes of rs1447295, AA+AC; rs6983561, CC+AC and CC; rs10090154, TT+TC; and rs16901979, AA were associated with PCa as well (P<0.01). No association was found between rs6983267, rs7837688 and PCa risk. In conclusions, variants including rs1447295, rs6983561, rs10090154 and rs16901979 on 8q24 might be associated with PCa risk in Chinese population, indicating these four variations may contribute risk to this disease. This meta-analysis was the first study to assess the role of 8q24 variants on PCa risk in Chinese population.
Keywords: Prostate cancer, 8q24, variant, meta-analysis
Introduction
Prostate cancer (PCa), a frequent noncutaneous cancer, is the third leading cancer for men in the world. The well-established risk factors for PCa are age, ethnicity, smoking, alcohol and family history [1]. Due to its high incidence and mortality, treatment costs, and lack of suitable therapy for any stage of this disease, PCa is becoming a significant public health issue. The incidence of PCa in Chinese population is much lower than that in Western men. In China, the incidence rate of PCa was 1.6/105 person year. While in the United States, PCa is more common among men with an estimated 233000 new cases and 29480 death in 2014 [2]. Even so, the occurrence of this disease has rapidly increased among Chinese men in recent years, especially in developed areas [3]. Thus, there is pressing obligation to explore the mechanism under PCa and develop new therapeutic strategy.
Nowadays, hereditary factors are generally believed to contribute to PCa etiology [4]. Genome-wide association studies have identified common variants on human chromosome 8q24 which are associated with increased the risk of PCa. 8q24, a highly conserved genomic region, contains at least three independent risk regions for PCa (region 2: 128.14-128.28, region 3: 128.47-128.54, and region 1: 128.54-128.62) [5]. Numerous researches have discussed the role of variants on 8q24 in PCa risk. Cropp et al. demonstrated that rs2124036 on 8q24 was significantly associated with PCa risk in African-Barbadian men in a Black Population study [6]. Antczak et al. showed that the rs188140481 allele conferred a moderate increase in the risk of PCa in Polish men [7]. Zhang et al. suggested that A allele of rs10505474 and rs7387328 on 8q24 were associated with PCa and cumulatively confer risk in the Northern Chinese Han population [8]. Oskina et al. identified that the A allele of rs1447295 and the T allele of rs10090154 were associated with PCa risk in the Russian population [9].
Although many studies proved that 8q24 variants were associated with PCa risk, the results still remain inconclusive. Furthermore, previous meta-analysis showed that the association between variants on 8q24 and PCa risk presented racial disparities [10]. Variants on 8q24 also have different effects on cancer development that depend on the tissue type [11]. Therefore, we conducted this meta-analysis to evaluate the role of 8q24 variants in PCa risk in Chinese population.
Materials and methods
Identification and eligibility of relevant studies
We systematically searched the online electronic databases of PubMed, Medline, CNKI (Chinese National Knowledge Infrastructure) and Wanfang to retrieve related articles published between 2000 and 2014. The following terms: “prostate cancer or prostate carcinoma”, “8q24”, “polymorphism or variant or mutation” and “Chinese population” as well as their combinations were used. The corresponding Chinese terms were used in the Chinese library. When the same authors or laboratory reported one gene polymorphism twice, only the most recent article was included.
Inclusion criteria
The studies must meet the following criteria: 1) case-control studies; 2) evaluating gene variants on 8q24 and PCa risk; 3) the participants should be Chinese population; 4) cases should be diagnosed with histologically confirmed PCa, the controls were age-matched healthy local residents; 5) the results were presented as odds ratio (OR) with a corresponding 95% confidence interval (CI); and 6) genotype distribution information in cases and controls were available to extract, and must be in Hardy-Weinberg equilibrium for a certain polymorphism in controls.
Exclusion criteria
The exclusion criteria were as follows: 1) articles concerned the non-Chinese population; 2) articles without control group or participants in control group were not health population; 3) controls were not age, race-matched; 4) studies were review articles or conference papers; and 5) information of genotype distribution was not available to extract.
Data extraction and quality assessment
Two investigators assessed the data collecting from eligible studies independently. Any disagreement was subsequently resolved by discussion with the third expert and then reached consensus on each item. The following information was sought from each article: first-author, year of publication, total numbers of cases and controls, and genotype distribution, respectively.
Statistical analysis
The strength of the association between polymorphisms on 8q24 and PCa risk was measured by pooled OR with its 95% CI. The significance of the pooled OR was determined by the Z test, and a P value less than 0.05 was considered significant. The allelic model (A vs. B: A represents the risk allele; B represents the non-risk allele) and genotype genetic models (dominant effect: AA+AB vs. BB; and recessive effect: AA vs. AB+BB;) were examined to evaluate the 8q24 polymorphisms and PCa risk. The heterogeneity among studies was estimated with the I2 test which used to evaluate the proportion of statistical heterogeneity and the Q-statistic test which used to define the degree of heterogeneity. The fixed-effect model was employed when the effects are assumed to be homogenous (the P-value more than 0.10 for the Q-test and I2 less than 50%), while the random-effect model is used when they are heterogeneous. Analyses were performed using the software Review Manager 5.2 (Oxford, England, UK). All the comparisons of genetic models were conducted according the description by Collaboration et al. [12]. All p-values were two-sided.
