Abstract
Recent integrative genomic approaches have defined molecular subgroups of medulloblastoma that are genetically and clinically distinct. Sonic hedgehog (Shh) medulloblastomas account for one-third of all cases and comprise the majority of infant and adult medulloblastomas. To discern molecular heterogeneity among Shh-medulloblastomas, we analyzed transcriptional profiles from four independent Shh-medulloblastoma expression datasets (n = 66). Unsupervised clustering analyses demonstrated a clear distinction between infant and adult Shh-medulloblastomas, which was reliably replicated across datasets. Comparison of transcriptomes from infant and adult Shh-medulloblastomas revealed deregulation of multiple gene families, including genes implicated in cellular development, synaptogenesis, and extracellular matrix maintenance. Furthermore, metastatic dissemination is a marker of poor prognosis in adult, but not in pediatric Shh-medulloblastomas. Children with desmoplastic Shh-medulloblastomas have a better prognosis than those with Shh-medulloblastomas and classic histology. Desmoplasia is not prognostic for adult Shh-medulloblastoma. Cytogenetic analysis of a large, non-overlapping cohort of Shh-medulloblastomas (n = 151) revealed significant over-representation of chromosome 10q deletion (P < 0.001) and MYCN amplification (P < 0.05) in pediatric Shh cases compared with adults. Adult Shh-medulloblastomas harboring chromosome 10q deletion, 2 gain, 17p deletion, 17q gain, and/or GLI2 amplification have a much worse prognosis as compared to pediatric cases exhibiting the same aberrations. Collectively, our data demonstrate that pediatric and adult Shh-medulloblastomas are clinically, transcriptionally, genetically, and prognostically distinct.
Keywords: Medulloblastoma, Sonic hedgehog, Molecular classification, Genomics
Introduction
The peak age of onset of medulloblastoma ranges from 7 to 9 years, but is also diagnosed from infancy through adulthood [2, 12, 16]. Current markers of poor prognosis for medulloblastoma include: patient age, extent of resection, and metastatic status [16]. Recent interest has focused on molecular markers of prognosis [5, 6, 18]. Transcriptomic technologies combined with unsupervised clustering methods have demonstrated that medulloblastomas can be segregated into distinct subgroups [1, 4, 10, 13, 14, 21].
The Shh subgroup is currently the best understood and most highly studied [8]. Germline mutations in PTCH1 (Gorlin Syndrome) [9] and SUFU [20], two negative regulators of the Shh pathway, predispose to medulloblastoma, and somatic mutations in these genes, as well as activating mutations in SMO are collectively found in 10–15% of sporadic medulloblastomas [8]. Curiously, Shh-medulloblastomas occur in a bimodal distribution, making up the majority of infant (≤3 years) and adult (≥16 years) medulloblastomas, but only a small fraction of childhood (4–15 years) tumors. The unusual bimodal age distribution of Shh-medulloblastomas suggests the existence of Shh-subgroup heterogeneity.
To discern the subgroups of Shh-medulloblastoma, we analyzed gene expression data for 33 Shh-medulloblastomas, and validated our findings in a non-overlapping cohort derived from three independent published datasets. We subsequently correlated cytogenetic events, clinical factors, and histology with survival on a cohort of 151 non-overlapping, well-documented Shh-medulloblastomas on a tissue microarray (TMA).
Materials and methods
Samples and data sets
All medulloblastoma cohorts analyzed in the study are summarized in Supplementary Table 1. Primary human Shh-medulloblastomas (n = 33) comprising our discovery cohort have been described previously and were derived from a larger original cohort of 103 primary cases representative of all medulloblastoma subgroups [14]. Previously published gene expression data used as the validation cohort were downloaded from the Gene Expression Omnibus (GEO) repository [(n = 62) [10]: GSE10327, (n = 40) [7]: GSE12992] and obtained directly from Dr. Nada Jabado (n = 12) [1]. This combined validation set consists of 114 primary medulloblastomas and was used as a resource to predict cases of the Shh subtype. For cytogenetic analysis, 151 non-overlapping Shh-medulloblastomas were studied as formalin-fixed paraffin embedded (FFPE) samples on a tissue microarray (TMA).
Copy number and expression array data were generated and analyzed as described previously [14, 15]. TMA content, construction, and FISH have been described previously [17].
Multicolor interphase FISH analysis was performed using commercial probe sets delineating the loci of interest (Vysis, USA): (1) centromere 2p11–q11 (spectrum orange) and 2p24/MYCN (spectrum green); (2) centromere 8p11–q11 (spectrum green) and 8q24/MYC (spectrum orange); (3) 17p13.3/LIS1 (spectrum orange), 17q21/RARA (spectrum green), and 6q23/MYB (spectrum aqua) (4) centromere 10p11–q11 (spectrum green) and 10q23/PTEN (spectrum orange), (5) 9q34/ASS/ABL (spectrum orange and aqua) and 22q11/BCR (spectrum green). For CDK6, GLI1 and GLI2 custom-made fluorescein isothiocyanate-labeled probes were used in combination with centromere 7p11–7q11, 12p11–q11, and 2p11–q11 probes (spectrum orange, Vysis), respectively.
