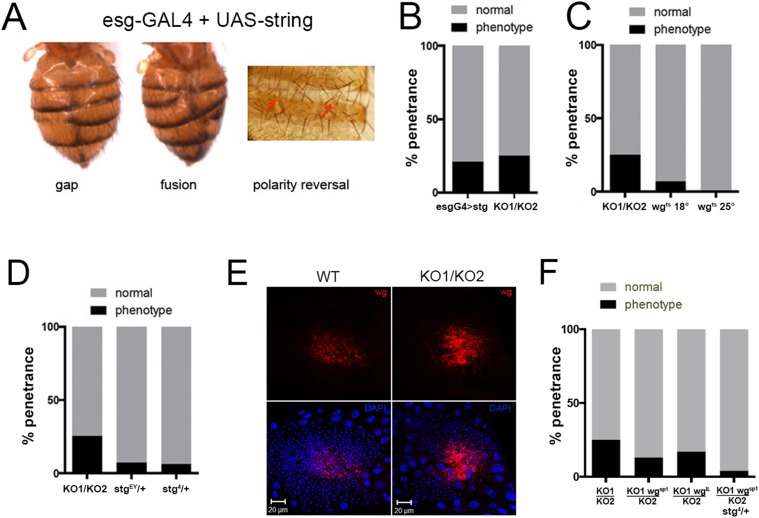
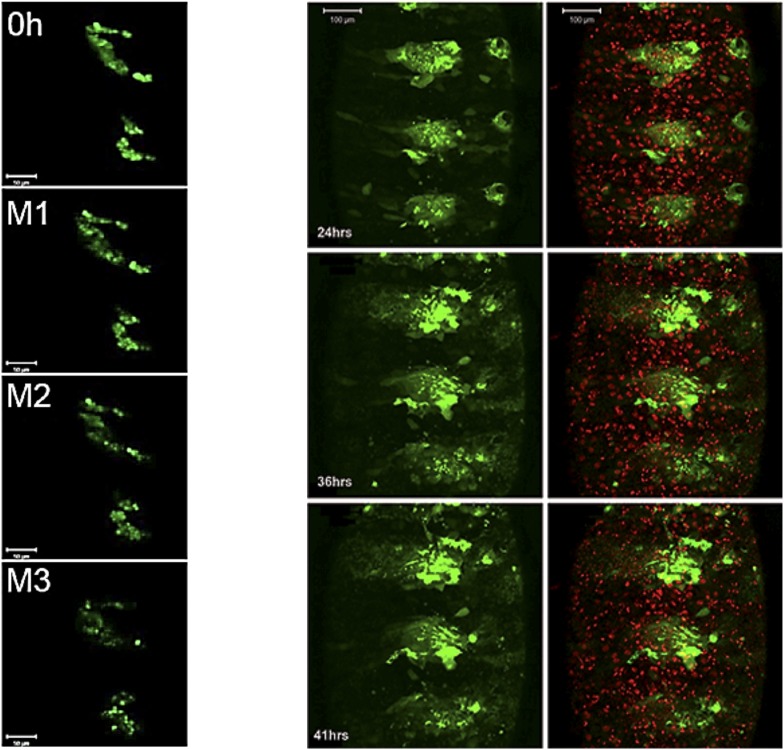
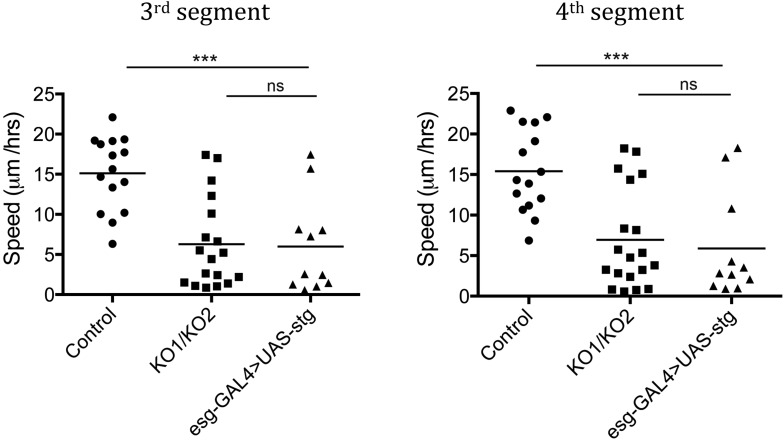
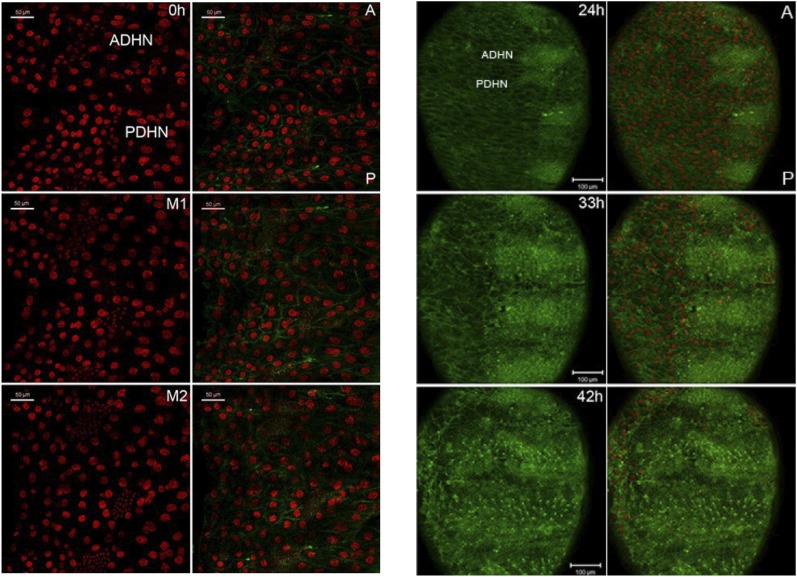
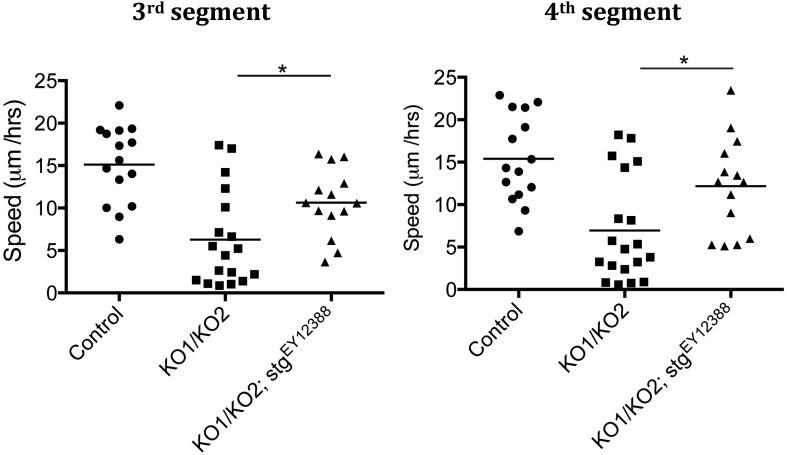
Figure 5. Overexpression of string and wg contributes to the miR-965 mutant phenotype.
(A) Dorsal views of abdomens from adult female esg-Gal4 UAS-string flies illustrating the segment gap, segment fusion and polarity reversal phenotypes. (B) Penetrance of abdominal defects of all classes in esg-Gal4 UAS-string vs mutant. esg-Gal4 UAS-string: n = 97/469; KO1/KO2 n = 110/446. p = 0.16 Fishers exact test. (C) Penetrance of abdominal defects in esg-Gal4 UAS-wgts flies reared at 18° and 25°C vs KO1/KO2. esg-Gal4 UAS-wgts reared at 18°C: n = 9/129; esg-Gal4 UAS-wgts at 25°C n = 1/254; KO1/KO2 n = 110/446. p = 0.014 comparing wgts at 18 vs 25°C, Fishers exact test. (D) Penetrance of abdominal defects comparing KO1/KO2 mutants with KO1/KO2 mutants carrying one copy of stringEY12388 or string4 alleles. p < 0.001 comparing KO1/KO2 to KO1/KO2; stgEY/+ or stg4/+ using Fisher's exact test. (E) Confocal micrographs showing dorsal histoblast nests of wild-type (WT) and miR-965 mutant (KO) at ∼24 hr APF labeled with anti-Wg (red). Nuclei were labeled with DAPI (blue). Scale bar: 20 µm. Anterior and dorsal histoblast nests in the miR-965 mutants were not yet fused at 24 hr APF, due to delayed migration. Images were captured using identical microscope settings. (F) Penetrance of abdominal segmentation defects comparing KO1/KO2 mutants with KO1/KO2 mutants carrying one copy of wgSP-1 or wgl-12 temperature sensitive alleles or carrying one copy of wgSP-1 and stg4 together. p < 0.05 comparing KO1/KO2 to KO1, wgSP-1/KO2 using Fisher's exact test. KO1/KO2 was not significantly different from KO1, wgI-12/KO2, perhaps because wgI-12 is a weaker, temperature sensitive allele. p < 0.001 comparing KO1/KO2 with KO1, wgSP-1/KO2; stg4/+ using Fisher's exact test.
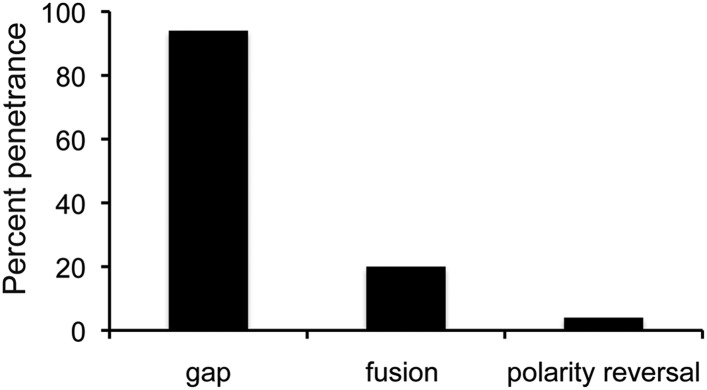
Figure 5—figure supplement 1. The proportion of flies with defects caused by string overexpression.