Results
Study inclusion
The literature search initially identified 106 studies that concerning the relationship between 8q24 polymorphisms and PCa risk. After applying the inclusion criteria, only 11 studies (6 in English [13-18] and 5 in Chinese [19-23]) were finally included. The study selection process was shown in Figure 1. Six SNPs was discussed, including rs1447295, rs6983267, rs6983561, rs7837688, rs10090154 and rs16901979. All of the included studies wereconducted in Chinese population. The sample size ranged from 80 to 1036. Table 1 presented the main characteristics and genotype information of included studies.
Figure 1.
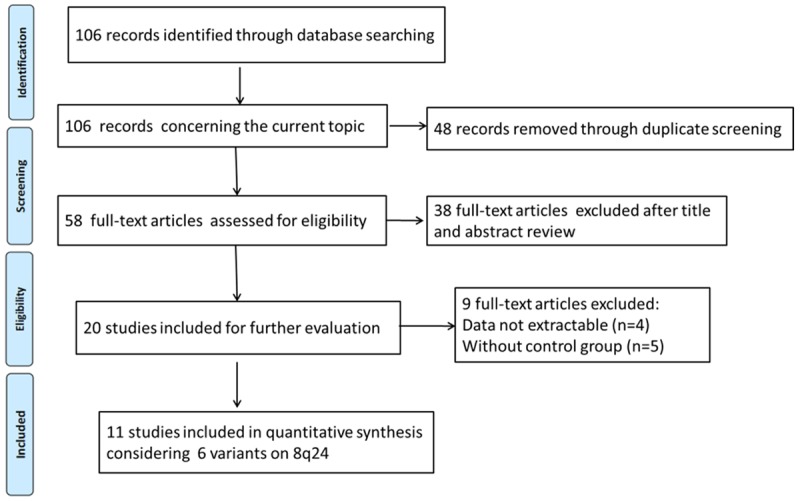
Flow chart of literature screening.
Table 1.
Main characteristics and genotype information of included studies
| First author | Year | Cases | Controls | Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| AA | AB | BB | A | B | AA | AB | BB | A | B | ||||
| rs1447295 (C/A) | AA | AC | CC | A | C | AA | AC | CC | A | C | |||
| Wang L | 2007 | 491 | 545 | 9 | 99 | 383 | 117 | 865 | 5 | 101 | 439 | 111 | 979 |
| Chen M | 2009 | 340 | 337 | 6 | 119 | 215 | 131 | 549 | 9 | 75 | 253 | 93 | 581 |
| Xie HJ | 2010 | 120 | 120 | 5 | 41 | 74 | 51 | 189 | 4 | 26 | 90 | 34 | 206 |
| Zheng SL | 2010 | 288 | 155 | 15 | 96 | 173 | 126 | 442 | 6 | 35 | 110 | 47 | 255 |
| Liu Y | 2011 | 40 | 40 | 22 | 7 | 11 | 51 | 29 | 20 | 15 | 5 | 55 | 25 |
| Chan J | 2013 | 289 | 144 | 17 | 92 | 180 | 126 | 452 | 5 | 44 | 94 | 54 | 232 |
| Zhao CX | 2013 | 289 | 288 | 8 | 108 | 161 | 124 | 430 | 4 | 86 | 197 | 94 | 480 |
| Zhang Z | 2014 | 123 | 137 | 4 | 45 | 74 | 53 | 193 | 2 | 44 | 91 | 48 | 226 |
| rs6983267 (T/G) | GG | GT | TT | G | T | GG | GT | TT | G | T | |||
| Zheng SL | 2010 | 288 | 155 | 62 | 134 | 86 | 258 | 306 | 29 | 72 | 51 | 130 | 174 |
| Liu Y | 2011 | 40 | 40 | 5 | 23 | 12 | 33 | 47 | 16 | 17 | 7 | 49 | 31 |
| Chan J | 2013 | 289 | 144 | 63 | 136 | 89 | 262 | 314 | 23 | 74 | 47 | 120 | 168 |
| Zhao CX | 2013 | 289 | 288 | 56 | 149 | 77 | 261 | 303 | 51 | 137 | 94 | 239 | 325 |
| Zhang Z | 2014 | 124 | 138 | 28 | 54 | 42 | 110 | 138 | 26 | 67 | 45 | 119 | 157 |
| rs6983561 (A/C) | CC | AC | AA | C | A | CC | AC | AA | C | A | |||
| Chen M | 2010 | 324 | 336 | 37 | 152 | 135 | 226 | 422 | 25 | 136 | 175 | 186 | 486 |
| Xie HJ | 2010 | 120 | 120 | 11 | 53 | 56 | 75 | 165 | 8 | 50 | 62 | 66 | 174 |
| Zheng SL | 2010 | 288 | 155 | 34 | 141 | 109 | 209 | 359 | 8 | 53 | 80 | 69 | 213 |
| Zhang YR | 2012 | 212 | 231 | 22 | 80 | 110 | 124 | 300 | 14 | 87 | 130 | 115 | 347 |
| rs7837688 (G/T) | TT | GT | GG | T | G | TT | GT | GG | T | G | |||
| Zhao CX | 2013 | 289 | 288 | 5 | 103 | 171 | 113 | 445 | 4 | 84 | 194 | 92 | 472 |
| Zhang Z | 2014 | 122 | 135 | 1 | 46 | 75 | 48 | 196 | 2 | 48 | 85 | 52 | 218 |
| rs10090154 (C/T) | TT | CT | CC | T | C | TT | CT | CC | T | C | |||
| Zheng SL | 2010 | 288 | 155 | 14 | 98 | 170 | 126 | 438 | 6 | 30 | 112 | 42 | 254 |
| Pu LM | 2011 | 124 | 111 | 1 | 48 | 74 | 50 | 196 | 1 | 32 | 63 | 34 | 158 |
| Zhao CX | 2013 | 289 | 288 | 5 | 106 | 168 | 116 | 442 | 4 | 73 | 203 | 81 | 479 |
