Abstract
Implementation of next-generation DNA sequencing (NGS) technology into routine diagnostic genome care requires strategic choices. Instead of theoretical discussions on the consequences of such choices, we compared NGS-based diagnostic practices in eight clinical genetic centers in the Netherlands, based on genetic testing of nine pre-selected patients with cardiomyopathy. We highlight critical implementation choices, including the specific contributions of laboratory and medical specialists, bioinformaticians and researchers to diagnostic genome care, and how these affect interpretation and reporting of variants. Reported pathogenic mutations were consistent for all but one patient. Of the two centers that were inconsistent in their diagnosis, one reported to have found ‘no causal variant', thereby underdiagnosing this patient. The other provided an alternative diagnosis, identifying another variant as causal than the other centers. Ethical and legal analysis showed that informed consent procedures in all centers were generally adequate for diagnostic NGS applications that target a limited set of genes, but not for exome- and genome-based diagnosis. We propose changes to further improve and align these procedures, taking into account the blurring boundary between diagnostics and research, and specific counseling options for exome- and genome-based diagnostics. We conclude that alternative diagnoses may infer a certain level of ‘greediness' to come to a positive diagnosis in interpreting sequencing results. Moreover, there is an increasing interdependence of clinic, diagnostics and research departments for comprehensive diagnostic genome care. Therefore, we invite clinical geneticists, physicians, researchers, bioinformatics experts and patients to reconsider their role and position in future diagnostic genome care.
Introduction
Clinical genetics is changing. Next-generation DNA sequencing (NGS) is slowly replacing traditional technologies for the diagnosis of genetic disorders. Instead of gene-by-gene approaches, large sets of genes can now be addressed in a single test. Moreover, scientific progress rapidly expands the group of ‘genetic disorders'. Compared with the previous ‘revolution' in molecular diagnostics – the introduction of genomic microarrays as a diagnostic tool 1 – NGS affects many more aspects of routine diagnostics.
The technology to sequence DNA has been under major development over the past 5 years. Excellent reviews have been published on the basic principles,2 the impact,3 and various applications4 of NGS. The steep decrease in run times and costs have turned reading an individual's DNA from a multi-annual, multi-laboratory undertaking into a feasible effort for individual laboratories within a reasonable time frame. Many individual genomes have now been sequenced, leading to increasingly comprehensive and specific maps of human genetic variation.5, 6, 7 Initial assessment of personal genome information proved highly valuable for disease characterization in individual patients.8, 9, 10, 11, 12 Moreover, exome sequencing revealed causal genetic mutations for rare congenital syndromes,13, 14, 15, 16, 17 intellectual disability,18, 19 autism 20 and schizophrenia.21 Nowadays, NGS technology is widely used in many areas of clinical genetic research, including genome-wide association studies for common disease.22, 23, 24
Many see the widespread application of NGS in research as a prelude to its broad acceptance as a diagnostic tool, thereby replacing most other molecular diagnostic technologies.25, 26, 27, 28 Indeed, a number of pioneering laboratories have already successfully implemented NGS-based gene panels in a diagnostic setting, but routine application in a widespread clinical context requires a further decrease in costs and run times.16, 29, 30, 31 The latest generation of NGS technologies – in various stages of development – holds great promise to bridge the cost-efficiency gap.32, 33, 34 Yet the biggest challenges are not in the technology itself, but in aspects that follow application of NGS, such as interpretation and exchange of data, informing patients appropriately and increased interdependencies of people involved in genome diagnostics.
Anticipating these developments, many clinical genetic centers (CGCs) and molecular genetic laboratories are preparing for a reconfiguration of their diagnostic process, and in doing so, make implementation choices based on local requirements and opportunities. Already, many molecular diagnostic laboratories have adopted NGS as the preferred technology for diagnosing an increasing number of diseases.16, 27, 30, 31, 35, 36, 37, 38 In the Netherlands, all CGCs have implemented one or more NGS-based diagnostic applications.39
To study the effect of implementation choices within each center, we set up a small-scale diagnostic NGS-based testing effort with all eight CGCs in the Netherlands. All centers received a request for diagnostic testing of nine patients who had been previously diagnosed for inherited forms of cardiomyopathy using traditional genetic technologies. Over the course of 6 months, each laboratory took the samples through their internal NGS-based testing facilities – which were set up as routine diagnostics or in a research context – and delivered a full diagnostic report for each patient. No prior criteria were set for sample intake, sequencing, data analysis or clinical interpretation; each laboratory was entirely autonomous in choosing its preferred approach. All centers drafted short summary reports of each step in the process, describing the key elements in their approach. The reports, combined with the diagnostic outcomes and underlying data provide valuable insights in implementation choices of NGS-based diagnostics infrastructure and their consequences. The overall aim of this study was therefore not to develop a standard diagnostic process or data analysis pipeline, nor to compare centers or platforms on performance or data quality. Rather, the set up of this study allowed for assessment of the existing variability in NGS-based diagnostics approaches, providing a starting point for discussions on future conditions for implementation procedures in the Netherlands and elsewhere.
Materials and Methods
Study design
All CGCs in the Netherlands received DNA samples and medical records from the same nine patients with cardiomyopathy. The centers were completely autonomous in applying their individual strategy for providing an NGS-based diagnosis; no restrictions or guidelines were set to the capture method, quality of sequencing, analysis tools and settings, or variant reporting. All centers provided summary reports of each phase, describing the approaches, the people in charge and general observations. Choices and procedures within each phase were specific for each CGC, and generally depended on internal organization, experience and expertize, and available laboratory, as well as bioinformatics infrastructure. Each CGC completed the diagnostic trajectory for the nine patients by providing a full diagnostic report for each patient, stating the identified causal variant (if present), other clinically relevant variants and suggestions for further testing.
For practical reasons, the individual patients were not seen by any of the clinical geneticists. Instead, we provided all CGCs with full (anonymized) medical records of all patients.
