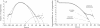Abstract
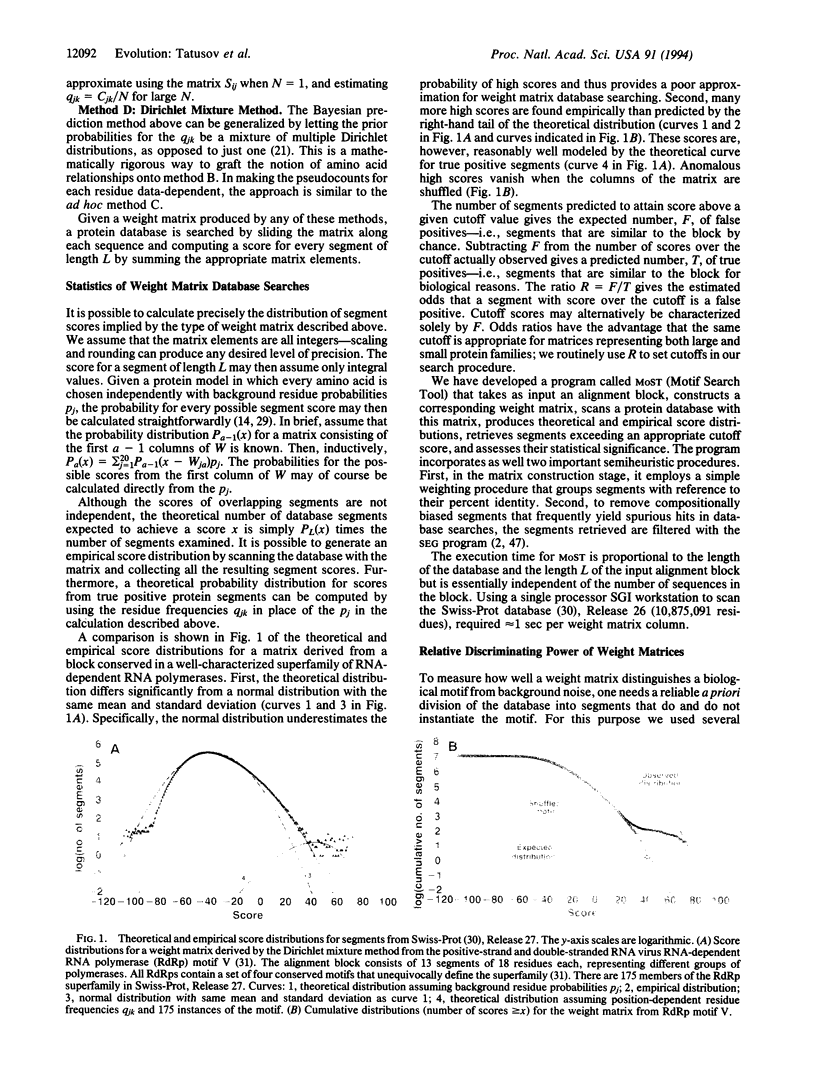
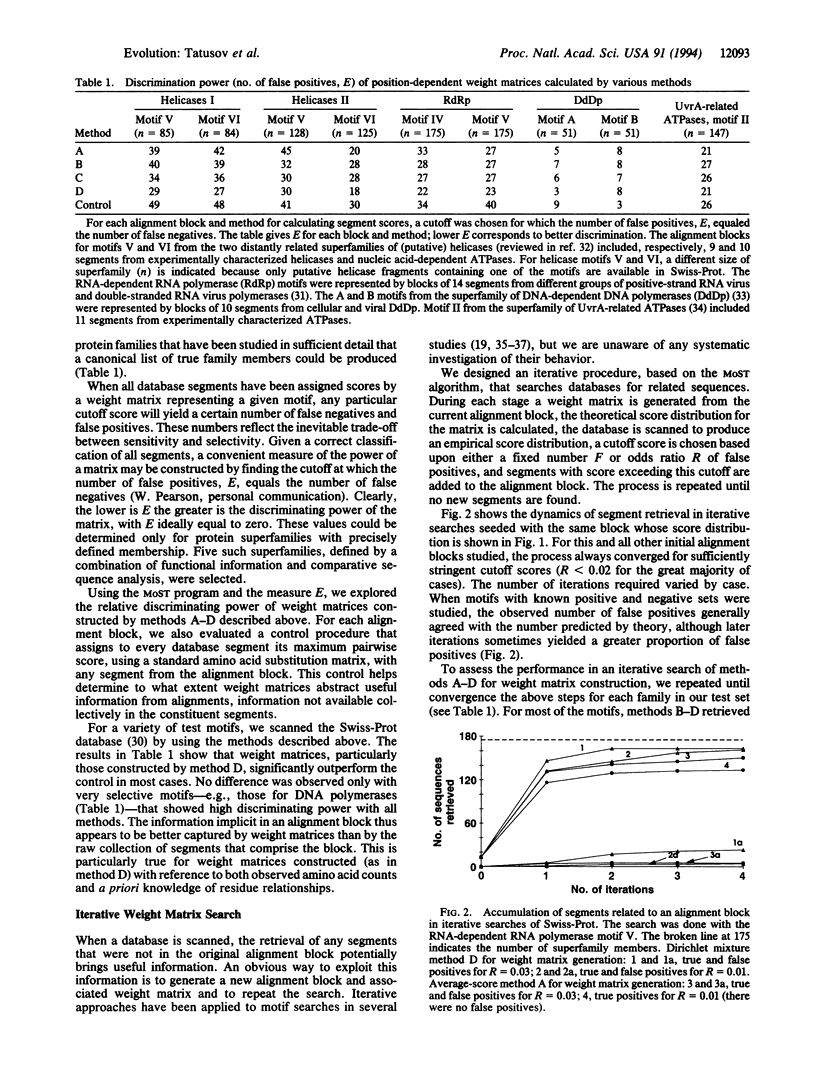
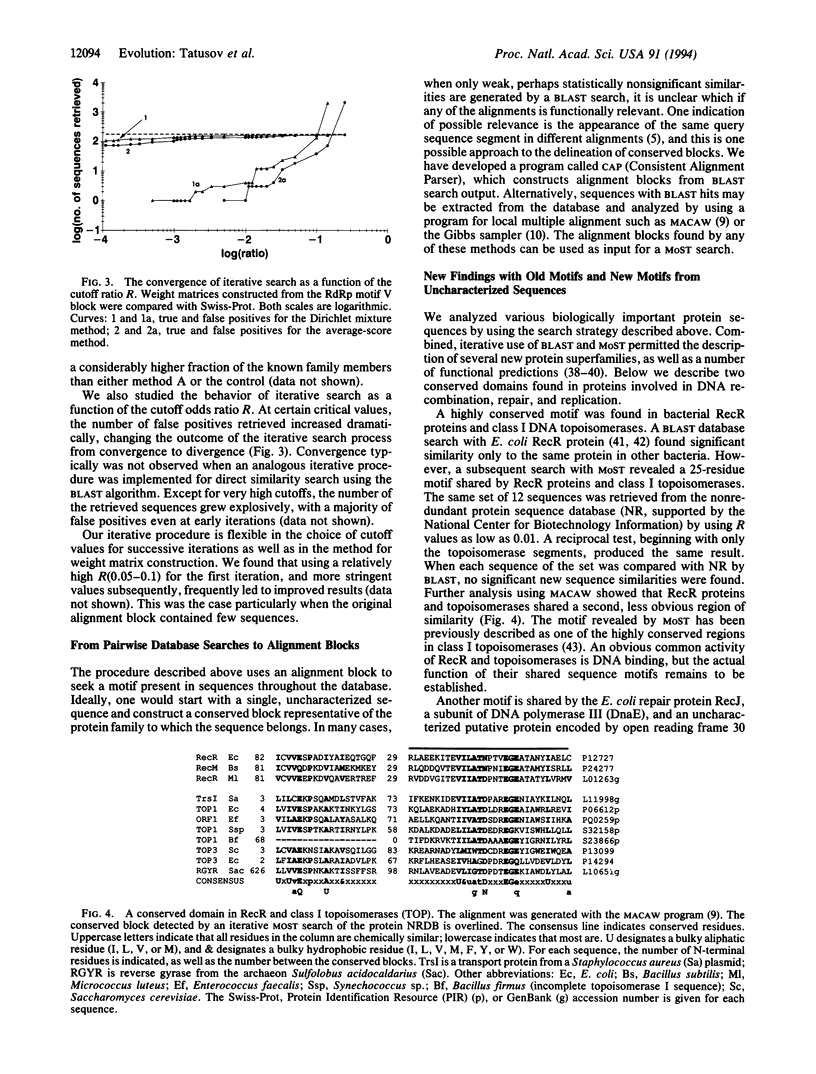
We describe an approach to analyzing protein sequence databases that, starting from a single uncharacterized sequence or group of related sequences, generates blocks of conserved segments. The procedure involves iterative database scans with an evolving position-dependent weight matrix constructed from a coevolving set of aligned conserved segments. For each iteration, the expected distribution of matrix scores under a random model is used to set a cutoff score for the inclusion of a segment in the next iteration. This cutoff may be calculated to allow the chance inclusion of either a fixed number or a fixed proportion of false positive segments. With sufficiently high cutoff scores, the procedure converged for all alignment blocks studied, with varying numbers of iterations required. Different methods for calculating weight matrices from alignment blocks were compared. The most effective of those tested was a logarithm-of-odds, Bayesian-based approach that used prior residue probabilities calculated from a mixture of Dirichlet distributions. The procedure described was used to detect novel conserved motifs of potential biological importance.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso J. C., Stiege A. C., Dobrinski B., Lurz R. Purification and properties of the RecR protein from Bacillus subtilis 168. J Biol Chem. 1993 Jan 15;268(2):1424–1429. [PubMed] [Google Scholar]
- Altschul S. F., Boguski M. S., Gish W., Wootton J. C. Issues in searching molecular sequence databases. Nat Genet. 1994 Feb;6(2):119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Lipman D. J. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood T. K., Findlay J. B. Design of a discriminating fingerprint for G-protein-coupled receptors. Protein Eng. 1993 Feb;6(2):167–176. doi: 10.1093/protein/6.2.167. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank, recent developments. Nucleic Acids Res. 1993 Jul 1;21(13):3093–3096. doi: 10.1093/nar/21.13.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 1993 Jul 1;21(13):3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Braithwaite D. K., Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993 Feb 25;21(4):787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M., Hughey R., Krogh A., Mian I. S., Sjölander K., Haussler D. Using Dirichlet mixture priors to derive hidden Markov models for protein families. Proc Int Conf Intell Syst Mol Biol. 1993;1:47–55. [PubMed] [Google Scholar]
- Confalonieri F., Elie C., Nadal M., de La Tour C., Forterre P., Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990 Sep 11;18(17):5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W., States D. J. Identification of protein coding regions by database similarity search. Nat Genet. 1993 Mar;3(3):266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J Mol Biol. 1990 Jun 20;213(4):583–591. doi: 10.1016/S0022-2836(05)80243-8. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M. Translational initiation factors IF-1 and eIF-2 alpha share an RNA-binding motif with prokaryotic ribosomal protein S1 and polynucleotide phosphorylase. Gene. 1992 Sep 21;119(1):107–111. doi: 10.1016/0378-1119(92)90073-x. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991 Dec 11;19(23):6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Performance evaluation of amino acid substitution matrices. Proteins. 1993 Sep;17(1):49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- Hodgman T. C. The elucidation of protein function by sequence motif analysis. Comput Appl Biosci. 1989 Feb;5(1):1–13. doi: 10.1093/bioinformatics/5.1.1. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992 Jun;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Karlin S., Altschul S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Bork P., Sander C. Yeast chromosome III: new gene functions. EMBO J. 1994 Feb 1;13(3):493–503. doi: 10.1002/j.1460-2075.1994.tb06287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28(5):375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. Prediction of an rRNA methyltransferase domain in human tumor-specific nucleolar protein P120. Nucleic Acids Res. 1994 Jul 11;22(13):2476–2478. doi: 10.1093/nar/22.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. E., Altschul S. F., Boguski M. S., Liu J. S., Neuwald A. F., Wootton J. C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993 Oct 8;262(5131):208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- Linderoth N. A., Julien B., Flick K. E., Calendar R., Christie G. E. Molecular cloning and characterization of bacteriophage P2 genes R and S involved in tail completion. Virology. 1994 May 1;200(2):347–359. doi: 10.1006/viro.1994.1199. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., Kolodner R. D. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D. Analysis of gene duplication repeats in the myosin rod. J Mol Biol. 1983 Sep 5;169(1):15–30. doi: 10.1016/s0022-2836(83)80173-9. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde K., Bork P. A fast, sensitive pattern-matching approach for protein sequences. Comput Appl Biosci. 1993 Apr;9(2):183–189. doi: 10.1093/bioinformatics/9.2.183. [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci U S A. 1990 Jan;87(1):118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Methods for calculating the probabilities of finding patterns in sequences. Comput Appl Biosci. 1989 Apr;5(2):89–96. doi: 10.1093/bioinformatics/5.2.89. [DOI] [PubMed] [Google Scholar]
- Stormo G. D. Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem. 1988;17:241–263. doi: 10.1146/annurev.bb.17.060188.001325. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Chi N. W., Kolodner R. D. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C. The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell. 1994 Jan 14;76(1):9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]