Abstract
The ability of skeletal muscle to hypertrophy in response to a growth stimulus is known to be compromised in older individuals. We hypothesized that a change in the expression of protein-encoding genes in response to a hypertrophic stimulus contributes to the blunted hypertrophy observed with aging. To test this hypothesis, we determined gene expression by microarray analysis of plantaris muscle from 5- and 25-mo-old mice subjected to 1, 3, 5, 7, 10, and 14 days of synergist ablation to induce hypertrophy. Overall, 1,607 genes were identified as being differentially expressed across the time course between young and old groups; however, the difference in gene expression was modest, with cluster analysis showing a similar pattern of expression between the two groups. Despite ribosome protein gene expression being higher in the aged group, ribosome biogenesis was significantly blunted in the skeletal muscle of aged mice compared with mice young in response to the hypertrophic stimulus (50% vs. 2.5-fold, respectively). The failure to upregulate pre-47S ribosomal RNA (rRNA) expression in muscle undergoing hypertrophy of old mice indicated that rDNA transcription by RNA polymerase I was impaired. Contrary to our hypothesis, the findings of the study suggest that impaired ribosome biogenesis was a primary factor underlying the blunted hypertrophic response observed in skeletal muscle of old mice rather than dramatic differences in the expression of protein-encoding genes. The diminished increase in total RNA, pre-47S rRNA, and 28S rRNA expression in aged muscle suggest that the primary dysfunction in ribosome biogenesis occurs at the level of rRNA transcription and processing.
Keywords: hypertrophy, age, skeletal muscle, microarray, ribosome biogenesis
human and rodent studies have reported that skeletal muscle hypertrophy is significantly diminished with old age (8, 14, 16, 26, 28, 30, 33). Although the underlying mechanism responsible for the blunted hypertrophic response in the elderly remains to be clearly defined, an alteration in protein metabolism is thought to be a primary factor (25). In particular, activation of protein synthesis by the mechanistic target of rapamycin (mTOR) signaling pathway has been shown to be attenuated in aged skeletal muscle subjected to a hypertrophic stimulus (9, 23, 31). The notion that blunted mTOR signaling contributes to a compromised hypertrophy with age is consistent with the pioneering work by Bodine and colleagues (3) showing that mTOR activity is absolutely necessary to mount a full hypertrophic response.
In an effort to identify other genes or pathways that might contribute to the age-related difference in skeletal muscle hypertrophy, microarray analyses were performed to identify changes in gene expression between young and old individuals in response to an acute bout of resistance exercise or following a training program (24, 27, 29). Although these studies were able to identify pathways that may have a role in the diminished hypertrophic response of the elderly, the power of these analyses was limited by the small number of time points, typically pre- and postmeasurements. Given the dynamic nature of gene expression during skeletal muscle hypertrophy, it is likely that this design limitation resulted in potentially important changes in gene expression being missed (5). This idea is supported by the finding that acute changes in gene expression are no longer observed following a resistance exercise training program and, therefore, may not truly reflect those biological processes governing hypertrophic growth (29).
The purpose of this study was to perform a comprehensive transcriptome analysis of old skeletal muscle undergoing hypertrophy in an effort to identify differentially expressed genes. Skeletal muscle hypertrophy was induced by synergist ablation with gene expression measured by microarray analysis after 1, 3, 5, 7, 10, and 14 days. To identify age-associated genes that were differentially expressed in response this hypertrophic stimulus, we compared this newly generated data set against our previously published transcriptome analysis in young skeletal muscle subjected to the identical perturbation (5). Despite observing only modest differences in gene expression between the two groups, pathways associated with the regulation of protein synthesis, metabolism, and immune response were highly enriched in old skeletal muscle. Somewhat paradoxical though, we found that ribosome biogenesis was significantly impaired at the level of ribosomal DNA (rDNA) transcription in aged skeletal muscle undergoing hypertrophy. These findings suggest that the blunted hypertrophic response in old skeletal muscle was primarily the result of a failure to sufficiently increase the translation capacity of the muscle more so than changes in gene expression.
METHODS
Animal care and use.
All experimental procedures performed in this study were approved by the University of Kentucky Institutional Animal Care and Use Committee. Male C57BL/6J mice, 5 mo of age (The Jackson Laboratory, Bar Harbor, ME) and 25 mo of age (National Institute on Aging, Bethesda, MD) were housed in a temperature- and humidity-controlled room and maintained on a 14:10-h light:dark cycle with food and water ad libitum.
Young (5 mo of age) and old (25 mo of age) mice were subjected to bilateral synergist ablation surgery to induce hypertrophy of the plantaris muscle as previously described (18). Briefly, following anesthetization with a mixture of 95% oxygen and 5% isoflurane, the soleus and the majority of the gastrocnemius muscles were surgically excised via an incision on the dorsal aspect of the hind limb. Particular attention was made to ensure neural and vascular supply to the plantaris muscle remained intact and undamaged. Animals that served as a control group underwent sham surgery without gastrocnemius and soleus muscle excision. Following recovery from surgery, mice were anesthetized as described above at the designated time point and plantaris muscles were excised, weighed, placed in RNAlater (Ambion, Austin, TX) and stored at 4°C. Plantaris muscle was collected at 1, 3, 5, 7, 10, and 14 days after the surgery (n = 6 per time point) during the same 4-h time period (10:00 A.M. to 2:00 P.M.) after the animals had been fed and were rested, thus ensuring a similar metabolic state between the groups. Plantaris muscles (n = 6) to serve as controls were collected from mice subjected to the sham synergist ablation surgery. Following collection of the plantaris muscles, mice were killed by cervical dislocation under anesthesia.