Patients
The patients had previously undergone genetic diagnostics for cardiomyopathy using traditional diagnostic methods. Causative mutations were identified and reported, but this was not disclosed to the receiving centers. DNA and appropriate consent were available for each patient, as well as overall approval of the medical ethics committee of the Academic Medical Center, Amsterdam, The Netherlands, for this multicenter trial. Whereas not specified beforehand, the genetic background of the patients was presumed not to be complex; causal nucleotide substitutions or indels in well-known cardiomyopathy genes were the cause of disease in all patients with a positive diagnosis. Full medical records – anonymised − for all patients were shared with all centers, containing family history, electrocardiogram results, previous tests and other relevant information.
DNA samples
DNA for all samples was centrally isolated by a single technician (to limit manipulation bias) from blood-derived cell lines and diluted to 500 ng/ul. Samples were split and a total of 10 μg of each sample was sent to every center. Individual 10 μg-portions were sent out to each center; 90 samples in total (the laboratories that applied an additional outsourcing strategy received two sets). From there, the individual laboratories started their internal diagnostic procedure. This procedure deviates slightly from routine diagnostics, where centers generally receive whole-blood samples for genetic testing. Initially, the underlying motivation for this approach was purely pragmatic – drawing 10 tubes of whole blood from each patient would be very invasive. However, in the context of the main objective of CARDIO – to identify new challenges associated with NGS-based diagnostics – one could take a broader perspective, and consider this set up a proxy for dealing with diagnostic requests from outside the own institute. Samples were not recognized (taken up) by all diagnostic centers and incorporated in the main flow as external requests, but rather considered as research samples.
Data analysis
The data obtained from the sequencing platforms were initially analyzed according to the available pipelines within the individual centers.
In the second phase, all data were centrally collected and annotated by five different annotation tools; commercial packages from Cartagenia (Leuven, Belgium), Ingenuity (Qiagen, Redwood City, CA, USA) and SoftGenetics (State College, PA, USA), and in-house developed tools from CGCs 4 and 8.
Data were submitted to the Leiden Open Variant Database (LOVD), under the following accession links: http://databases.lovd.nl/shared/individuals/
Patient 1: 00016140
Patient 2: 00016141
Patient 3: 00016142
Patient 4: 00016143
Patient 5: 00016144
Patient 6: 00016145
Patient 7: 00016146
Patient 8: 00016147
Patient 9: 00016148
Informed consent forms
All laboratories provided additional information on their local informed consent procedures. As these procedures could not be simulated (the laboratories only received DNA and medical records; they did not see patients themselves), the laboratories responded to hypothetical questions on the consent that would be required from the patients in a routine setting. In a pre-sequencing report, the laboratories indicated whether or not they would have the patient's signed consent for the NGS-based diagnostic test that they were about to perform.
Results
Following the objective of this study − to analyze the choices that each CGC made to reach a genetic diagnosis for nine patients − the results do not comprise a performance comparison of sequencing platforms or bioinformatics pipelines based on technical parameters. Moreover, all patients had already received a definite diagnosis, and therefore the nature and type of mutations identified are irrelevant in the context of this study. Instead, the results focus on the collected information that reflects the considerations for the interpretation and reporting of variants − including personal communication. On the basis of the collected quantitative and qualitative data we identified critical issues and challenges for future diagnostic genome care.
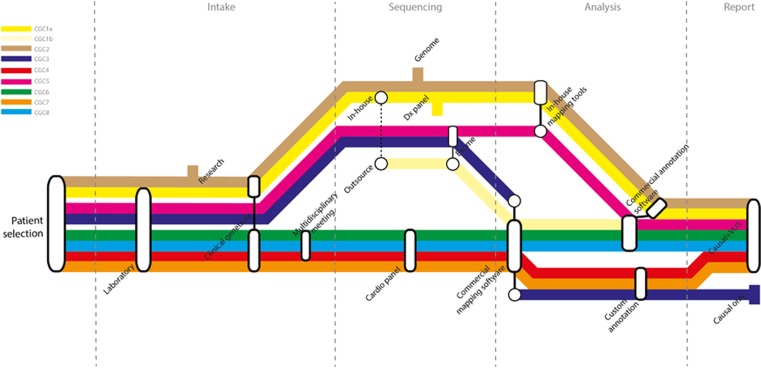
The centers were completely autonomous in their choices for sample intake, sequencing approach, data analysis and clinical interpretation, which resulted in a wide routing spectrum for the patient samples across centers (Figure 1). Critical choices in each phase of the diagnostic process – intake, sequencing, data analysis and interpretation, and reporting − determined the route in each center.
Figure 1.
Overview of the diagnostic process. The diagnostic process of nine samples with cardiomyopathies varied substantially among CGCs. Each colored line represents the process the nine patients went through in a particular CGC. The ‘metro stops' represent choices that the various CGCs made in the intake, sequencing, analysis and reporting phase.