Biostatistics and bioinformatics
Hierarchical clustering (HCL), non-negative matrix factorization (NMF) clustering, principal component analysis (PCA), subclass mapping (SubMap), class prediction (PAM) and Gene Set Enrichment Analysis (GSEA) were carried out as described [14]. For unsupervised HCL, PCA, and NMF of Shh cases, a variance filter (high standard deviation, SD) was used to select a subset of variant genes in the datasets prior to clustering. For HCL and PCA, 1,450 high SD genes were ultimately selected on the basis of cluster stability, whereas NMF was performed using 1,000–5,000 high SD genes with comparable results. Significant genes between classes were identified using t test statistics. Categorical clinical and pathological parameters were compared between groups with Fisher’s exact test. Distribution of survival times was estimated using Kaplan–Meier estimates. The log-rank test was used to compare survival curves between groups.
Results
Age dependent molecular and clinical heterogeneity among Shh-medulloblastomas
In our previous analysis of medulloblastoma subgroups, we confirmed a bimodal age distribution for Shh-medulloblastomas, accounting for 65 and 71% of infant and adult medulloblastomas, respectively (Fig. 1a) [15]. To examine possible heterogeneity among Shh-medulloblastomas, we performed unsupervised HCL of 33 Shh-medulloblastomas to reveal three statistically robust ‘clusters’ of nearly equal proportion (Fig. 1b). Interestingly, 9/10 adult cases clustered together (cluster 2) and independently of pediatric cases, whereas the two remaining clusters included both infant and childhood medulloblastomas (clusters 1 and 3).
Fig. 1.
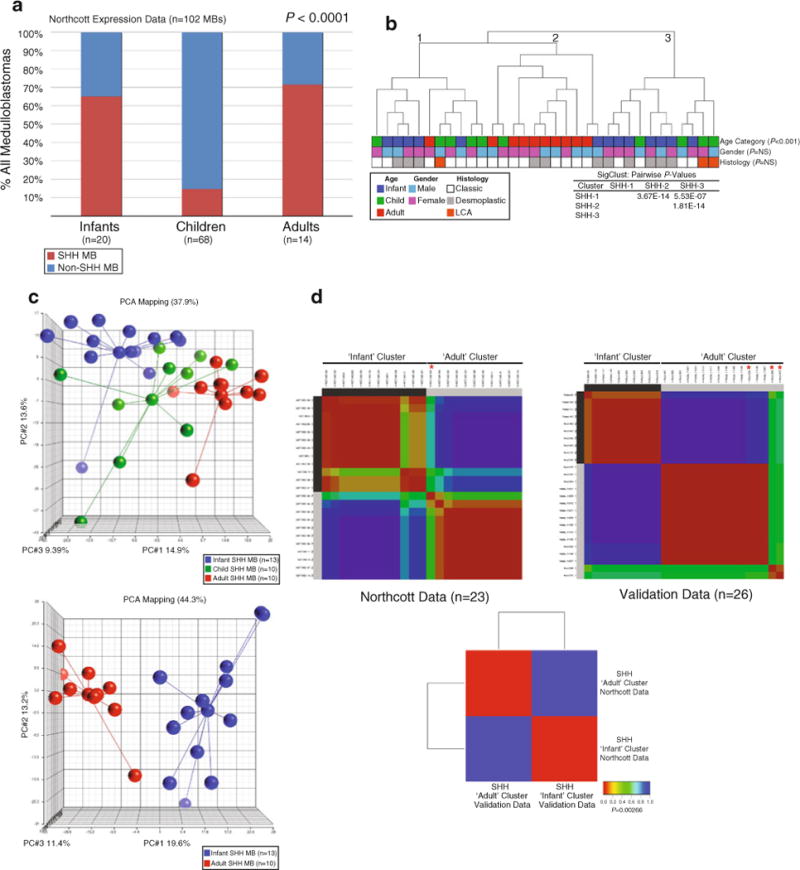
Transcriptional heterogeneity among Shh-medulloblastomas. a Frequency plot of Shh-medulloblastoma incidence reveals a bimodal age distribution. Shh-medulloblastomas from our expression cohort (n = 33) are plotted as a percentage of the total number of tumors (n = 102) according to their respective age category: infants (≤3 years), children (4–15 years), and adults (≥16 years). b Unsupervised HCL of Shh-medulloblastomas identifies three distinct clusters. HCL was performed using 1,450 high SD genes. Pairwise significance values for the three sample clusters as determined using SigClust are shown in the legend. Patient age, gender, and histology are listed below the dendrogram. c Upper panel PCA of Shh-medulloblastomas reveals separation of samples according to patient age category. PCA was performed using the same 1,450 high SD genes utilized in b. Lower panel PCA of infant and adult Shh-medulloblastomas demonstrates clear separation between the two age groups. d Left panel Unsupervised NMF clustering of infant and adult Shh-medulloblastomas identifies two stable classes in the dataset that are indicative of patient age category. The ‘infant’ cluster is comprised solely of infant cases, whereas the ‘adult’ cluster consists of all analyzed adult cases and one infant case (asterisk, *). Right panel NMF clustering performed on a validation dataset of 26 Shh-medulloblastomas recapitulates the ‘infant’ and ‘adult’ clusters identified in our dataset. Infant cases clustering with the ‘adult’ cluster are marked with an asterisk (*). Lower panel SubMap analysis comparing the two Shh subclasses in the current dataset (‘Northcott Data’) to those present in the validation cohort (‘Validation Data’) demonstrates that they are transcriptionally equivalent