RNA isolation.
Total RNA was prepared from plantaris muscle using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's directions. RNA samples were treated with TURBO DNase (Ambion) to remove genomic DNA contamination. Total RNA concentration and purity was assessed by measuring the optical density (230, 260, and 280 nm) with a Nanodrop 1000 Spectrophotometer (ThermoFisher Scientific, Wilmington, DE). RNA integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA); the average RNA integrity number (RIN) value for all samples was 9.12 ± 0.17 (scale 1-10) indicating high-quality RNA with minimal degradation products.
Microarray analysis.
Microarray analysis was performed at the University of Kentucky Microarray Core Facility according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Gene expression was measured using the Mouse Gene 1.1 ST chip, which provides coverage of 28,000 protein-coding transcripts and 7,000 noncoding transcripts of which ∼2,000 are long, intergenic noncoding transcripts. We previous published (5) a microarray analysis of plantaris muscle of young mice undergoing hypertrophy and used that information in the current study to compare it with data generated from the plantaris muscle of old mice. As in the earlier study, two gene chips were processed at each time point from 250 ng of total RNA. Total RNA was derived from a pooled sample of either the right or left plantaris muscle from six animals. We pooled RNA samples on the basis of experimental results reported by Kendziorski et al. (15), who showed that gene expression from a pooled RNA sample is similar to the average from the individual samples comprising the pooled sample. To minimize variability due to systematic biases (such as dye effects, hybridization artifacts, or both) the chips for both the young and old samples were hybridized at the same time with the resulting probe signal for each transcript summarized using repeated-measures ANOVA, and the quantiles were normalized using the Affymetrix Expression console software. Furthermore, these normalized data sets were then all uploaded to the Partek Genomics Suite so that the data set from young animals was reanalyzed against the data set from the old animals. At this step, we did not set a lower cutoff for the signal intensity to avoid excluding low-expressing genes that might show a significant age-associated upregulation in response to synergist ablation. Data were log-transformed and duplicate probes sets for the same gene were removed, with the probe set demonstrating the highest signal intensity being retained in the analysis. To facilitate downstream pathway analysis, only the probe sets with annotation information were included. Following processing, 21,735 genes were exported and used for further analysis. Gene expression data have been made available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) for the hypertrophy studies of young (GSE47098) and old (GSE67160) animals.
Identification of differentially expressed genes.
Detection of differential gene expression profiles was performed using R-based Bioconductor statistical software, version 2.6 (11). To detect gene expression differences between the young and old groups, data were analyzed using the maSigPro package (7). This package is specifically designed to identify differential expression profiles across experimental groups in time-course microarray data. It utilizes a regression-based analysis that allows for time to be maintained as an independent variable. First, genes exhibiting significantly different profiles across the time course were detected using the least-squares technique to determine the parameters of the general regression model. Any gene with different profiles between the 5- and 25-mo-old groups will show some statistically significant coefficient, and its corresponding regression model will be statistically significant. The P value associated to the F-statistic in the general regression model was used to select significant genes between age groups. Second, once gene models showing significant differences were identified, backward stepwise regression was performed on this set of genes to identify the conditions for which genes shows statistically significant profile changes.
Cluster and pathway analysis.
Ingenuity Pathway Analysis (IPA) was used to identify those pathways that were significantly enriched across all differentially expressed genes, restricting it to those pathways associated with whole tissues (analysis conducted in June 2014). To elucidate how these biological pathways were affected by age, genes were categorized on the basis of their response to the synergist ablation (i.e., upregulated or downregulated) and the mean fold change at each time point was then generated for both the upregulated and downregulated genes. Hierarchical clustering was performed on all differentially expressed genes on the basis of similar expression profiles across time.
RT-PCR analysis.
Complementary DNA was generated from 250 ng of total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen/Life Technologies, Grand Island, NY) using a combination of random hexamers and oligo(dT) primers. Quantitative PCR for each candidate gene was performed using KiCqStart SYBR Green qPCR ReadyMix (Sigma-Aldrich, St. Louis, MO) with the following cycle conditions: 95°C for 3 min, 40 cycles at 95°C for 30 s, and at 60°C for 60 s. Primer sequences are available upon request. All transcripts were normalized to the geometric mean of Gapdh and Rn7sk. These genes were selected on the basis of having the lowest variability (0.24% and 0.34%, respectively) across age and treatment as determined by the microarray analysis.
Statistical analysis.
The muscle wet weight, total RNA concentration, and quantitative PCR data were analyzed by two-way ANOVA followed by Tukey's post hoc test with significance set at P ≤ 0.05. Significant gene profiles were determined using polynomial regression (degree = 3) using a false discovery rate-corrected P value of 0.01 to determine statistical significance. For stepwise regression analysis the P value for the regression coefficients was set to α = 0.01. Only genes that had a clear trend and fit the model with an R2 value of >0.7 were used for further analysis. A right-tailed Fisher's exact test was used to determine the top statistically significant canonical pathways from IPA. For this analysis, P < 0.0001 was considered statistically significant.