Sequencing approach
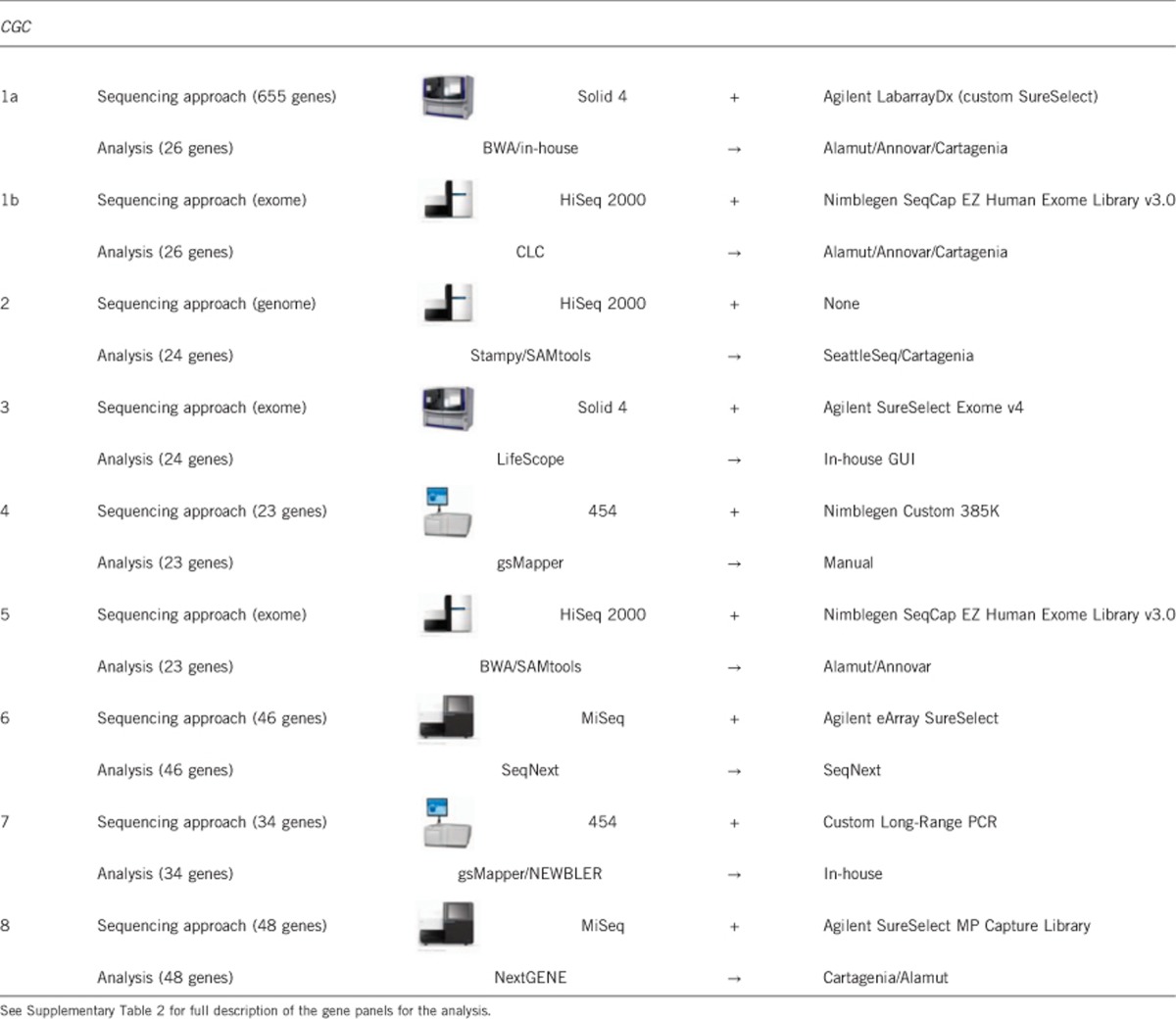
The choice for a particular sequencing approach generally depended on the available in-house infrastructure within each center. CGCs 1 and 2 both applied an ‘outsourcing' strategy; they outsourced part of the diagnostic trajectory to a service provider as an addition to their in-house strategies. CGC 1 treated the ‘in-house' and ‘outsourcing' approaches as separate trajectories, that is enrichment, sequencing and primary data analysis were done independently in-house (Figure 1; CGC1a) and at the service provider (CGC1b). Downstream analysis and interpretation were done in-house for both trajectories, according to the NEN-ISO 15189/CCKL accreditation guidelines. CGC 2 only outsourced the sample preparation step and subsequently combined the corresponding in-house and outsourced samples for whole-genome sequencing (WGS; because of budgetary restrictions). Therefore the outsourcing strategy of CGC 2 was rather an internal-technical diagnostic validation strategy, and was considered as a single strategy for the rest of the study. The study thus comprised nine distinct combinations of enrichment protocols and sequencing platforms (Table 1); four cardiomyopathy-targeted gene panels (23−48 genes; CGCs 4, 6, 7 and 8), one array-based panel of 655 diagnostically interpretable genes (CGC 1), three whole-exome sequencing (WES) approaches (CGCs 1b, 3 and 5) and one WGS approach (CGC 2). Overall, we observed a 10−20 times average coverage difference (30–500x) between the various approaches (Supplementary Table 1). Because our objective was not to compare platforms, we did not collect other parameters, such as coverage per target base and non-covered target bases.
Table 1. Overview of the sequencing approach of the participating laboratories.
Data analyses and interpretation
The subsequent phase − data analysis − revealed two main sources of variation. First, the number of genes included in the analysis panel varied considerably among centers, ranging from 23 to 48 (Table 1 and Supplementary Table 2). We set no prior criteria for the genes to be included in the analysis, nor for the type of variants to be considered. CGCs applying broad-enrichment or WGS approaches (CGCs 1, 2, 3 and 5) all restricted their analyses to existing panels of cardiomyopathy-related genes, provided to them by one of the other centers. This is in accordance with existing informed consent procedures, which are based on a policy of non-active screening for variants unrelated to the disease (unsolicited findings). Whereas the average number of (identified) variants per patient ranged from 1315 to 4.2 million, the diagnostic interpretation was restricted to the ±50 variants implicated in cardiomyopathy.
Second, there was considerable variety in the tools that were used for analysis and primary interpretation of the data. To reduce the number of variants to manageable proportions (up to 500 000-fold reductions), the CGCs used custom in-house analysis pipelines (CGCs 2 and 7), commercially available software packages (CGCs 6 and 8), or a combination of both (CGCs 1, 3, 4 and 5; Table 1). As a consequence, there was a wide variety in resulting file formats for mapped, called and annotated data. This became especially relevant when we set out to perform a coordinated analysis of all data from this study. Even files that appeared in a 'standard' format (like .BAM or .VCF) had center-specific content, which required extensive reformatting for inclusion in the central analysis.