Application of PCA to our Shh-medulloblastoma expression series revealed notable separation of cases according to patient age category with childhood Shh-medulloblastomas existing as a ‘molecular intermediate’ of the more extreme infant and adult cases (Fig. 1c, upper panel). As the vast majority of Shh-medulloblastomas occur in infants and adults (Fig. 1a), we decided to focus our molecular analysis on these two classes. PCA of infant (n = 13) and adult (n = 10) Shh-medulloblastomas demonstrated notable separation, and further established the homogeneity within the adult cases (Fig. 1c, lower panel). Unsupervised consensus NMF clustering of expression data from infant (n = 13) and adult (n = 10) cases identified two significant classes in the matrix essentially corresponding to an ‘infant cluster’ and an ‘adult cluster’ (Fig. 1d, Northcott Data, left panel). For validation, we analyzed gene expression data from three independent published datasets. Class prediction analysis was performed using our previously published data as a ‘training’ dataset and three combined datasets [1, 7, 10] (n = 114) as the ‘test’ dataset (data not shown). This analysis revealed 33 validation cases predicted to be of the Shh subtype (data not shown). NMF clustering of infant (n = 13) and adult (n = 13) Shh-medulloblastomas from this validation cohort recapitulated the results obtained above (Fig. 1d, Validation Data, right panel). SubMap analysis using a gene set common to both expression array platforms revealed statistically significant concordance (P = 0.00266) between the two data series’, further supporting our hypothesis that infant and adult Shh-medulloblastomas are molecularly distinct (Fig. 1d, lower panel).
A combination of t test statistics and GSEA confirmed differentially regulated genes and gene sets discriminating adult Shh-medulloblastomas from infant cases (Supplementary Table 2). Among the most highly up-regulated genes identified in adult Shh-medulloblastoma were members of the homeobox (HOX) family, as well as HOX subfamily genes BARHL1 and LHX2 (Fig. 2a). Infant cases showed elevated expression of transcriptional regulators functioning in neuronal development: ID2, ZIC5, and ZIC2. GSEA implicated processes related to tissue development and synaptogenesis as two of the most highly significant themes in adult Shh-medulloblastoma (Table 1; Fig. 2b). Gene sets related to extracellular matrix (ECM) function were highly enriched in pediatric Shh-medulloblastoma (Table 2; Fig. 2c). To identify genes consistently differentially expressed across datasets, we performed an overlap analysis between significant genes identified in our dataset with those in the validation cohort. This approach revealed consistent upregulation of genes involved in cellular and organismal development (i.e. HOX family genes) in adult Shh-medulloblastomas (Fig. 2d). Despite exhibiting common activation of the Shh pathway, infant and adult Shh-medulloblastoma can be discriminated on the basis of their transcriptional programs, and exhibit intertumoral heterogeneity.
Fig. 2.