| Zhang Z | 2014 | 123 | 131 | 1 | 48 | 74 | 50 | 196 | 2 | 39 | 90 | 43 | 219 |
| rs16901979 (C/A) | AA | AC | CC | A | C | AA | AC | CC | A | C | |||
| Chen M | 2010 | 331 | 335 | 35 | 148 | 148 | 218 | 444 | 24 | 138 | 173 | 186 | 484 |
| Xie HJ | 2010 | 120 | 120 | 10 | 56 | 54 | 76 | 164 | 8 | 54 | 58 | 70 | 170 |
| Zheng SL | 2010 | 288 | 155 | 34 | 139 | 110 | 207 | 359 | 8 | 52 | 85 | 68 | 222 |
| Chan J | 2013 | 289 | 144 | 31 | 119 | 139 | 181 | 397 | 12 | 68 | 64 | 92 | 196 |
A = risk allele; B = non-risk allele; AA = homozygous risk alleles; AB = heterozygous; BB = homozygous non-risk alleles.
Meta-analysis of 8q24 variants on PCa risk in Chinese population
Table 2 listed the results of allelic effect, dominant effect and recessive effect of all the six 8q24 polymorphisms on PCa risk.
Table 2.
Meta-analysis of 8q24 variants on prostate cancer risk in Chinese population
| SNPs | N | Risk allele | Comparison | Z test of studies | P-value for homogeneity | Model | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| OR (95% CI) | P | Ph | I2 | |||||
| rs1447295 | 8 | A | A vs. C | 1.35 (1.19, 1.53) | <0.00001 | 0.59 | 0% | F |
| AA+AC vs. CC | 1.42 (1.23, 1.64) | <0.00001 | 0.13 | 37% | F | |||
| AA vs. AC+CC | 1.40 (0.96, 2.05) | 0.08 | 0.84 | 0% | F | |||
| rs6983267 | 5 | G | G vs. T | 1.05 (0.85, 1.29) | 0.67 | 0.08 | 52% | R |
| GG+GT vs. TT | 1.12 (0.91, 1.37) | 0.30 | 0.46 | 0% | F | |||
| GG vs. GT+TT | 1.06 (0.72, 1.58) | 0.76 | 0.05 | 58% | R | |||
| rs6983561 | 4 | C | C vs. A | 1.41 (1.21, 1.63) | <0.00001 | 0.32 | 15% | F |
| CC+AC vs. AA | 1.49 (1.23, 1.80) | <0.0001 | 0.20 | 36% | F | |||
| CC vs. AC+AA | 1.74 (1.23, 2.47) | 0.002 | 0.87 | 0% | F | |||
| rs7837688 | 2 | T | T vs. G | 1.21 (0.94, 1.55) | 0.14 | 0.38 | 0% | F |
| TT+GT vs. GG | 1.28 (0.96, 1.70) | 0.09 | 0.39 | 0% | F | |||
| TT vs. GT+GG | 1.03 (0.33, 3.24) | 0.95 | 0.55 | 0% | F | |||
| rs10090154 | 4 | T | T vs. C | 1.48 (1.22, 1.80) | <0.00001 | 0.59 | 0% | F |
| TT+TC vs. CC | 1.67 (1.34, 2.09) | <0.00001 | 0.55 | 0% | F | |||
| TT vs. TC+CC | 1.11 (0.55, 2.27) | 0.77 | 0.92 | 0% | F | |||
| rs16901979 | 4 | A | A vs. C | 1.28 (0.98, 1.67) | 0.07 | 0.03 | 67% | R |
| AA+AC vs. CC | 1.31 (0.90, 1.92) | 0.16 | 0.01 | 73% | R | |||
| AA vs. AC+CC | 1.58 (1.11, 2.24) | 0.01 | 0.71 | 0% | F | |||
SNP, single nucleotide polymorphism; N, number of included studies for a certain variant; Ph, p-value of heterogeneity; F, fixed-effect model; R, random-effect model.
rs1447295
Eight studies that met the inclusion criteria were retrieved, including 1964 PCa cases and 1760 controls. No significant heterogeneity was found among studies and the fixed-effect model was used. Overall, our result discovered that the frequency of A risk allele was higher in cases than that in controls (19.8% vs. 15.2%), showing a significant association between the A allele and PCa risk (OR=1.35, 95% CI=1.19-1.53, P<0.00001) as shown in Figure 2. This significant relationship was also seen in dominant effect (AA+AC vs. CC: OR=1.42, 95% CI=1.23-1.64, P<0.00001), while was not found in receive effect (AA vs. AC+CC: OR=1.40, 95% CI=0.96-2.05, P=0.08).