RESULTS AND DISCUSSION
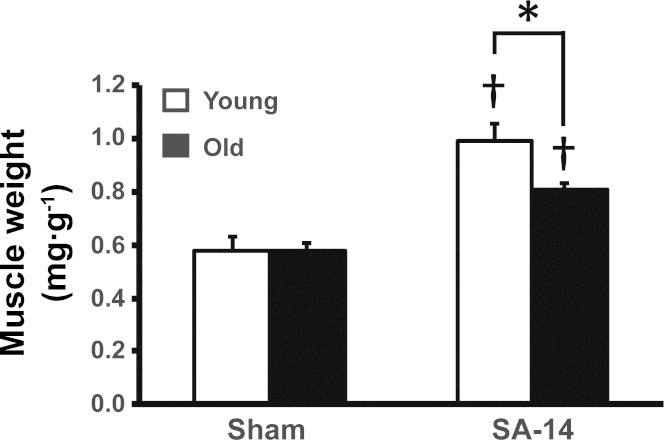
In response to 14 days of synergist ablation, plantaris muscle weight was significantly increased in both young and old mice; however, the increase in normalized plantaris muscle weight was significantly less (40% vs. 71%, respectively) in old mice than in young mice (Fig. 1). Importantly, there was no difference in the body weight of young and old sham-operated mice and following 14 days of synergist ablation. These findings are consistent with previous observations in aged animals showing blunted hypertrophic growth following mechanical overload ranging from 7 days to 8 wk (2, 4, 6, 8, 22, 31).
Fig. 1.
Aged skeletal muscle demonstrates blunted hypertrophic growth following mechanical overload. Normalized (to body weight) plantaris muscle weight from 25-mo-old mice showed impaired hypertrophy compared with young mice following 14 days of synergist ablation (SA-14). *Significant age-effect; †significant increase relative to sham (P < 0.05).
In an effort to identify differentially expressed genes that may contribute to the blunted hypertrophic response observed in old skeletal muscle, we performed a time-course microarray analysis of gene expression following 1, 3, 5, 7, 10, and 14 days of synergist ablation in young and old mice. Regression analysis using the two data sets identified 1,607 genes that were differentially expressed across the time course between the young and old groups (see Supplemental Table 1 online).
Table 1.
Most enriched biological pathways among differentially expressed genes
| Ingenuity Canonical Pathways | P | Ratio | Genes in Pathway |
|---|---|---|---|
| EIF2 signaling | 6.69E-09 | 2.04E-01 | RPL11, RPL24, EIF2B4, Rpl36a, HRAS, EIF2B2, Ppp1 ml, RPL18A, RPL7A, RPS13, MAPK3, PAIP1, RPL19, RPS3, RPS5, RPL18, RPL31, RPL13, AKT2, RPS19, EIF3J, RPL7L1, RPL12, RPL23, EIF3E, RPLP0, RPL10A, RPS4Y1, RPS26, PIK3R6, RPS15, RPL10, RPS27A, RPL5, EIF3L, RPL13A, RPSA |
| Mitochondrial dysfunction | 4.35E-07 | 1.89E-01 | NDUFA4, SDHB, NDUFA9, ATP5H, Cox6c, UQCR11, ACO2, TRAK1, NCSTN, NDUFA1, PDHA1, SOD2, ATP5J2, NDUFS2, NDUFB6, ATP5F1, ATP5J, NDUFAF1, CYCS, GLRX2, SURF1, COX7A1, APP, FIS1, PRDX3, NDUFV2, NDUFA11, UQCR10, NDUFA6, NDUFB7, SDHD, NDUFB2 |
| CTLA4 signaling in cytotoxic T-lymphocytes | 3.53E-06 | 2.27E-01 | AP2M1, AKT2, CD3E, HLA-A, AP1M1, CLTC, CLTB, TRG, CD8B, PPP2R5A, AP2S1, AP1S1, CD3G, LCK, PPP2R1A, AP1G2, PTPN11, LAT, PIK3R6, PTPN22 |
| Mechanistic target of rapamycinm (mTOR) signaling | 3.82E-06 | 1.72E-01 | ULK1, PLD2, PRKAB1, HRAS, RHOD, RPS13, MAPK3, TSC2, RPS6KB2, PRKAA2, FIGF, RPS3, RPS5, AKT2, PLD3, STK11, RPS19, EIF3J, EIF3E, PPP2R5A, PLD4, RPS4Y1, PPP2R1A, RPS26, PIK3R6, RPS15, PRR5, RPS27A, RPS6KA1, FNBP1, EIF3L, RPSA |
| Oxidative phosphorylation | 7.91E-06 | 2.04E-01 | NDUFA4, ATP5J, SDHB, ATP5H, NDUFA9, Cox6c, UQCR11, CYCS, SURF1, COX7A1, NDUFA1, NDUFV2, NDUFA11, NDUFA6, UQCR10, NDUFB7, ATP5J2, NDUFB6, NDUFS2, SDHD, ATP5F1, NDUFB2 |