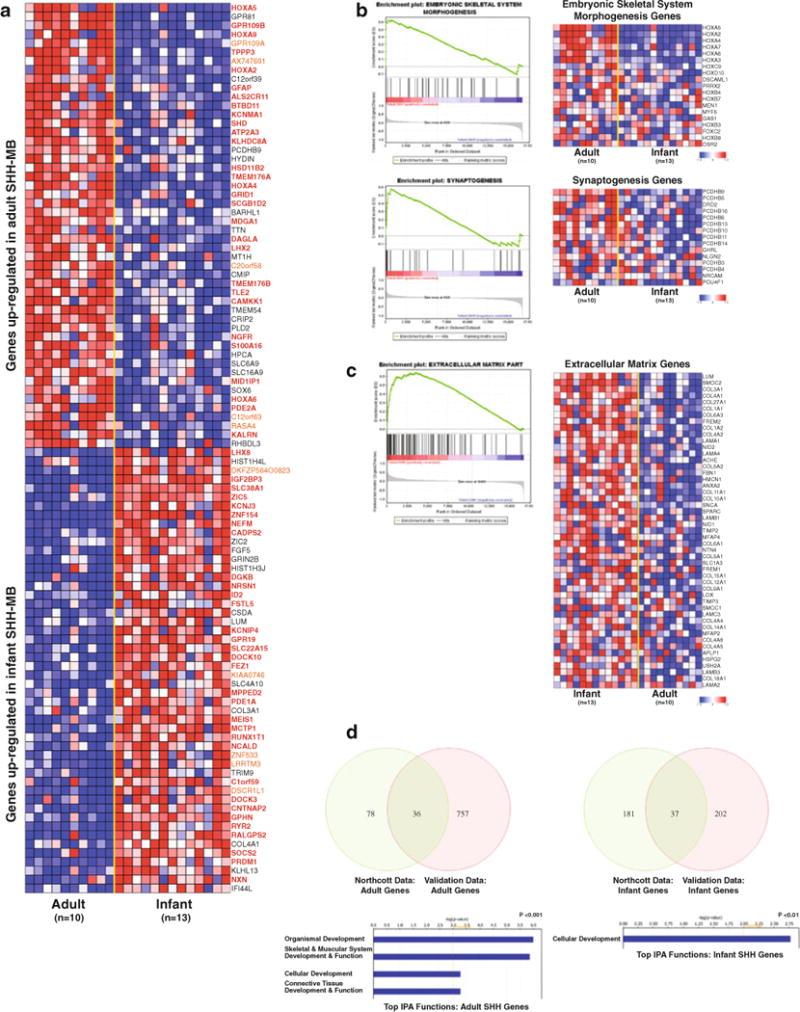
Infant and adult Shh-medulloblastomas exhibit distinct transcriptional profiles. a Heatmap showing the top 100 differentially expressed genes between infant (n = 13) and adult (n = 10) Shh-medulloblastoma as determined using t test statistics applied to our expression cohort. Genes determined to be significantly differentially expressed in the same class (i.e. upregulated in adult Shh-medulloblastoma) in the validation cohort (n = 26) are highlighted in bold/red, whereas genes not present on the array platform used in the validation cohort are highlighted in orange. b GSEA enrichment plots and heatmaps showing enrichment of genes involved in skeletal system morphogenesis (upper panel) and synaptogenesis (lower panel) are significantly correlated with adult Shh-medulloblastoma. c GSEA enrichment plot and heatmap demonstrating a positive correlation between infant Shh-medulloblastoma and genes involved in ECM function. d Significant functional themes identified among adult and infant Shh-medulloblastomas. Venn diagrams illustrate the overlap of significant genes between our Shh expression dataset (‘Northcott Data’) and the validation cohort (‘Validation Data’). Genes identified as significant across all datasets were input according to age category into IPA to determine top functional themes
Table 1.
Top 10 gene sets enriched in adult Shh-MB
| Gene set | Gene set size | NES | Nominal p value | FDR q value |
|---|---|---|---|---|
| Embryonic skeletal system morphogenesis | 38 | 2.1728 | 0.0000 | 0.0298 |
| Skeletal system morphogenesis | 59 | 2.1332 | 0.0000 | 0.0302 |
| Calcium-dependent cell–cell adhesion | 20 | 2.0455 | 0.0000 | 0.0695 |
| Telencephalon development | 31 | 2.0073 | 0.0000 | 0.0763 |
| Myosin binding | 16 | 1.8817 | 0.0000 | 0.2459 |
| Embryonic skeletal system development | 48 | 1.8509 | 0.0000 | 0.2773 |
| Visual behavior | 17 | 1.8200 | 0.0036 | 0.3176 |
| Cardiac muscle tissue morphogenesis | 22 | 1.8175 | 0.0072 | 0.2833 |
| Synaptogenesis | 24 | 1.8149 | 0.0109 | 0.2581 |
| Anterior/posterior pattern formation | 85 | 1.7954 | 0.0000 | 0.2761 |
Table 2.