Figure 2.
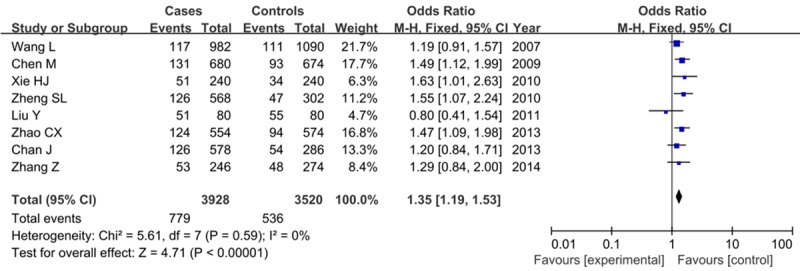
Forest plot on the association for allelic model (A vs. C) of rs1447295 on 8q24 and risk of PCa in a fixed-effects model.
rs6983267
We identified five studies that reported genotype frequencies of rs6983267, containing 1016 cases and 756 controls. No significant association was found between rs6983267 risk allele and PCa risk under any genetic models (G vs. T: OR=1.05, 95% CI=0.85-1.29, P=0.67; GG+GT vs. TT: OR=1.12, 95% CI=0.91-1.37, P=0.30; GG vs. GT+TT: OR=1.06, 95% CI=0.72-1.58, P=0.76).
rs6983561
Four articles were screened out concerning the relationship between rs6983561 and PCa risk, and including 940 cases and 828 controls. The C allele rate was higher in patients than that in controls (33.7% vs. 26.3%), and genotyping studies indicated that the C allele was associated with a higher risk for PCa (C vs. A: OR=1.41, 95% CI=1.21-1.63, P<0.00001). The CC+AC genotype in dominant effect and CC genotype in receive effect were significant increased the PCa risk as well (CC+AC vs. AA: OR=1.49, 95% CI=1.23-1.80, P<0.0001; CC vs. AC+AA: OR=1.74, 95% CI=1.23-2.47, P=0.002). Figure 3 showed the results of 8q24 rs6983561 on PCa risk.
Figure 3.
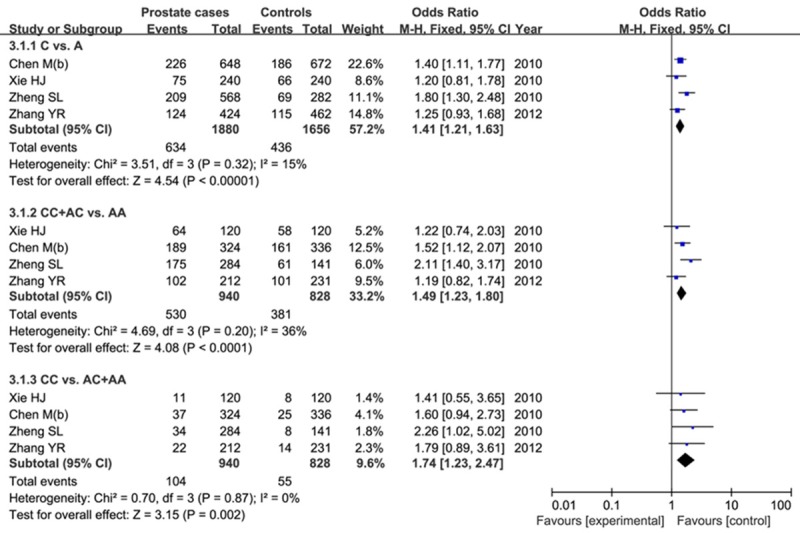
Forest plot of the results of 8q24 rs6983561 on PCa risk.
rs7837688
For rs7837688 variant, two studies were assessed, including 401 cases and 417 controls. Overall, our study did not find a significant association between this SNP and PCa risk in any genetic models in a fixed-effect model (T vs. G: OR=1.21, 95% CI=0.94-1.55, P=0.14; TT+GT vs. GG: OR=1.28, 95% CI=0.96-1.70, P=0.09; TT vs. GT+GG: OR=1.03, 95% CI=0.33-3.24, P=0.95).
rs10090154
This SNP was genotyped in men with PCa in 4 studies, including 807 cases and 655 controls. We found a significantly higher frequency of the C risk allele in cases than in controls (21.2% vs. 15.3%). The results indicated a significant association between the rs10090154 risk allele and PCa risk in allele effect and dominant effect (T vs. C: OR=1.48, 95% CI=1.22-1.80, P<0.00001; TT+TC vs. CC: OR=1.67, 95% CI=1.34-2.09, P<0.00001) as shown in Figure 4. However, no significant relationship was found between TT genotype in receive effect and PCa risk (OR=1.11, 95% CI=0.55-2.27, P=0.77).
Figure 4.