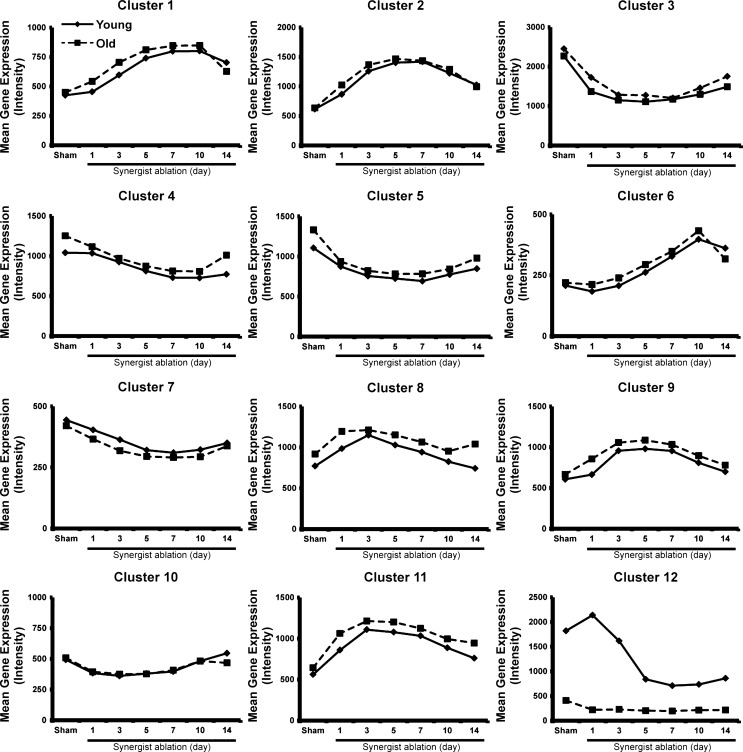
We next used cluster analysis to determine how age affected the temporal pattern of expression of differentially expressed genes. We found that 12 clusters provided a sufficient number of discrete clusters with enough genes in each cluster to allow a biologically meaningful pathway analysis (Fig. 2). Cluster analysis showed that the pattern of expression of differentially expressed genes was remarkably similar between young and old groups, with the one exception being cluster 12 (Fig. 2). This cluster contained genes that were expressed at a much lower level in old muscle across the time course and showed a different response to synergist ablation compared with young muscle. Within this cluster, about one-third of the genes were from either the major urinary protein family (Mup1-4, 7, 11, 19, and 20) or the serine (or cysteine) peptidase inhibitor family (Serpina1a, b, c, and e) (see Supplement Table 1).
Fig. 2.
Hierarchical clustering of differentially expressed genes in response to synergist ablation. The expression pattern of genes within cluster 12 demonstrated the most significant effect of age during hypertrophic growth. The majority of genes within this cluster belonged to either the major urinary protein family or the serine (or cysteine) peptidase inhibitor family. Solid lines indicate young animals; dashed lines indicate old animals.
Although few studies have examined the effect of the Mup genes on skeletal muscle function, increasing levels of circulating Mup1 have been reported to enhance whole-body energy expenditure that was associated with the activation of Akt signaling and mitochondrial biogenesis in skeletal muscle (13). Thus elevated expression of these genes in young skeletal muscle suggests a possible role in regulating the metabolic adaptations that occur with synergist ablation; however, it should be noted that the human genome does not harbor any functional Mup genes, thus limiting the potential translational significance of the finding.
Pathway analysis of the differentially expressed gene set revealed the primary biological processes affected by age were the regulation of protein synthesis (EIF2 and mTOR signaling), metabolism (oxidative phosphorylation and mitochondrial dysfunction), and immune response (T-cell signaling) (Table 1). These pathways have been implicated in the physiological adaptation to chronic mechanical overload (1), with mTOR signaling identified as the most enriched pathway in a human transcriptome analysis of skeletal muscle in response to resistance exercise (24), thus giving us confidence that the informatics approach we used effectively captured genes involved in the hypertrophic response. We focused on the differentially expressed genes associated with EIF2 and mTOR signaling pathways given the findings from previous studies showing that the mTOR pathway was misregulated in aging muscle subjected to a hypertrophic stimulus (9, 10, 23, 31).