Top 10 gene sets enriched in infant Shh-MB
| Gene set | Gene set size | NES | Nominal p value | FDR q value |
|---|---|---|---|---|
| Extracellular matrix part | 90 | 2.0634 | 0.0000 | 0.0050 |
| Collagen | 33 | 2.0172 | 0.0000 | 0.0060 |
| Extracellular matrix structural constituent | 60 | 1.9823 | 0.0000 | 0.0110 |
| MHC protein complex | 30 | 1.9506 | 0.0000 | 0.0190 |
| Signaling in immune system | 169 | 1.9464 | 0.0000 | 0.0160 |
| Antigen processing and presentation | 47 | 1.9447 | 0.0000 | 0.0135 |
| Cell surface interactions at the vascular wall | 53 | 1.9367 | 0.0000 | 0.0142 |
| HSA04514_cell_adhesion_ molecules | 122 | 1.8978 | 0.0000 | 0.0267 |
| Proteinaceous extracellular matrix | 269 | 1.8974 | 0.0000 | 0.0243 |
| Integrin binding | 47 | 1.8939 | 0.0000 | 0.0237 |
Pediatric and adult Shh-medulloblastoma cytogenetics
We analyzed high-resolution SNP copy number profiles for the Shh cases in our cohort (n = 33, Supplementary Fig. 1a) [15]. We observed a trend towards a higher incidence of chromosome 2 copy number gain (Supplementary Fig. 1b) and chromosome 10 deletion (Supplementary Fig. 1c) in pediatric (≤15 years) cases compared to adults (≥16 years). Indeed, 10q deletion is not seen in any of the adult cases, suggesting that it is an infant-specific event.
To gain an improved understanding of clinical and cytogenetic heterogeneity among different age categories of Shh-medulloblastoma, we analyzed a large cohort of 151 well-documented Shh cases on a TMA (Fig. 3a) [11]. Adult Shh-medulloblastomas (n = 96) were predominantly male, localized to cerebellar hemispheres, and non-metastatic, whereas pediatric Shh-medulloblastomas (n = 55) occurred equally in males and females, were predominantly localized to the vermis, and were more frequently metastatic (Fig. 3a).
Fig. 3.
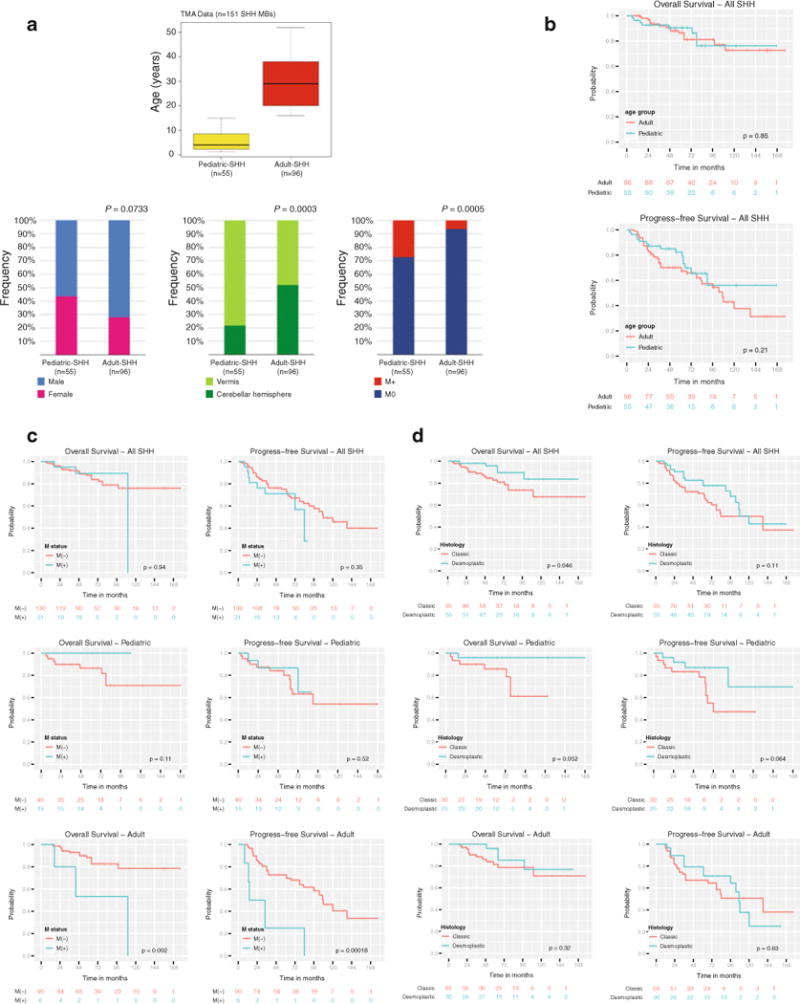
Prognostic significance of clinicopathological features in Shh-medulloblastoma. a Age distribution of pediatric (n = 55) and adult (n = 96) Shh-medulloblastomas in our MB TMA cohort (n = 151). Lower panels Gender, tumor location, and metastatic frequency in our MB TMA cohort. b OS and PFS probabilities for pediatric and adult Shh-medulloblastomas included in the cohort. c OS and PFS probabilities for Shh-medulloblastomas stratified by metastatic status. d OS and PFS probabilities for Shh-medulloblastomas stratified by histological subtype (classic vs. desmoplastic)
We used FISH to interrogate the copy number status of specific genomic loci in our medulloblastoma TMA cohort. Loci analyzed included the known medulloblastoma oncogenes MYC, MYCN, GLI2, and CDK6, as well as chromosomal locations known to be targeted in medulloblastoma (Table 3; Supplementary Fig. 1d). Multiple statistically significant aberrations are over-represented in pediatric Shh-medulloblastomas compared with adults, with MYCN amplification (P = 0.0298) and chromosome 10q deletion (P = 0.0002) being the most notable (Table 3). Although not statistically significant, there was also a higher incidence of chromosome 2 gain (27.3 vs. 15.6%) and 9q deletion (56.4 vs. 37.5%) in the pediatric cases compared to adults (Table 3).