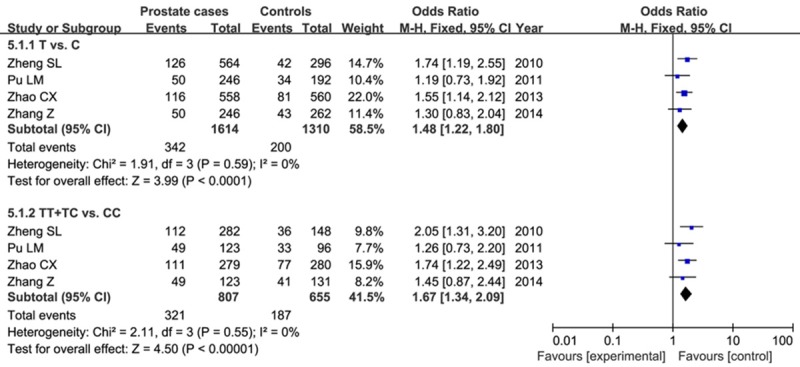
Forest plot of the role of T allele in allelic model and TT+TC in dominant model on PCa risk.
rs16901979
Four studies assessed rs16901979 in Chinese population, containing 1023 patients and 744 controls. Our meta-analysis found that rs16901979 AA genotype significantly increased the risk of PCa in receive model (AA vs. AC+CC: OR=1.58, 95% CI=1.11-2.24, P=0.01) in fixed-effect model as shown in Figure 5. However, this relationship was not found in other genetic models in random-effect model (A vs. C: OR=1.28, 95% CI=0.98-1.67, P=0.07; AA+AC vs. CC: OR=1.31, 95% CI=0.90-2.24, P=0.16).
Figure 5.
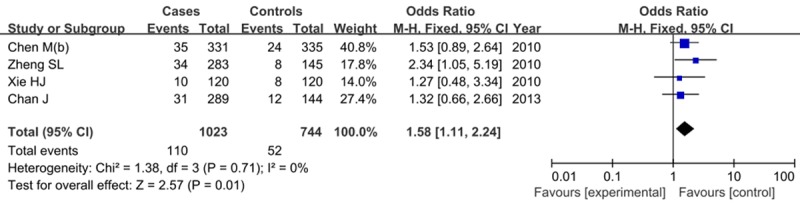
Forest plot of AA genotype in recessive model of rs16901979 on PCa risk.
Publication bias
Each study in any comparisons was deleted once a time, and the ORs were not significantly changed, indicating no publication bias was presented. Furthermore, the funnel plot did not show an asymmetry as shown in Figure 6.
Figure 6.
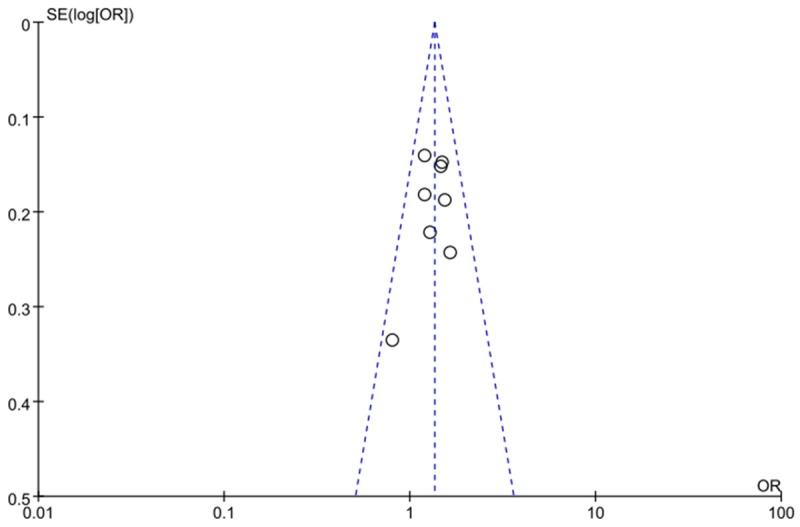
Funnel plot on the association for allelic model (A vs. C) of rs1447295 on 8q24 and PCa risk in a fixed-effects model.
Discussion
Human chromosome 8q24 is a risk locus for many cancers, and is currently considered as the most important susceptibility region for PCa risk. But the results from previous studies still remained objectionable between different populations. In this meta-analysis, we evaluated the association between six single nucleotide polymorphism (SNPs) on 8q24 and PCa risk in men of Chinese population. The result showed that the A allele in rs1447295, C allele in rs6983561 and T allele in rs10090154 significantly increased the risk of PCa. Furthermore, genotypes of AA+AC in rs1447295, CC+AC and CC in rs6983561, TT+TC in rs10090154 and AA in rs16901979 were associated with PCa risk as well. No significant association was found between rs6983267, rs7837688 and PCa risk. This is the first study to systematically evaluate the role of 8q24 variants on PCa risk in Chinese population.
The mechanism of 8q24 affect the process of PCa is still poorly understood. Evidences have shown that this risk region may function as a regulatory hub by physical interactions with multiple genes important for prostate carcinogenesis such as PVT1 (a host gene for several miRNAs), FAM84B and GSDMC [24]. 8q24 cancer-associated variants increase prostate enhancer activity whose expression mimics that of the nearby MYC (a transcriptional activator controlling cell growth, apoptosis, differentiation and other cellular responses) proto-oncogene in vivo [25,26]. The 8q24 risk locus is proposed to operate through a common mechanism-as tissue-specific enhancers of MYC [26]. The region surrounding rs378854 on 8q24 is proved to be interacted with the MYC and PVT1 promoters [27]. 8q24 amplification is associated with MYC expression and PCa progression and is an independent predictor of recurrence after radical prostatectomy [28]. Furthermore, a rare variant rs188140481, which destroys a FoxA1 site at 8q24, is associated with PCa risk [29].