Central collection of all data required repeated requests and depended much on the willingness of individuals to upload data, indicating that most centers are not optimally set up for sharing data. Owing to its total size (±1.5 TB), the files from CGC 2 (whole-genome data) were transferred via an external hard-drive, instead of an upload to a central server. Eventually, we collected all data in workable formats, and performed two subsequent studies:
A proof-of-concept to explore possibilities for using centralised data infrastructure. CGC 2 and CGC 6 set up a collaboration with the Dutch Health Hub (www.dutchhealthhub.nl) to test its storage, analysis and sharing capabilities. On the basis of the proof-of-concept study we identified two important challenges. First, centralizing data infrastructure requires cooperation of many people within the individual organizations. Second, doing the two phases in parallel – technical development and scientific implementation – results in miscommunication and requires strict management of expectations.
A proof-of-principle on three commercially available data analysis packages. We tested the capacities of three off-the-shelf software packages – Bench Lab NGS (Cartagenia; www.cartagenia.com), Variant Analysis (Ingenuity; www.ingenuity.com) and Geneticist Assistant (Softgenetics/BioKé www.softgenetics.com) – to consistently analyze data and filter variants. All packages consistently identified the 8 causal variants in all data sets, but the number of variants included as clinically significant differed between the various tools. Further in-depth analysis on the nature of these differences was not performed as this is beyond the scope of this study.
Diagnoses
Despite the variety in approach and analyses, the primary molecular diagnoses – as indicated in the diagnostic letters that would in normal practice be sent to the responsible clinician – were largely consistent over the eight centers. Patient 9 formed an exception to this consistency, for which six CGCs reported a causal mutation in MYBPC3 (Table 2). This MYBPC3 mutation was not identified by CGC 1 and CGC 8, owing to low sequence coverage in the exon 26 region of this gene, where the mutation was located. The subsequent interpretation and diagnosis for patient 9 differed considerably between these two centers. CGC 8 – a center for which cardiomyopathy diagnostics is a key strategy program – reported to have found ‘no causal variant' in patient 9. CGC 8 thus provided an underdiagnosis based on the NGS results. Conversely, CGC 1 – with less focus on diagnosis of cardiomyopathies – reported that ‘the diagnosis of Fabry disease' was confirmed in patient 9, based on the detection of another mutation in GLA. This center thus provided an alternative diagnosis for patient 9.
Table 2. Diagnosis for 9 CARDIO patientsa.
| Patient | CGC 1 | CGC 2 | CGC 3 | CGC 4 | CGC 5 | CGC 6 | CGC 7 | CGC 8 |
|---|---|---|---|---|---|---|---|---|
| 1 | TNNI3 | TNNI3 | TNNI3 | TNNI3 | TNNI3 | TNNI3 | TNNI3 | TNNI3 |
| 2 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 |
| 3 | MYH7 | MYH7 | MYH7 | MYH7 | MYH7 | MYH7 | MYH7 | MYH7 |
| 4 | − | − | − | − | − | − | MYH7 | − |
| 5 | LMNA | LMNA | LMNA | LMNA | LMNA | LMNA | LMNA | NA |
| 6 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 |
| 7 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 |
| 8 | PLN | PLN | PLN | PLN | PLN | PLN | PLN | PLN |
| 9 | GLA | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | MYBPC3 | − |
Each center independently provided a diagnosis for each patient based on the most likely gene mutation. The table cells indicate in which gene the particular center identified the causal variant for the corresponding patient. Discordant diagnoses are indicated in bold. Only one center reported a causal mutation for patient 4. NA, not assessed; ‘−‘, no causal variant reported.
Notably, only CGCs 1, 4 and 7 confirmed all reported mutations by Sanger sequencing; the other centers indicated they would do so in a routine diagnostic setting before reporting. Some centers would also apply additional Sanger-based amplicon sequencing in routine diagnostics, to fill any potential gaps that their NGS-based approach may leave. After collection of the results from all centers, but before disclosing the expected results, CGC 8 applied such gap-filling approach in a second stage and indeed identified the causal mutation in MYBPC3 of patient 9.
For patient 4, only CGC 7 reported a causal mutation in MYH7; all other centers did identify the mutation in this gene, but did not report it as causal. Despite the absence of a definite diagnosis, the CGCs that applied a broad sequencing approach did not expand their analyses beyond the known cardiomyopathy genes for this patient.
In addition to the causal variants (Table 2), seven of the eight centers reported variants of unknown significance (VUSs) in their diagnostic letters (Supplementary Table 3). No two centers provided identical reports for all patients. The largest source of variation is the inclusion or exclusion of TTN40 in the analysis panel. CGCs 6 and 8 include TTN in their routine-analysis pipeline, and thus reported in total 11 and 10 VUSs for this gene, respectively (Supplementary Table 4). The reports of the centers that reported VUSs were largely consistent for patients 2, 4 and 9.
The contribution of different experts
In addition to the final diagnostic outcomes, we also considered the process that led to the diagnosis, and the people involved. Within each CGC, responsibility − throughout the procedure − was given to three to five different people (Table 3). The study coordinator only communicated with a single local-contact person per CGC; the latter was responsible for the intake and further processing of the samples and medical records within his or her CGC. Two observations can be made from the reports. First, the CGCs varied considerably in the people that were involved in each phase (Figure 1 and Table 3). For example, clinical geneticists, laboratory specialists, cardiologists or a combination of these evaluated the medical records. The choice for a certain sequencing and analysis approach was irrespective of disease subtype or expected genetic complexity of the individual patients. This is inherent to the set up of the study and how it was perceived, but it also indicates the connections between clinic, laboratory and research. CGCs 4, 6 and 8 brought the medical records into a multidisciplinary meeting (in which lab specialists, clinical geneticists and cardiologists discuss individual patients). CGCs 3 and 5 kept the medical records within the laboratory, and the contacts of the other centers automatically forwarded the records to the clinical geneticist, without specific request (Table 3). CGC 7 did not have the need to involve a multidisciplinary meeting for their diagnostics because of their experience in cardiomyopathy diagnosis.