Table 3.
Cytogenetics of adult and pediatric Shh-MB and association with survival
| Variable | % Adult SHH | % Pediatric SHH | Raw P value | Adjusted P value | Log-rank P value (OS) all SHH | Log-rank P value (OS) adult SHH | Log-rank P value (OS) pediatric SHH |
|---|---|---|---|---|---|---|---|
| MYC amplification | 0.0 (0/96) | 0.0 (0/55) | N/A | N/A | N/A | N/A | N/A |
| CDK6 amplification | 2.1 (2/96) | 3.6 (2/55) | 0.6224 | 1.0000 | N/A | N/A | N/A |
| Chr6q loss | 3.1 (3/96) | 0.0 (0/55) | 0.5541 | 1.0000 | N/A | N/A | N/A |
| Chr9q loss | 37.5 (36/96) | 56.4 (31/55) | 0.0280 | 0.1960 | 0.8038 | 0.4478 | 0.5142 |
| MYCN amplification | 1.0 (1/96) | 12.7 (7/55) | 0.0037 | 0.0298 | 0.282 | N/A | N/A |
| GLI2 amplification | 6.2 (6/96) | 7.3 (4/55) | 1.0000 | 1.0000 | 0.0000 | 0.0000 | 0.5422 |
| Chr2 gain | 15.6 (15/96) | 27.3 (15/55) | 0.0938 | 0.5629 | 0.0008 | 0.0012 | 0.1286 |
| Chr10q loss | 19.8 (19/96) | 54.5 (30/55) | 0.0000 | 0.0002 | 0.0004 | 0.0000 | 0.1887 |
| Chr17p loss | 19.8 (19/96) | 12.7 (7/55) | 0.3708 | 1.0000 | 0.0000 | 0.0000 | 0.2201 |
| Chr17q gain | 16.7 (16/96) | 18.2 (10/55) | 0.8256 | 1.0000 | 0.0000 | 0.0000 | 0.0783 |
Age-related prognostic significance of clinical, histological and cytogenetic variables in Shh-medulloblastoma
Examination of progression-free survival (PFS) and overall survival (OS) probabilities comparing pediatric and adult Shh-medulloblastomas showed no significant difference in patient outcome, although there was a trend towards a higher probability of relapse (PFS) in adults (Fig. 3b, median follow-up = 68.0 months, 0.95LCL = 63.0, 0.95LCL = 78.0). Although leptomeningeal dissemination is much more common in pediatric Shh-medulloblastoma as compared to adults (27 vs. 6%, P < 0.001, Fig. 3a), the presence of metastases is prognostic in adults (P = 0.002), but not in pediatric cases (P = 0.11) (Fig. 3c). Conversely, pediatric desmoplastic Shh-medulloblastomas have a better prognosis than classical histology Shh-medulloblastomas (P = 0.052), whereas in adults, histology has no detectable effect on prognosis (P = 0.32) (Fig. 3d). Chromosome 10q deletion and survival probability was shown to be highly significant (log-rank P = 0.0004, Table 3; Fig. 4a) with adult Shh-medulloblastomas harboring 10q loss exhibiting particularly poor PFS and OS probabilities (log-rank P < 0.0001, Fig. 4a). Although chromosome 10q deletion is more prevalent in pediatric Shh cases as compared to adults, it appears to predict a worse outcome in adults, suggesting that the critical gene(s) targeted by 10q deletion may differ between the two populations. Chromosome 2 gain, 17p loss, and 17q gain all demonstrate prognostic significance in Shh-medulloblastoma (log-rank P < 0.001, Table 3, Supplementary Fig. 2a–c), with the most adverse effect on OS observed in adults (log-rank P < 0.01, data not shown). GLI2 amplification is highly correlated with OS probability (log-rank P < 0.0001, Table 3; Fig. 4b), particularly in adults (log-rank P < 0.0001, Fig. 4b). The disparate effects of metastases, histology, and cytogenetics between adult and pediatric Shh-medulloblastoma support our hypothesis that they are distinct disorders.
Fig. 4.