Many SNPs are proved to be associated with PCa risk. The rs6983267 in region 3 of the chromosome 8q24 appears to be a prominent risk factor for PCa [30]. A new low-frequency variant, rs188140481 [A] at 8q24 has been identified to be associated with PCa in European populations [31]. This variant also conferred greater risk and its carriers were 6.73-fold more likely to develop PCa than non-carriers [32]. Zhao et al. discovered that the rs4242382-A variation might be associated with increased PCa susceptibility and might be a useful risk biomarker for PCa in multi-ethnic populations [33]. Chung et al. suggested that SNPs between rs1456315 and rs7463708 to be most significantly associated with PC susceptibility [34]. Okobia et al. found that SNP rs16901979 in region 2 was associated with significantly increased risk of PCa with the risk stronger in men with early-onset PCa [35]. Hui et al. showed that the loci including rs10086908, rs1016343, and rs6983561 at 8q24 could be associated with PCa in Jing-jin residents in northern China [36].
Previous studies have also shown that 8q24 variants increased the risk of other cancers. rs35252396 [CG] located at 8q24.21, was proved to be significantly associated with renal cell carcinoma and had an average risk allele frequency in controls of 46% [37]. Bladder cancer risk was associated specifically with variation in the discrete 8q24 region containing rs9642880 [38]. Three polymorphic sites at chromosome 8q24 (rs7837328, rs10808555, rs6983267) had been associated with risk for colorectal adenomas and colorectal epithelial cell proliferation [39]. The SNP rs10505477 might contribute to the survival of gastric cancer and be a potential prognostic biomarker of gastric cancer [40]. Rs13281615, rs6983267 polymorphisms in 8q24 may contribute to susceptibility to breast cancer risk [41].
Several limitations were presented in the meta-analysis. Firstly, the number of included studies was small, for example, only two articles were retrieved for rs7837688. Secondly, SNP-SNP interaction should be considered. Zheng et al. have discussed the cumulative association of SNPs on 8q24, and the results showed that when the most significant SNPs from different regions were included in a multivariate analysis, each SNP remained significant after adjustment for other SNPs and family history [42]. Thirdly, we only assessed the Chinese population, while other population should be considered as well. Lastly, other risk factors such as smoking, age should be included in the future research.
In conclusion, our results showed that rs1447295 A allele, rs6983561 C allele and rs10090154 T allele are significantly associated with increased the risk of PCa. No significant association was found between rs6983267, rs7837688 and PCa risk. Further studies are needed to systematically evaluate 8q24 variants on PCa risk among all populations.
Disclosure of conflict of interest
None.
References
- 1.Plata Bello A, Concepcion Masip T. Prostate cancer epidemiology. Arch Esp Urol. 2014;67:373–382. [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Hsing AW, Tsao L, Devesa SS. International trends and patterns of prostate cancer incidence and mortality. Int J Cancer. 2000;85:60–67. doi: 10.1002/(sici)1097-0215(20000101)85:1<60::aid-ijc11>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;1:R103–121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 5.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, Whittemore AS, Gayther SA, Giles GG, Guy M, Edwards SM, Morrison J, Donovan JL, Hamdy FC, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Brown PM, Hopper JL, Neal DE, Pharoah PD, Ponder BA, Eeles RA, Easton DF, Dunning AM. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cropp CD, Robbins CM, Sheng X, Hennis AJ, Carpten JD, Waterman L, Worrell R, Schwantes-An TH, Trent JM, Haiman CA, Leske MC, Wu SY, Bailey-Wilson JE, Nemesure B. 8q24 risk alleles and prostate cancer in African-Barbadian men. Prostate. 2014;74:1579–1588. doi: 10.1002/pros.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antczak A, Wokolorczyk D, Kluzniak W, Kashyap A, Jakubowska A, Gronwald J, Huzarski T, Byrski T, Debniak T, Masojc B, Gorski B, Gromowski T, Golab A, Sikorski A, Slojewski M, Gliniewicz B, Borkowski T, Borkowski A, Przybyla J, Sosnowski M, Malkiewicz B, Zdrojowy R, Sikorska-Radek P, Matych J, Wilkosz J, Rozanski W, Kis J, Bar K, Janiszewska H, Stawicka M, Milecki P, Lubinski J, Narod SA, Cybulski C. The variant allele of the rs188140481 polymorphism confers a moderate increase in the risk of prostate cancer in Polish men. Eur J Cancer Prev. 2015;24:122–7. doi: 10.1097/CEJ.0000000000000079. [DOI] [PubMed] [Google Scholar]