Table 3. Responsibles for various aspects of diagnostic process.
| Main contact | Medical record | Sequencing, including sample prep | Data analysis, including filtering | Interpretation and reporta | Counselingb | |
|---|---|---|---|---|---|---|
| CGC1 | LAB | CLINGEN | LAB | BOINF (diagnostics) | LAB | NA |
| CGC2 | Postdoc | CLINGEN | Sequencing facility | BOINF (research) | LAB and CLINGEN | NA |
| CGC3 | LAB | LAB | LAB | BOINF (diagnostics) | CLINGEN | NA |
| CGC4 | LAB | Multidisciplinary meeting | LAB | LAB | LAB and CLINGEN | NA |
| CGC5 | LAB | LAB | LAB | BOINF (diagnostics) | LAB | CARDIO |
| CGC6 | LAB | Multidisciplinary meeting | Sequencing facility | LAB | LAB | NA |
| CGC7 | LAB | LAB and CLINGEN | LAB | BOINF (diagnostics) | LAB | NA |
| CGC8 | LAB | Multidisciplinary meeting | LAB | BOINF (diagnostics) | LAB | NA |
Abbreviations: BIOINF, bioinformatician; CARDIO, cardiologist; CLINGEN, clinical geneticist; LAB, laboratory specialist clinical genetics; NA, not assessed.
This involves the selection of causal variants and relevant variants of unknown significance to be reported back to the referring specialists. Reports (also referred to as ‘outcome letters') comprise full interpretation of all variants reported.
Counseling was not literally done as a part of this study, but CGC 5 provided a summary of the counseling that would be done for each patient.
In the data analysis phase, six of eight CGCs involved a bioinformatician to analyze the data, and in five CGCs this bioinformatician was specifically appointed within the diagnostic unit. In practice, however, most of them work in a research environment, to develop pipelines and adapt them to novel technologies, as well as diagnostic needs. The bioinformatics experts thus work on both sides of the traditional divide between diagnostic care and genetic research.
To obtain insight in the consent procedures in each CGC, we collected the forms that each CGC would normally use to obtain consent from patients to undergo NGS-based diagnostics. In subsequent interviews, we obtained additional information on the application of a particular consent procedure. We found that informed consent procedures in all CGCs are generally adequate for diagnostic NGS applications that target a limited set of genes (CGCs 1, 4, 6, 7 and 8; Table 4). Where appropriate, the consent forms contained information on the approach taken, on unsolicited findings and actions in diagnostic and research contexts. Conversely, the available consent procedures for exome and genome sequencing (CGCs 2, 3 and 5) generated debate across centers. In total we identified seven characteristics that are important in providing comprehensive information to patients (Table 4).
Table 4. Informed consent elementsa.
| CGC | Sequencing approach (targeted or broad) | Distinction diagnostics − research | Information sheet | Distinction minors − adults | Unsolicited findings | Option ‘right-not-to-know' |
|---|---|---|---|---|---|---|
| 1 | Yes | Yes | Yes | No | Yes | Yes |
| 2 | Yes | No | Yes | No | Yes | No |
| 3 | Yes | No | Yes | No | Yes | No |
| 4 | Yes | Yes | No | Yes | No | No |
| 5 | Yes | No | Yes | No | Yes | No |
| 6 | Yes | Yes | Yes | No | Yes | No |
| 7 | Yes | Yes | No | Yes | No | No |
| 8 | Yes | Yes | No | Yes | No | No |
Overview of the elements mentioned (‘yes') and not mentioned (‘no') in the informed consent forms of the various centers. Note, the informed consent procedures were under considerable debate during this study, and may thus have changed accordingly.
Subsequent to gathering data from all CGCs, we organized a series of workshops with representatives from all the participating centers, to discuss the various strategies used in the participating centers, and aimed at obtaining consensus at a single strategy. Main conclusions from these workshops were:
The classical divide between research and diagnostics is becoming more blurred when using NGS. To control responsible integration of research and diagnostics, CGCs should develop a framework that manages data access responsibly, i.e. respecting the rights of patients and optimal care, while not hampering research needed to obtain answers to the questions of these same patients and families.
Analysis of limited gene panels (regardless of the sequencing approach) can generally be done under existing consent procedures and is essentially not different from evaluation of genes using traditional Sanger sequencing. For broader analysis using untargeted analysis of NGS results (eg screening for de novo mutations in WES/WGS data) patients should be specifically counseled, and be asked to consent specifically for this procedure (including the risk for unsolicited findings).
Discussion
The results of our study highlight some of the challenges that the ‘genetics clinic of the future' faces. CGCs across the Netherlands obtained largely consistent NGS-based diagnoses for nine patients, even without prior standardization or detailed prior agreements. Nevertheless, the centers were faced with a number of choices that become increasingly important for high-quality clinical genomic care. Some of these are well-known and have been discussed extensively, such as increasing data volumes, informed consent procedures and identification of variants with unknown clinical relevance.17, 25, 26, 28, 30, 41 In our study we focused on the less-obvious challenges: interpreting and reporting genetic variants, incorporating multidimensional diagnoses and defining roles and responsibilities in the genetics clinic of the future.
The interpretation challenge
Diagnoses were consistent across centers for all but patient 9. For this patient, six centers identified a pathogenic MYBPC3 mutation, and provided a diagnosis of ‘hypertrophic cardiomyopathy'. Conversely, CGC 1 and 8 failed to identify the MYBPC3 mutation by their sequencing approach, and reported a diagnosis of ‘Fabry's disease', and a negative diagnosis, respectively. The central conclusion from this observation is not that CGC 1 and CGC 8 provide lower quality sequencing than the other centers. Rather, it is more interesting to consider the implicit implementation choices that reflect the difference in diagnostic outcome. Like CGC 1 and 8 in our study, many CGCs apply complementary Sanger-based sequencing in a routine diagnostic setting to assure sufficient coverage for all relevant regions. Indeed, after the NGS-based results were collected, CGC 8 ‘closed the gaps' and confirmed the presence of the MYBPC3 mutation. The necessity to apply such ‘closing-the-gaps' strategy may reflect the phase of evolution that NGS-based diagnostics is currently going through,39 but it is unlikely to be sustainable for future purposes. As the number of patients and disease types that apply for NGS-based diagnostics increases, Sanger-based complementation may put a disproportionate burden on time and budget of individual CGCs. There is thus an urgent need to develop strategies that are not dependent on additional Sanger sequencing. This may even hold for applying Sanger sequencing for validation purposes, although this aspect was not considered in our study. Of note, the MYBPC3 mutation was present in the exome data from the outsourcing trajectory (CGC1b), but as the latter was considered a research setting, CGC 1 did not include this in the diagnostic report.