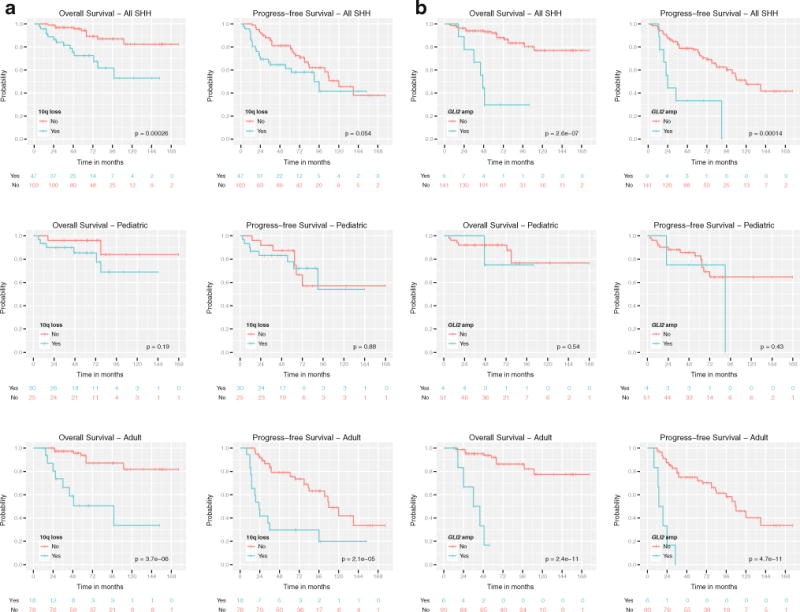
Prognostic significance of chromosome 10q deletion and GLI2 amplification among Shh-medulloblastomas. a OS and PFS probabilities for Shh-medulloblastomas stratified by chromosome 10q deletion status. b OS and PFS probabilities for Shh-medulloblastomas stratified by GLI2 amplification status
Discussion
Now that several subgroups of medulloblastoma have been demonstrated [4, 10, 14, 21], focus will turn to determination of subgroup characterization and heterogeneity. Our unbiased, unsupervised approach to multiple non-overlapping cohorts of Shh-medulloblastomas demonstrates considerable heterogeneity, particularly between adult and non-adult cases. A full and statistically significant characterization of all Shh-medulloblastoma subgroups will require analysis of larger cohorts. The clinical, pathological, and cytogenetic differences between adult and pediatric Shh-medulloblastomas are clinically relevant. It is very notable that infant and adult medulloblastomas are treated very differently, and that differences in prognosis could relate to treatment effects as well as underlying biology. In particular, the lack of prognostic significance for metastatic status in pediatric Shh-medulloblastoma requires validation, as it has important implications for treatment intensity of children with M+ Shh-medulloblastoma, and as it diverges from prior publications on non-sub-grouped infant medulloblastoma. However, prior publications demonstrating a prognostic effect of M status in infant medulloblastoma may have included infants with Group C medulloblastoma, who have both a high incidence of metastasis, and a very poor prognosis. SMO inhibitors have recently been shown to have dramatic, albeit transient efficacy in cases of Shh-medulloblastoma [3, 19]. More durable responses to SMO inhibitors will likely require the use of concurrent therapies, the targets for which might be suggested by over-represented pathways identified above [3]. The clinically prognostic, and age group specific cytogenetic aberrations that we identify could be used for treatment stratification in the setting of clinical trials. Future, multidimensional studies, prospective when possible, profiling much larger cohorts of Shh-medulloblastoma will likely be necessary to fully appreciate the molecular and clinical diversity of Shh-medulloblastoma.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-011-0846-7) contains supplementary material, which is available to authorized users.
Contributor Information
Paul A. Northcott, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada Program in Developmental and Stem Cell Biology, Hospital for Sick Children, Toronto, ON, Canada.
Thomas Hielscher, Division of Biostatistics, German Cancer Research Center, (DKFZ), Heidelberg, Germany.
Adrian Dubuc, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada; Program in Developmental and Stem Cell Biology, Hospital for Sick Children, Toronto, ON, Canada; Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada.
Stephen Mack, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada; Program in Developmental and Stem Cell Biology, Hospital for Sick Children, Toronto, ON, Canada; Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada.
David Shih, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada; Program in Developmental and Stem Cell Biology, Hospital for Sick Children, Toronto, ON, Canada; Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada.
Marc Remke, Division Molecular Genetics, German Cancer Research Center, (DKFZ), Heidelberg, Germany; Department of Pediatric Oncology, Hematology, Immunology, University of Heidelberg, Heidelberg, Germany.
Hani Al-Halabi, Department of Radiation Oncology, Montreal General Hospital, McGill University Health Centre, Montreal, QC, Canada.
Steffen Albrecht, Department of Pathology, Montreal Children’s Hospital, McGill University Health Centre, Montreal, QC, Canada.
Nada Jabado, Departments of Pediatrics and Human Genetics, McGill University Health Centre, Montreal, QC, Canada.