- 8.Zhang LL, Sun L, Zhu XQ, Xu Y, Yang K, Yang F, Yang YG, Chen GQ, Fu JC, Zheng CG, Li Y, Mu XQ, Shi XH, Zhao F, Wang F, Yang Z, Wang BY. rs10505474 and rs7837328 at 8q24 cumulatively confer risk of prostate cancer in Northern Han Chinese. Asian Pac J Cancer Prev. 2014;15:3129–3132. doi: 10.7314/apjcp.2014.15.7.3129. [DOI] [PubMed] [Google Scholar]
- 9.Oskina NA, Boyarskikh UA, Lazarev AF, Petrova VD, Ganov DI, Tonacheva OG, Lifshits GI, Filipenko ML. A replication study examining association of rs6983267, rs10090154, and rs1447295 common single nucleotide polymorphisms in 8q24 region with prostate cancer in Siberians. Urol Oncol. 2014;32:37.e7–12. doi: 10.1016/j.urolonc.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Troutman SM, Sissung TM, Cropp CD, Venzon DJ, Spencer SD, Adesunloye BA, Huang X, Karzai FH, Price DK, Figg WD. Racial disparities in the association between variants on 8q24 and prostate cancer: a systematic review and meta-analysis. Oncologist. 2012;17:312–320. doi: 10.1634/theoncologist.2011-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collaboration C, Review Manager (RevMan)[Computer Program] Version 5.2. Copenhagen: The Nordic Cochrane Centre; 2012. [Google Scholar]
- 13.Wang L, McDonnell SK, Slusser JP, Hebbring SJ, Cunningham JM, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Huang YC, Ko IL, Yang S, Chang YH, Huang WJ, Chen YM. The rs1447295 at 8q24 is a risk variant for prostate cancer in Taiwanese men. Urology. 2009;74:698–701. doi: 10.1016/j.urology.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 15.Chen M, Huang YC, Yang S, Hsu JM, Chang YH, Huang WJ, Chen YM. Common variants at 8q24 are associated with prostate cancer risk in Taiwanese men. Prostate. 2010;70:502–507. doi: 10.1002/pros.21084. [DOI] [PubMed] [Google Scholar]
- 16.Zheng SL, Hsing AW, Sun J, Chu LW, Yu K, Li G, Gao Z, Kim ST, Isaacs WB, Shen MC, Gao YT, Hoover RN, Xu J. Association of 17 prostate cancer susceptibility loci with prostate cancer risk in Chinese men. Prostate. 2010;70:425–432. doi: 10.1002/pros.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao CX, Liu M, Wang JY, Xu Y, Wei D, Yang K, Yang Z. Association of 8 loci on chromosome 8q24 with prostate carcinoma risk in northern Chinese men. Asian Pac J Cancer Prev. 2013;14:6733–6738. doi: 10.7314/apjcp.2013.14.11.6733. [DOI] [PubMed] [Google Scholar]
- 18.Chan JY, Li H, Singh O, Mahajan A, Ramasamy S, Subramaniyan K, Kanesvaran R, Sim HG, Chong TW, Teo YY, Chia SE, Tan MH, Chowbay B. 8q24 and 17q prostate cancer susceptibility loci in a multiethnic Asian cohort. Urol Oncol. 2013;31:1553–1560. doi: 10.1016/j.urolonc.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Wang J, Shi X, Yang Z, Huo Z. The association between EEFSEC, 8q24 and prostate cancer risk in Chinese Han Populations. Journal of Ningxia Medical University. 2012;34:981–985. [Google Scholar]
- 20.Zhang Z, Wang J, Wei D, Yang Z. Association study of 4 single nucleotide polymorphisms in 8q24 region and prostate cancer. Journal of Ningxia Medical University. 2014;36:608–614. [Google Scholar]
- 21.Pu L, Wei D, LIU M, Chen X, Wang J. The association between two single nucleotide polymorphisms and risk of prostate cancer in the northern Chinese population. China Oncology. 2011;21:688–695. [Google Scholar]
- 22.Xie H, Xu Y. Master’s dissertation. Tianjin Medical University; 2010. Association study between single nucleotide polymorphisms on 8q24 with prostate cancer risk in Chinese Han Population. [Google Scholar]
- 23.Liu Y, Tian X. Master’s dissertation. Tianjin Medical University; 2011. A study of relevance between single nucleotide polymorphism on 8q24 with prostate cancer risk in Chinese Tianjin Population. [Google Scholar]
- 24.Du M, Yuan T, Schilter KF, Dittmar RL, Mackinnon A, Huang X, Tschannen M, Worthey E, Jacob H, Xia S, Gao J, Tillmans L, Lu Y, Liu P, Thibodeau SN, Wang L. Prostate cancer risk locus at 8q24 as a regulatory hub by physical interactions with multiple genomic loci across the genome. Hum Mol Genet. 2015;24:154–66. doi: 10.1093/hmg/ddu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, Caswell JL, Beckwith CA, Hills A, Macconaill L, Coetzee GA, Regan MM, Freedman ML. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci U S A. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer KB, Maia AT, O’Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, Ambs S, Prokunina-Olsson L, Carroll J, Ponder BA. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7:e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, Cathelineau X, Cussenot O. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol. 2013;44:1617–1623. doi: 10.1016/j.humpath.