In addition to various sequencing outcomes, we observed substantial differences in interpretation of similar sequencing outcomes. For example, the CGCs varied substantially in their judgment on the contribution of the GLA variant to the disease in patient 9. Apart from CGC 1, four other centers (CGCs 4–7) observed the GLA variant, but these all classified it as ‘variant of unknown significance' (VUS). The origin of such interpretation differences is hard to retrieve. One possible explanation may stem from variation in bioinformatic approaches. Many factors – in-house experience, literature, population frequencies, clinical findings, public and in-house databases – may determine how variants are prioritized. Moreover, experience with interpreting particular variants (eg, in the GLA gene) may determine the diagnostic outcome. Whereas all centers generally follow the same best practice guidelines on ‘Unclassified Variants' and ‘Next Generation Sequencing' (Clinical Molecular Genetics Society), these cannot assure absolute consistency among centers. Another explanation may be that the diagnosis for this patient is more complex than originally thought. Compound genetic effects are known to occur, especially in cardiomyopathies, so they could have had a role in this particular patient.42, 43 Indeed, CGC 7 speculated on a possible compound genetic effect of the GLA variant in addition to the causal MYBPC3 mutation, but could not report this with certainty.
From these results it becomes clear that the main challenge for interpreting and reporting variants is not in the technology. Rather, meeting the interpretation challenge requires a number of non-technological issues to be solved. First, interpretation could be significantly improved by sophisticated and transparent exchange of variants associated with clinical indications. Several databases exist that document clinical variation, such as the Human Gene Mutation Database (HGMD), ClinVar and the LOVD,44, 45 and managed data access models are being developed.46 However, the strict divide between diagnostic care and research seriously hampers effective data exchange. Whereas many foresee a blurring of boundaries between research, clinic and society, the ethical and legal issues are of major concern.46, 47 An inherent requirement for data exchange is the harmonization of data formats. The different approaches of the CGCs resulted in a variety of data file formats that could not be automatically exchanged. Central analysis of data from all CGCs required substantial file conversion, whereas accepting that some information (eg second allele information) could not be retrieved. Data-file validation by format validation tools may reduce the heterogeneity, making data sharing and sustainable archiving easier.
Second, assessment of the effect of individual variants is only useful if they are considered in their biological context. Many centers use recurrence in patients with similar phenotypes as clinical validation, whereas application of functional assays – preferably in a diagnostic context – would be more appropriate for comprehensive interpretation. NGS technologies have considerably increased the number of variants that are being detected in a person's genome, but the (putative) function for most of these remains largely unknown.48, 49 In silico functional prediction programs are widely available,50, 51 but the nuances that are inherent to any algorithm and the lack of standards are difficult to work with in a clinical context.25 Functional assays and allele- and/or gene-specific animal models do already exist for a small subset of disorders (eg, metabolic, mitochondrial and congenital disorders).52, 53 However, most clinical laboratories cannot easily incorporate such tests, as they are expensive, labor intensive, difficult to standardize and automate, challenging to interpret, time-consuming and outside the scope of existing regulatory guidelines. Incorporating functional annotation into routine clinical interpretation of results thus requires stronger ties between clinical testing and functional modeling laboratories, consensus guidelines for functional modeling, and introduction of functionally annotated variants into electronic medical records of patients.
Finally, there is a clear need for extensive clinical phenotyping of patients.54 In our study, we provided all centers with full medical records for all nine patients. Two centers (CGCs 1 and 2) actually used these in pre-test clinical assessment, but none of the centers adapted their sequencing and analysis strategy according to the information in the medical records. Instead, the medical records were mostly used for interpretation after testing. This may be partly inherent to the set up of our study, but it may also reflect that the inclusion of clinical features is shifting from the front of the diagnostic process, to the end.
A ‘genetic diagnosis' has many dimensions
Before the introduction of NGS, there was broad consensus on what was to be considered a ‘genetic diagnosis'. It referred to the outcome of genetic testing in a symptomatic individual, and could be either positive (‘we found a cause for disease') or negative (‘no variant'), with an occasional ‘variant of unknown significance' (VUS). Alternative diagnoses – in addition to underdiagnoses – go beyond what is traditionally considered ‘a genetic diagnosis'. What a genetic diagnosis may be in the future – a multidimensional and non-static outcome of a genetic testing procedure – is currently not so often taken into account in the implementation of NGS in routine diagnostics.
Alternative diagnoses may impose many issues, of which ‘greedy interpretation' may be particularly challenging. As we observed for the diagnosis of patient 9, there may be a certain level of greediness to find a genetic cause for disease, especially in cases where known disease genes provide no definite answer. This is also the primary motivation for broad interrogation of exomes or genomes of patients for which diagnosis remains unclear. Despite the negative diagnosis for patient 4, none of the centers applying exome- and genome-wide approaches ‘opened up' the exome or genome for this patient. This may have be owing to the impossibility to do a trio-analysis, which is generally required for relevant interpretation of variants. Also, the appropriate consent for performing exome- or genome-wide analysis may have been lacking, or it may have been unclear which consent to follow; the one that the patients originally signed, or the one that centers generally apply for NGS-based diagnostics. Fact is, that exome- or genome-wide analyses increase the number of potential disease-related variants, which may further induce greediness in interpreting the data. Such level of greediness is necessary to provide the highest quality of genome care, but there is thus far no obvious strategy available for controlling it.