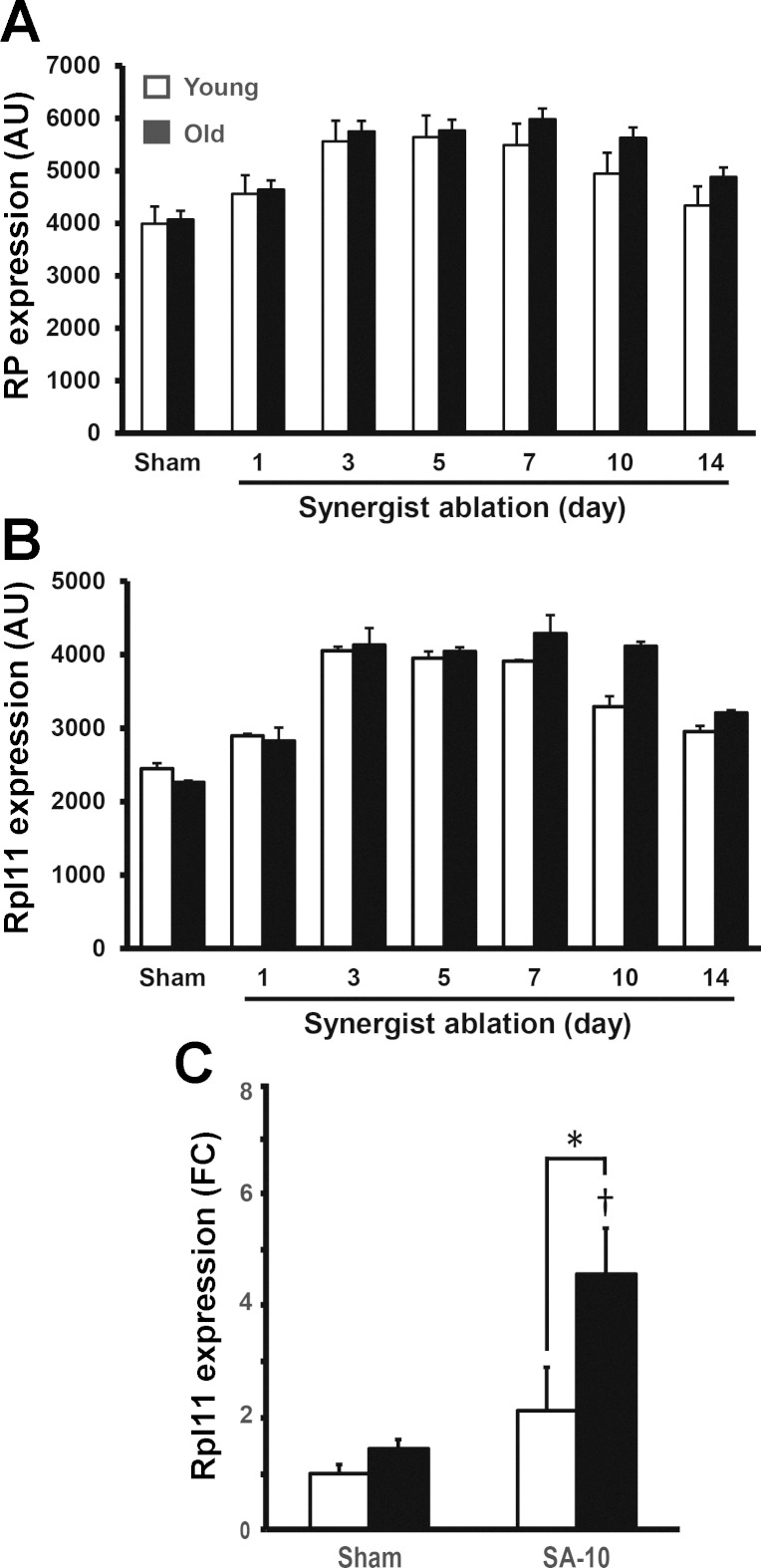
Of the differentially expressed genes associated with EIF2 and mTOR signaling, almost half (24/54) encoded ribosomal proteins (Table 1). Expression of these ribosomal protein genes increased in a similar manner until day 5 in both age groups, after which expression remained higher in the old group (Fig. 3A). The ribosomal protein genes showing the greatest age-depended differences were Rpl24, Rps19, Rpl10a, Rpl13 and, in particular, Rpl11 (Fig. 3B and Supplement Table 1). RT-PCR confirmed that Rpl11 expression was significantly increased by 4.6-fold in the old mice at day 10 but was unchanged from baseline at this time point in the young group (Fig. 3C). These results are consistent with findings from a recent human study that found increases in lean mass were negatively correlated with expression of 30 ribosomal protein genes (9).
Fig. 3.
Increased expression of ribosomal proteins during hypertrophy with old age. A: geometric mean expression of all differentially expressed ribosomal protein genes following synergist ablation. B: Rpl11 increases earlier during mechanical overload in both young and old animals; however, the aged animals maintain higher expression levels at the later time points following synergist ablation. C: RT-PCR confirmed a significant increase in Rpl11 expression in old animals following 10 days of synergist ablation (SA-10), which did not occur in young animals. *Significant age-effect; †significant increase relative to sham (P < 0.05).
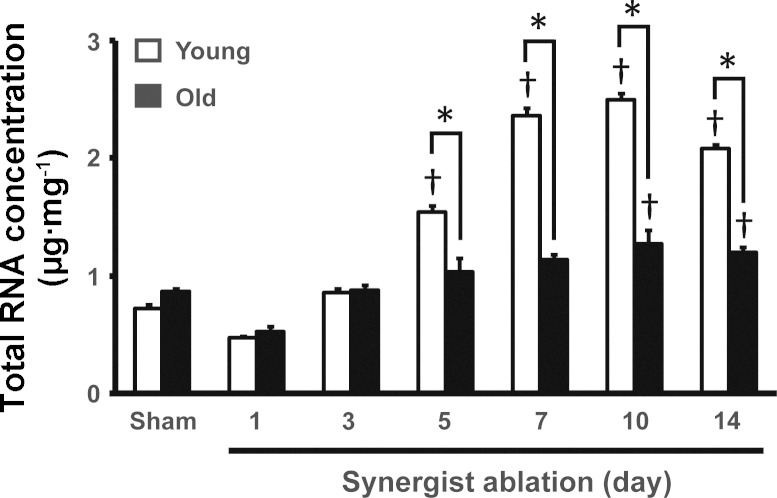
Given the higher expression of ribosomal protein genes in old skeletal muscle in response to synergist ablation, we were curious to see what affect this might have on ribosome biogenesis. We and others have reported that synergist ablation-induced muscle hypertrophy is associated with a significant increase in ribosome biogenesis (12, 19, 32). To determine whether ribosome biogenesis was altered in response to synergist ablation in old mice, we measured total RNA per unit of muscle mass at each time point; given that 85% of total RNA is ribosomal RNA (rRNA), total RNA per unit of tissue is considered a measurement of ribosome content (34). There was no difference in the ribosome concentration of plantaris muscle of young and old animals that underwent sham operation animals because total RNA per unit of muscle was the same between these two groups (Fig. 4). In response to synergist ablation, total RNA concentration of plantaris muscle of young mice significantly increased from day 5 through day 14, with an approximately 2.5-fold peak expression occurring at days 7 and 10 (Fig. 4). In contrast, total RNA concentration of the plantaris muscle of old mice showed a modest ∼50% increase at days 10 and 14 that was significantly less compared with young mice (Fig. 4).