Charles G. Eberhart, Departments of Pathology, Ophthalmology and Oncology, Johns Hopkins University, Baltimore, MD, USA
Wieslawa Grajkowska, Department of Pathology, Children’s Memorial Health Institute, Warsaw, Poland.
William A. Weiss, Departments of Neurology, Pediatrics, and Neurological Surgery, UCSF, San Francisco, CA, USA
Steven C. Clifford, Northern Institute for Cancer Research, Newcastle University, Newcastle, UK
Eric Bouffet, Neuro-oncology Program, Division of Haematology/Oncology, The Hospital for Sick Children, Toronto, Canada.
James T. Rutka, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada; Division of Neurosurgery, Hospital for Sick Children, Toronto, ON, Canada.
Andrey Korshunov, Clinical Cooperation Unit Neuropathology (G-380), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Stefan Pfister, Division Molecular Genetics, German Cancer Research Center, (DKFZ), Heidelberg, Germany; Department of Pediatric Oncology, Hematology, Immunology, University of Heidelberg, Heidelberg, Germany.
Michael D. Taylor, Email: mdtaylor@sickkids.ca, The Arthur and Sonia Labatt Brain Tumour Research Center, Hospital for Sick Children, Toronto, ON, Canada; Program in Developmental and Stem Cell Biology, Hospital for Sick Children, Toronto, ON, Canada; Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada; Division of Neurosurgery, Hospital for Sick Children, Toronto, ON, Canada.
References
- 1.Al-Halabi H, Nantel A, Klekner A, Guiot MC, Albrecht S, Hauser P, Garami M, Bognar L, Kavan P, Gerges N, Shirinian M, Roberge D, Muanza T, Jabado N. Preponderance of sonic hedgehog pathway activation characterizes adult medulloblastoma. Acta Neuropathol. 2011;121:229–239. doi: 10.1007/s00401-010-0780-0. [DOI] [PubMed] [Google Scholar]
- 2.Brandes AA, Franceschi E, Tosoni A, Reni M, Gatta G, Vecht C, Kortmann RD. Adult neuroectodermal tumors of posterior fossa (medulloblastoma) and of supratentorial sites (stPNET) Crit Rev Oncol Hematol. 2009;71:165–179. doi: 10.1016/j.critrevonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, Yu Q, Ostrom L, Fordjour P, Anderson DL, Monahan JE, Kelleher JF, Peukert S, Pan S, Wu X, Maira SM, Garcia-Echeverria C, Briggs KJ, Watkins DN, Yao YM, Lengauer C, Warmuth M, Sellers WR, Dorsch M. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, Lau CC, Olson JM, Gilbertson RJ, Gajjar A, Delattre O, Kool M, Ligon K, Meyerson M, Mesirov JP, Pomeroy SL. Integrative Genomic Analysis of Medulloblastoma Identifies a Molecular Subgroup That Drives Poor Clinical Outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberhart CG. Molecular diagnostics in embryonal brain tumors. Brain Pathol. 2011;21:96–104. doi: 10.1111/j.1750-3639.2010.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, Manie E, Raquin MA, Bours D, Carpentier S, Barillot E, Grill J, Doz F, Puget S, Janoueix-Lerosey I, Delattre O. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218:86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 8.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 9.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 10.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, Mrsic A, Ylstra B, Grajkowska W, Hartmann W, Pietsch T, Ellison D, Clifford SC, Versteeg R. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korshunov A, Remke M, Werft W, Benner A, Ryzhova M, Witt H, Sturm D, Wittmann A, Schottler A, Felsberg J, Reifenberger G, Rutkowski S, Scheurlen W, Kulozik AE, von Deimling A, Lichter P, Pfister SM. Adult and pediatric medulloblastomas are genetically distinct and require different algorithms for molecular risk stratification. J Clin Oncol. 2010;28:3054–3060. doi: 10.1200/JCO.2009.25.7121. [DOI] [PubMed] [Google Scholar]
- 12.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra YS, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Packer RJ, Vezina G. Management of and prognosis with medulloblastoma: therapy at a crossroads. Arch Neurol. 2008;65:1419–1424. doi: 10.1001/archneur.65.11.1419. [DOI] [PubMed] [Google Scholar]
- 17.Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, Wittmann A, Devens F, Gerber NU, Joos S, Kulozik A, Reifenberger G, Rutkowski S, Wiestler OD, Radlwimmer B, Scheurlen W, Lichter P, Korshunov A. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 18.Pfister SM, Korshunov A, Kool M, Hasselblatt M, Eberhart C, Taylor MD. Molecular diagnostics of CNS embryonal tumors. Acta Neuropathol. 2010;120:553–566. doi: 10.1007/s00401-010-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Stavrou T, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 21.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A, Gilbertson RJ. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