The widespread implementation of NGS in routine diagnostics adds new dimensions to the concept of ‘genetic diagnosis'. Especially in the context of interrogating whole exomes or even whole genomes, and with increasing knowledge about the effect of genomic variation, identifying the multiple dimensions of a genetic diagnosis becomes urgent, especially if other variants than those related to the clinical phenotype are to be considered as part of comprehensive diagnosis.55
Positions and responsibilities in diagnostic genome care
Our results do not only show the effect of NGS on the diagnostic outcome, but also on the organization of diagnostic genome care in its entirety. It is expected that the absolute number of requests for NGS-based testing in classical diagnostic disease areas will increase significantly.17 Moreover, there is an ongoing broadening of genes and disease areas that are eligible for diagnostic testing.18 Obtaining a high-quality diagnosis can thus no longer be the sole responsibility of clinical geneticists, but requires intensive interaction between laboratory specialists, clinical geneticists, other medical specialists and bioinformaticians/data analists. Especially the latter have become crucial; building the appropriate analysis pipelines and assuring the quality and versioning of the tools is in their hands. Primary caretakers and laboratory specialists typically have to trust the judgment of bioinformaticians on certain relevant details, often without full comprehension. Whereas trusting (appropriately validated and certified) ‘black boxes' is not a new phenomenon in the diagnostic context – capillary sequencing similarly comprised automated analysis and visualization steps – the appreciation of bioinformatics knowledge may be exemplary for the changing context in which data analysis occurs.
These developments also raise questions about the information flow between patient, treating physician and testing laboratory. Until recently, information exchange has been asymmetrical. Whereas models for ‘shared decision making' have been in practice for many years, the inherent complexity of genetic information makes supplying it a largely paternalistic undertaking.56 Moreover, diagnostic reports are typically textual documents – and thus based on fractional and point-in-time data, ill-suited for computer-based decision support.57 Implementation and use of electronic health records, the call for sharing data from both funders and patients, and the increasing public interest in genetic services warrants novel approaches to dynamic and automated reporting of genomic knowledge.58, 59, 60, 61, 62
Finally, our results further contribute to the obvious challenges of NGS-based diagnostics. Regardless of costs and throughput of sequencing (which are generally expected to be overcome sooner or later), NGS requires substantial investments in data infrastructure,63 and likely expands the working field of diagnostics. Whereas our ability to interpret genomic data does not instantly improve with NGS – the biology remains complex – the ability to observe the entire genome at ultimate nucleotide resolution with predefined hypotheses on pathogenic origin opens up powerful new clinical opportunities. In addition to efficient continuation of traditional diagnostics, NGS proves very effective in providing diagnosis for previously unsolved rare cases 9, 64 and in supporting appropriate treatment.65 For instance, clinical information from medical records previously determined which genes should be (individually) tested for and in which order; now it mainly serves to correctly filter and interpret genetic variants. Obviously, this may also affect professional roles in relation to genetic care;66, 67 genetic testing can be expected to be requested much more often as these increasingly contribute to clinical diagnosis and decision making.
Acknowledgments
This research was supported by the NGI Booster Grant (050-040-210) for the Netherlands Consortium for Personalized Genome Diagnostics. We thank the Dutch Society for Human Genetics (VKGN) and the Dutch Society for Clinical Genetic Laboratory Diagnostics (VKGL) for their input and support.
Disclaimer
DB and AC are employed by BaseClear, a for-profit service provider for NGS technology. WD and WE are employed by Agilent Technologies Netherlands B.V., a for-profit provider of life science laboratory products and services. SvH is employed by Illumina Inc., a for-profit developer of NGS equipment. HL is employed by Roche Diagnostics, a for-profit provider of diagnostic laboratory products and services. MR is employed by Life Technologies, a for-profit developer of NGS equipment. WvW is employed by ServiceXS, a for-profit service provider for NGS technology.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- de Vries BBA, Pfundt R, Leisink M et al: Diagnostic genome profiling in mental retardation. Am J Hum Genet 2005; 77: 606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER: The impact of next-generation sequencing technology on genetics. Trends Genet 2008; 24: 133–141. [DOI] [PubMed] [Google Scholar]
- Biesecker LG: Hypothesis-generating research and predictive medicine. Genome Res 2013; 23: 1051–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA: Disease gene identification strategies for exome sequencing. Eur J Human Genet 2012; 20: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, Wijmenga C, Slagboom EP et al: The genome of the Netherlands: design, and project goals. Eur J Hum Genet 2014; 22: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Genome of the Netherlands C: whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014; 46: 818–825. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD et al. 1000 Genomes Project Consortium.: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC et al: The diploid genome sequence of an individual human. PLoS Biol 2007; 5: e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C et al: Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 2010; 362: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL et al: Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 2009; 19: 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R et al: The diploid genome sequence of an Asian individual. Nature 2008; 456: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M et al: The complete genome of an individual by massively parallel DNA sequencing. Nature 2008; 452: 872–876. [DOI] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C et al: Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 2010; 42: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Gilissen C et al: De novo mutations of SETBP1 cause Schinzel−Giedion syndrome. Nat Genet 2010; 42: 483–485. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ et al: Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature Genet 2010; 42: 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD et al: Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med 2011; 13: 255–262. [DOI] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Lupski JR, Gibbs RA: Human genome sequencing in health and disease. Ann Rev Med 2012; 63: 35–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG: De novo mutations in human genetic disease. Nat Rev Genet 2012; 13: 565–575. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W et al: Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P et al: Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet 2011; 43: 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bras J, Guerreiro R, Hardy J: Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci 2012; 13: 453–464. [DOI] [PubMed] [Google Scholar]