Fig. 4.
Increased ribosome content during hypertrophy is blunted in aged animals. Total RNA content, of which >85% is ribosomal RNA, increased in response to synergist ablation (SA), indicating increased ribosome biogenesis. Conversely, skeletal muscle of old animals demonstrated a compromised ability to increase total RNA content in response to synergist ablation. Old animals demonstrated a delayed increase in RNA content, not occurring until at SA10 and SA14. Furthermore, at SA5 through SA14, RNA content was significantly lower in the old animals relative to the young animals. *Significant age-effect at that time-point; †significant increase from sham (P < 0.05).
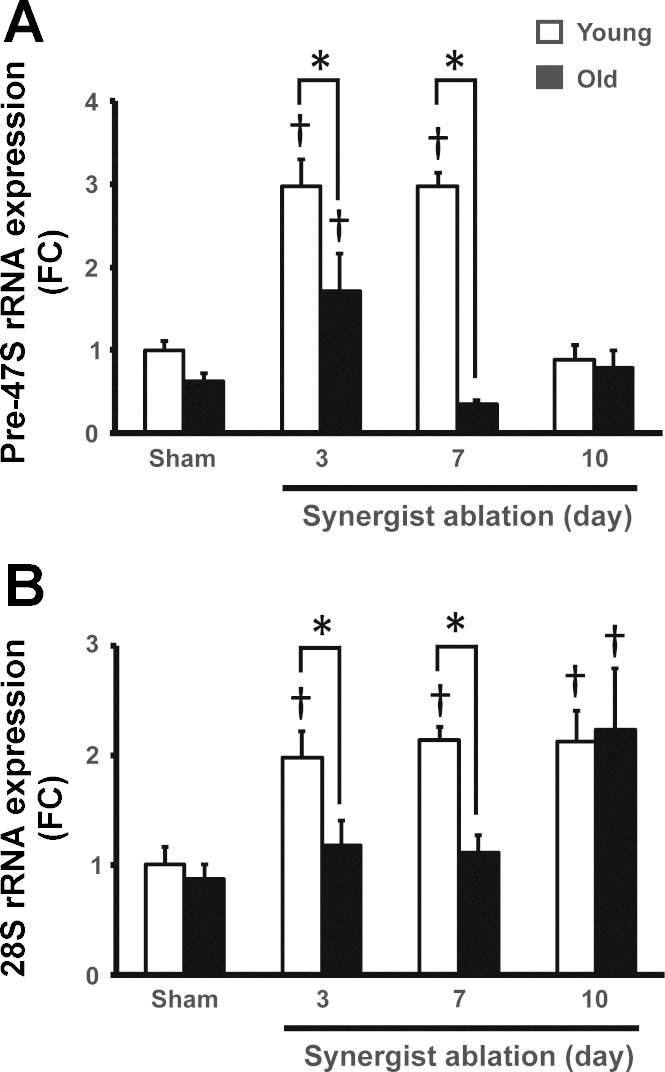
The lower ribosome content of plantaris muscle from old animals in response to synergist ablation might be caused by a failure to fully activate rDNA transcription in response to synergist ablation. To determine whether rDNA transcription was altered in plantaris muscle of old animals, we measured pre-47S rRNA abundance in sham-operated animals and after 3, 7, and 10 days of synergist ablation in both young and old mice. We chose to focus on these time points because day 3 was the last time point before total RNA concentration diverged between the two groups and days 7 and 10 because it was when total RNA concentration peaked in the young group. There was no difference in pre-47S rRNA expression between young and old sham-operated groups suggesting that rDNA transcription at baseline was not affected by age (Fig. 5A). Following 3 days of synergist ablation, there was a significant increase in pre-47S rRNA expression in both young and old groups, though the increase in the young group was significantly greater than in the old (3-fold vs. 1.7-fold, respectively). Of note, the increased expression of pre-47S rRNA observed at day 3 in both groups preceded the increase in ribosome content as assessed by total RNA concentration (compare Fig. 4 and 5A). Although the current study does not provide a mechanism to explain the delay between 47S pre-rRNA expression and total RNA accumulation, Nagatomo et al. (21) reported that an acute increase in 60S ribosome formation preceded an increase in total RNA at day 5 in response to cardiac pressure overload. The authors proposed that this delay in total RNA accumulation was a reflection of ribosome half-life of 10–12 days. Given that this was observed in the heart, it is reasonable to speculate that a similar process may be occurring in skeletal muscle in response to mechanical overload induced by synergist ablation and might be affected by age. At day 7, pre-47S rRNA expression in the young group remained significantly elevated by 3-fold, whereas pre-47S rRNA expression returned to baseline level in the old group (Fig. 5A).