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Hazelett DJ, Coetzee SG, Coetzee GA. A rare variant, which destroys a FoxA1 site at 8q24, is associated with prostate cancer risk. Cell Cycle. 2013;12:379–380. doi: 10.4161/cc.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Francisco IF, Rojas PA, Torres-Estay V, Smalley S, Cerda-Infante J, Montecinos VP, Hurtado C, Godoy AS. Association of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic population. J Cell Mol Med. 2014;18:125–133. doi: 10.1111/jcmm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, Benediktsdottir KR, Sigurdsson A, Magnusson OT, Gudjonsson SA, Magnusdottir DN, Johannsdottir H, Helgadottir HT, Stacey SN, Jonasdottir A, Olafsdottir SB, Thorleifsson G, Jonasson JG, Tryggvadottir L, Navarrete S, Fuertes F, Helfand BT, Hu Q, Csiki IE, Mates IN, Jinga V, Aben KK, van Oort IM, Vermeulen SH, Donovan JL, Hamdy FC, Ng CF, Chiu PK, Lau KM, Ng MC, Gulcher JR, Kong A, Catalona WJ, Mayordomo JI, Einarsson GV, Barkardottir RB, Jonsson E, Mates D, Neal DE, Kiemeney LA, Thorsteinsdottir U, Rafnar T, Stefansson K. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grin B, Loeb S, Roehl K, Cooper PR, Catalona WJ, Helfand BT. A rare 8q24 single nucleotide polymorphism (SNP) predisposes North American men to prostate cancer and possibly more aggressive disease. BJU Int. 2015;115:101–5. doi: 10.1111/bju.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao CX, Liu M, Xu Y, Yang K, Wei D, Shi XH, Yang F, Zhang YG, Wang X, Liang SY, Zhao F, Zhang YR, Wang NN, Chen X, Sun L, Zhu XQ, Yuan HP, Zhu L, Yang YG, Tang L, Jiao HY, Huo ZH, Wang JY, Yang Z. 8q24 rs4242382 polymorphism is a risk factor for prostate cancer among multi-ethnic populations: evidence from clinical detection in China and a meta-analysis. Asian Pac J Cancer Prev. 2014;15:8311–8317. doi: 10.7314/apjcp.2014.15.19.8311. [DOI] [PubMed] [Google Scholar]
- 34.Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T, Tsunoda T, Daigo Y, Matsuda K, Kamatani N, Nakamura Y, Kubo M. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 35.Okobia MN, Zmuda JM, Ferrell RE, Patrick AL, Bunker CH. Chromosome 8q24 variants are associated with prostate cancer risk in a high risk population of African ancestry. Prostate. 2011;71:1054–1063. doi: 10.1002/pros.21320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui J, Xu Y, Yang K, Liu M, Wei D, Zhang Y, Shi XH, Yang F, Wang N, Wang X, Liang S, Chen X, Sun L, Zhu X, Zhu L, Yang Y, Tang L, Yang Z, Wang J. Study of genetic variants of 8q21 and 8q24 associated with prostate cancer in Jing-Jin residents in northern China. Clin Lab. 2014;60:645–652. doi: 10.7754/clin.lab.2013.130624. [DOI] [PubMed] [Google Scholar]
- 37.Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Petursdottir V, Hardarson S, Gudjonsson SA, Johannsdottir H, Helgadottir HT, Stacey SN, Magnusson OT, Helgason H, Panadero A, van der Zanden LF, Aben KK, Vermeulen SH, Oosterwijk E, Kong A, Mayordomo JI, Sverrisdottir A, Jonsson E, Gudbjartsson T, Einarsson GV, Kiemeney LA, Thorsteinsdottir U, Rafnar T, Stefansson K. A common variant at 8q24.21 is associated with renal cell cancer. Nat Commun. 2013;4:2776. doi: 10.1038/ncomms3776. [DOI] [PubMed] [Google Scholar]
- 38.Cortessis VK, Yuan JM, Van Den Berg D, Jiang X, Gago-Dominguez M, Stern MC, Castelao JE, Xiang YB, Gao YT, Pike MC, Conti DV. Risk of urinary bladder cancer is associated with 8q24 variant rs9642880 [T] in multiple racial/ethnic groups: results from the Los Angeles-Shanghai case-control study. Cancer Epidemiol Biomarkers Prev. 2010;19:3150–3156. doi: 10.1158/1055-9965.EPI-10-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B, Thyagarajan B, Gross MD, Goodman M, Sun YV, Bostick RM. Genetic variants at chromosome 8q24, colorectal epithelial cell proliferation, and risk for incident, sporadic colorectal adenomas. Mol Carcinog. 2014;53(Suppl 1):E187–192. doi: 10.1002/mc.22047. [DOI] [PubMed] [Google Scholar]
- 40.Ma G, Gu D, Lv C, Chu H, Xu Z, Tong N, Wang M, Tang C, Xu Y, Zhang Z, Wang B, Chen J. Genetic variant in 8q24 is associated with prognosis for gastric cancer in a Chinese population. J Gastroenterol Hepatol. 2015;30:689–95. doi: 10.1111/jgh.12801. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Yi P, Chen W, Ming J, Zhu B, Li Z, Shen N, Shi W, Ke J, Zhao Q, Lu X, Xun X, Liu L, Song R, Guo H, Zhong R, Liang L, Huang T, Miao X. Association between polymorphisms within the susceptibility region 8q24 and breast cancer in a Chinese population. Tumour Biol. 2014;35:2649–2654. doi: 10.1007/s13277-013-1348-0. [DOI] [PubMed] [Google Scholar]
- 42.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Balter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Gronberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]