- Faye LL, Machiela MJ, Kraft P, Bull SB, Sun L: Re-ranking sequencing variants in the post-GWAS era for accurate causal variant identification. PLoS Genet 2013; 9: e1003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, Thorleifsson G, Beer NL et al: Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet 2014; 46: 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis SH, Katsanis N: Molecular genetic testing and the future of clinical genomics. Nat Rev Genet 2013; 14: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC: Diagnostic exome sequencing−are we there yet? N Engl J Med 2012; 367: 1951–1953. [DOI] [PubMed] [Google Scholar]
- Nelen M, Veltman JA: Genome and exome sequencing in the clinic: unbiased genomic approaches with a high diagnostic yield. Pharmacogenomics 2012; 13: 511–514. [DOI] [PubMed] [Google Scholar]
- Samani NJ, Tomaszewski M, Schunkert H: The personal genome—the future of personalised medicine? Lancet 2010; 375: 1497–1498. [DOI] [PubMed] [Google Scholar]
- Mardis ER: A decade's perspective on DNA sequencing technology. Nature 2011; 470: 198–203. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BWM et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Fokstuen S, Lyle R, Munoz A et al: A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Hum Mutat 2008; 29: 879–885. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J et al: Real-time DNA sequencing from single polymerase molecules. Science 2009; 323: 133–138. [DOI] [PubMed] [Google Scholar]
- Maitra RD, Kim J, Dunbar WB: Recent advances in nanopore sequencing. Electrophoresis 2012; 33: 3418–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H: Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 2009; 4: 265–270. [DOI] [PubMed] [Google Scholar]
- Neveling K, Collin RWJ, Gilissen C et al: Next-generation genetic testing for retinitis pigmentosa. Hum Mutat 2012; 33: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y et al: Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet 2012; 49: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee EW, Hoppman-Chaney NL, Ferber MJ: Expanding DNA diagnostic panel testing: is more better? Expert Rev Mol Diagn 2011; 11: 703–709. [DOI] [PubMed] [Google Scholar]
- Shanks ME, Downes SM, Copley RR et al: Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet 2013; 21: 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MM, Van der Zwaag B, Jongbloed JDH et al: Best practice guidelines for the use of next-generation sequencing applications in genome diagnostics: A National Collaborative Study of Dutch Genome Diagnostic Laboratories. Hum Mutat 2013; 34: 1313–1321. [DOI] [PubMed] [Google Scholar]
- Winkens RA, Ament AJ, Pop P, Reniers PH, Grol RP, Knottnerus JA: Routine individual feedback on requests for diagnostic tests: an economic evaluation. Med Decis Making 1996; 16: 309–314. [DOI] [PubMed] [Google Scholar]
- Ormond KE, Wheeler MT, Hudgins L et al: Challenges in the clinical application of whole-genome sequencing. Lancet 2010; 375: 1749–1751. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Srivastava D: Genetics of human cardiovascular disease. Cell 2012; 148: 1242–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M, Semsarian C: Multiple mutations in genetic cardiovascular disease: a marker of disease severity? Circ Cardiovasc Genet 2009;2:182–190. [DOI] [PubMed] [Google Scholar]
- Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT: LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 2011; 32: 557–563. [DOI] [PubMed] [Google Scholar]
- Stenson P, Mort M, Ball E et al: The Human Gene Mutation Database: 2008 update. Genome Med 2009; 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddyman D, Smee C, Griffin H, Kaye J: Implementing a successful data-management framework: the UK10K managed access model. Genome Med 2013; 5: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez LL, Brooks LD, Greenberg JH, Green ED: The complexities of genomic identifiability. Science 2013; 339: 275–276. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A et al: A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012; 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Shendure J: Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet 2011; 12: 628–640. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Meth 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdayer C, Dehainault C, Mattler C et al: Evaluation of in silico splice tools for decision-making in molecular diagnosis. Human Mutat 2008; 29: 975–982. [DOI] [PubMed] [Google Scholar]
- Harakalova M, van Harssel JJT, Terhal PA et al: Dominant missense mutations in ABCC9 cause Cantu syndrome. Nat Genet 2012; 44: 793–796. [DOI] [PubMed] [Google Scholar]
- Zaghloul NA, Liu Y, Gerdes JM et al: Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet‚ÄìBiedl syndrome. Proc Natl Acad Sci USA 2010;107:10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Mundlos S: The human phenotype ontology. Clin Genet 2010; 77: 525–534. [DOI] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB et al: ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 2008; 10: 294–300. [DOI] [PubMed] [Google Scholar]
- Rigter T, van Aart CJA, Elting MW, Waisfisz Q, Cornel MC, Henneman L: Informed consent for exome sequencing in diagnostics: exploring first experiences and views of professionals and patients. Clin Genet 2014; 85: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masys DR, Jarvik GP, Abernethy NF et al: Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform 2012; 45: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Kay S, Marley T, Hardiker NR, Cimino JJ: Incorporating personalized gene sequence variants, molecular genetics knowledge, and health knowledge into an EHR prototype based on the continuity of care record standard. J Biomed Inform 2012; 45: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Khoury MJ, Evans JP: Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med 2011; 13: 499–504. [DOI] [PubMed] [Google Scholar]
- Church G, Heeney C, Hawkins N et al: Public access to genome-wide data: five views on balancing research with privacy and protection. PLoS Genet 2009; 5: e1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J, Heeney C, Hawkins N, de Vries J, Boddington P: Data sharing in genomics−re-shaping scientific practice. Nat Rev Genet 2009; 10: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K: Public expectations for return of results from large-cohort genetic research. Am J Bioethics 2008; 8: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB: The real cost of sequencing: higher than you think!. Genome Biol 2011; 12: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC: Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum Mol Genet 2010; 19: 4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, McLeod HL, Weinshilboum RM: Genomics and drug response. N Engl J Med 2011; 364: 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Cuppen E, Vrijenhoek T: Challenges for implementing next-generation sequencing-based genome diagnostics: its also the people, not just the machines. Personalized Med 2013; 10: 473–484. [DOI] [PubMed] [Google Scholar]
- Qureshi N, Modell B, Modell M: Timeline: raising the profile of genetics in primary care. Nat Rev Genet 2004; 5: 783–790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.