Fig. 5.
Increased pre-47S and 28S rRNA expression during muscle hypertrophy are attenuated in old animals. A: pre-47S rRNA expression, a readout of polymerase I activity, increased in response to synergist ablation in young animals after 3 and 7 days of synergist ablation with old animals only showing a modest increase at day 3. Furthermore, pre-47S expression was significantly higher in young animals compared with old animals at both days 3 and 7 of synergist ablation. B: 28S rRNA expression increased in response to synergist ablation at from day 3 through day 10 in young animals, whereas it increased in old animals only at day 10. 28S expression was significantly attenuated at days 3 and 7 in old animals. *Significant age-effect at that time-point; †significant increase from sham (P < 0.05).
Using the same model of hypertrophy as we did, von Walden et al. (32) also reported an increase in pre-47S expression after 3 days of synergist ablation which, in contrast to our findings, returned to baseline by day 7. In both studies, however, pre-47S expression paralleled the change in total RNA, further lending support to the idea that the rate of rDNA transcription dictates ribosome biogenesis. These results suggest that polymerase I (Pol I) activity is compromised in aged animals during hypertrophic growth, which may underlie an impaired ability in increase ribosome biogenesis, potentially limiting growth.
To determine whether processing of pre-47S rRNA was altered in the muscle of old animals undergoing hypertrophy, we measured the 28S rRNA expression. There was no difference in 28S rRNA expression in young or old sham-operated groups, indicating that pre-47S processing was unaffected by age in resting skeletal muscle (Fig. 5B). In good agreement with pre-47S rRNA expression, 28S rRNA expression was significantly increased in response to synergist ablation at days 3, 7, and 10 in the young group; in contrast, 28S rRNA expression remained unchanged in the old group until day 10 (Fig. 5B). These findings suggest that rDNA transcription and processing had become uncoupled in the muscle of old animals during hypertrophic growth.
On the basis of our findings, we present a novel notion that dysfunctional ribosome biogenesis is one the primary factor inhibiting muscle hypertrophy in aged animals. We suggest that in young animals, the expression levels of the core components of mature ribosomes promotes a cellular environment that is permissive for maximizing ribosome biogenesis during muscle hypertrophy, which does not occur in aged animals. The attenuated increase in total RNA, pre-47S, and 28S content in muscle from old mice suggests that the primary dysfunction is occurring at the level of rDNA transcription and/or processing. Because the synergist ablation model of hypertrophy induces such a robust hypertrophic growth, it would be of interest to determine whether ribosome biogenesis would be altered in a more physiological model that induced a more modest level of hypertrophy with age.
Results from the current study suggest that although age-dependent differences exist across in the transcriptome profile between the skeletal muscle of young and old animals during hypertrophy, the differences appear to be modest. Moreover, the response of individual genes to the mechanical overload resulting from synergist ablation appear to be conserved across age groups, with little evidence to suggest that disparities in transcript levels or misexpression are driving the impaired hypertrophic growth in aged animals. Furthermore, it is important to factor in acute gene expression changes vs. chronic changes, because we observed that most of the changes occur early in the time course. Recently, Nader and colleagues (20) challenged the idea that using acute changes in gene expression to better understand the mechanisms regulating muscle hypertrophy are of limited value because most of these changes are not observed with chronic overload. Thus the findings from this study provide support for the hypothesis that attenuated translational capacity is one of the primary factors governing the growth of skeletal muscle with age. To this end, many studies have focused on examining the translational efficacy of muscle and how this process is affected by age, but the role of age on translational capacity has been largely unexplored. Moreover, studies indicate that even acute differences in protein synthesis, via increased translational efficacy, are not sufficient to predict the degree of muscle hypertrophy (17). These findings further argue for an important role for ribosome biogenesis in regulating chronic changes in muscle size. By utilizing multiple time points during hypertrophic growth, our data indicate that one key determinant of translational capacity (i.e., ribosome biogenesis) is severely impaired in aged skeletal muscle, and this appears to be at the level of rDNA transcription.
Conclusion.
Currently, the regulation of rDNA transcription and ribosome biogenesis in skeletal muscle adaptation is poorly understood, with the majority of studies focusing on assessing the activation of downstream components of the mTORC1 pathway as a readout of ribosome biogenesis. We propose that the regulation of ribosome biogenesis, in particular, the regulation of rDNA transcription, may provide additional insights into the regulation of protein synthesis during skeletal muscle hypertrophy and its importance with aging.
GRANTS
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR-061939 to J.J. McCarthy and K.A. Esser, and by a grant from Merck to K.A. Esser. This work was conducted while J.D. Lee was being supported by Ellison Medical Foundation/American Federation of Aging Research Fellowship EPD 12102.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.A.E. and J.J.M. conception and design of research; J.D.L. and J.H.E. performed experiments; T.J.K., J.D.L., J.H.E., and T.C. analyzed data; T.J.K. interpreted results of experiments; T.C. prepared figures; T.J.K. drafted manuscript; J.D.L., T.C., K.A.E., and J.J.M. edited and revised manuscript; T.J.K., J.H.E., T.C., K.A.E., and J.J.M. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank personnel at the University of Kentucky Microarray Core Facility, specifically Dr. Kuey-Chu Chen and Donna Wall, for assistance with microarray sample preparation and analysis. We thank Dr. Sarah White for assistance with statistical analysis.
REFERENCES
- 1.Adams GR, Bamman MM. Characterization and regulation of mechanical loading-induced compensatory muscle hypertrophy. Comp Physiol 2: 2829–2870, 2012. [DOI] [PubMed] [Google Scholar]
- 2.Blough ER, Linderman JK. Lack of skeletal muscle hypertrophy in very aged male Fischer 344 × Brown Norway rats. J Appl Physiol 88: 1265–1270, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019, 2001. [DOI] [PubMed] [Google Scholar]
- 4.Carson JA, Alway SE, Yamaguchi M. Time course of hypertrophic adaptations of the anterior latissimus dorsi muscle to stretch overload in aged Japanese quail. J Gerontol A Biol Sci Med Sci 50: B391–B398, 1995. [DOI] [PubMed] [Google Scholar]
- 5.Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J Appl Physiol 115: 1065–1074, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chale-Rush A, Morris EP, Kendall TL, Brooks NE, Fielding RA. Effects of chronic overload on muscle hypertrophy and mTOR signaling in young adult and aged rats. J Gerontol A Biol Sci Med Sci 64: 1232–1239, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conesa A, Nueda MJ, Ferrer A, Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 22: 1096–1102, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Degens H, Alway SE. Skeletal muscle function and hypertrophy are diminished in old age. Muscle Nerve 27: 339–347, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Dhanani S, Volpi E, Rasmussen BB. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet Muscle 1: 11, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funai K, Parkington JD, Carambula S, Fielding RA. Age-associated decrease in contraction-induced activation of downstream targets of Akt/mTor signaling in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 290: R1080–R1086, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol 589: 5485–5501, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui X, Zhu W, Wang Y, Lam KS, Zhang J, Wu D, Kraegen EW, Li Y, Xu A. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem 284: 14050–14057, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwee DT, Bodine SC. Age-related deficit in load-induced skeletal muscle growth. J Gerontol A Biol Sci Med Sci 64: 618–628, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA 102: 4252–4257, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol 101: 531–544, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM. Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol 107: 1655–1662, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 102: 306–313, 2007. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, Van Zant G, Campbell KS, Esser KA, Dupont-Versteegden EE, Peterson CA. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138: 3657–3666, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol 116: 693–702, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Nagatomo Y, Carabello BA, Hamawaki M, Nemoto S, Matsuo T, McDermott PJ. Translational mechanisms accelerate the rate of protein synthesis during canine pressure-overload hypertrophy. Am J Physiol Heart Circ Physiol 277: H2176–H2184, 1999. [DOI] [PubMed] [Google Scholar]
- 22.Owino V, Yang SY, Goldspink G. Age-related loss of skeletal muscle function and the inability to express the autocrine form of insulin-like growth factor-1 (MGF) in response to mechanical overload. FEBS Lett 505: 259–263, 2001. [DOI] [PubMed] [Google Scholar]
- 23.Parkington JD, LeBrasseur NK, Siebert AP, Fielding RA. Contraction-mediated mTOR, p70S6k, and ERK1/2 phosphorylation in aged skeletal muscle. J Appl Physiol 97: 243–248, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Phillips BE, Williams JP, Gustafsson T, Bouchard C, Rankinen T, Knudsen S, Smith K, Timmons JA, Atherton PJ. Molecular networks of human muscle adaptation to exercise and age. PLoS Genet 9: e1003389, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips SM. Physiologic and molecular bases of muscle hypertrophy and atrophy: impact of resistance exercise on human skeletal muscle (protein and exercise dose effects). Appl Physiol Nutr Metab 34: 403–410, 2009. [DOI] [PubMed] [Google Scholar]
- 26.Raue U, Slivka D, Minchev K, Trappe S. Improvements in whole muscle and myocellular function are limited with high-intensity resistance training in octogenarian women. J Appl Physiol 106: 1611–1617, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S. Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol 112: 1625–1636, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slivka D, Raue U, Hollon C, Minchev K, Trappe S. Single muscle fiber adaptations to resistance training in old (>80 yr) men: evidence for limited skeletal muscle plasticity. Am J Physiol Regul Integr Comp Physiol 295: R273–R280, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thalacker-Mercer AE, Dell'Italia LJ, Cui X, Cross JM, Bamman MM. Differential genomic responses in old vs. young humans despite similar levels of modest muscle damage after resistance loading. Physiol Genomics 40: 141–149, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomson DM, Gordon SE. Diminished overload-induced hypertrophy in aged fast-twitch skeletal muscle is associated with AMPK hyperphosphorylation. J Appl Physiol 98: 557–564, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Thomson DM, Gordon SE. Impaired overload-induced muscle growth is associated with diminished translational signalling in aged rat fast-twitch skeletal muscle. J Physiol 574: 291–305, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol 302: C1523–C1530, 2012. [DOI] [PubMed] [Google Scholar]
- 33.Welle S, Totterman S, Thornton C. Effect of age on muscle hypertrophy induced by resistance training. J Gerontol A Biol Sci Med Sci 51: M270–M275, 1996. [DOI] [PubMed] [Google Scholar]
- 34.Zak R, Rabinowitz M, Platt C. Ribonucleic acids associated with myofibrils. Biochemistry 6: 2493–2499, 1967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.