Abstract
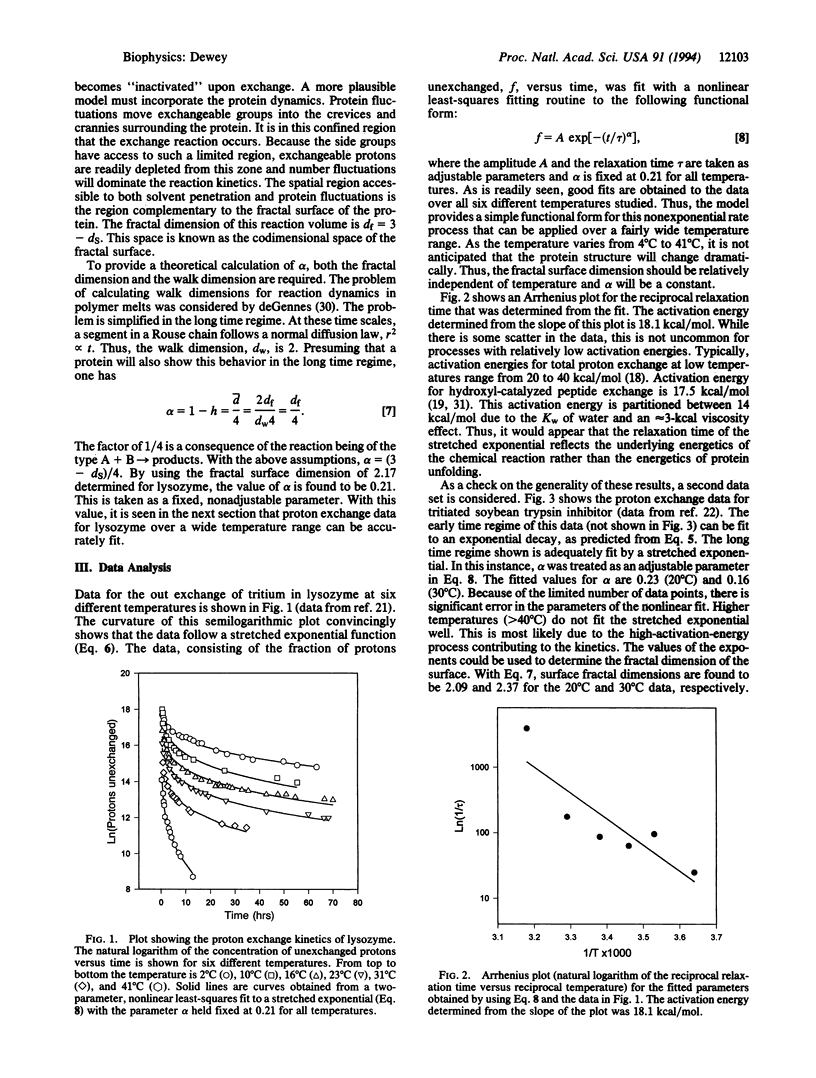
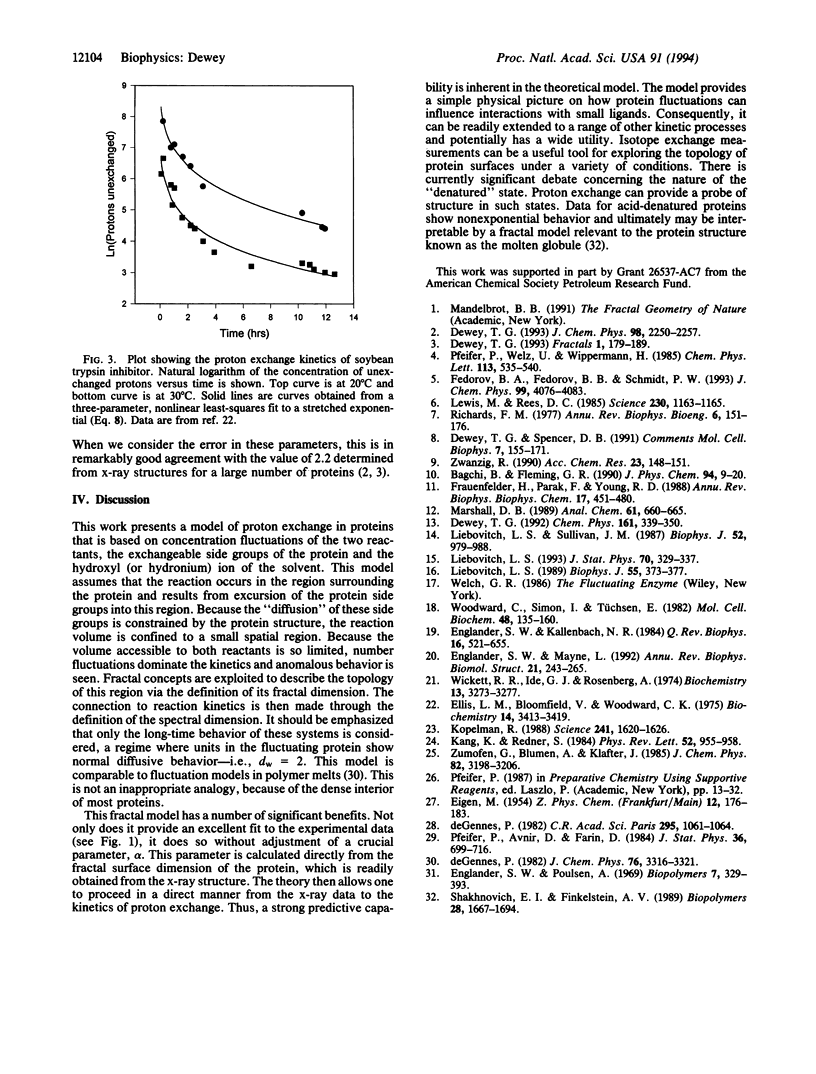
Experimental data for the exchange of protons in tritiated lysozyme is reexamined by using a fractal model. The fraction of protons unexchanged, f, is seen to follow a stretched exponential, f infinity exp[(-t/tau)alpha], in the long time limit. Data over a range of temperatures are considered, and accurate fits are obtained with a single, unadjusted scaling exponent, alpha. The time constant, tau, follows an Arrhenius law and gives an activation energy comparable to that obtained for free peptide exchange. A model is proposed where proton exchange occurs as a result of solvent reacting with protein side groups in a restricted volume surrounding the protein. Dynamic fluctuations of the protein allow the protonated groups to enter this volume. Solvent also penetrates this volume, allowing proton exchange to occur. The fluctuations of reactants in this restricted volume dominate the kinetics and result in anomalous behavior. The topology of this reaction volume can be characterized by its fractal dimension. The fractal dimension of the space excluded by the protein is equal to 3-ds, where ds is the fractal dimension of the protein surface. The dimensionality of this "reaction space" can be used to predict the value of the exponent alpha. When the problem is treated as a reaction of the type A + B-->C in a confined region, the exponent is given by alpha = (3-ds)/4. By using the value of 2.17 previously established for the surface dimension of lysozyme [Pfeifer, P., Welz, U. & Wippermann, H. (1985) Chem. Phys. Lett. 113, 535-540], a corresponding alpha of 0.21 is calculated. Data for lysozyme at six different temperatures could be accurately fit by using this unadjusted value for alpha. These results show how the surface morphology of a protein influences diffusional processes of small molecules associating with the protein.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ellis L. M., Bloomfield V. A., Woodward C. K. Hydrogen-tritium exchange kinetics of soybean trypsin inhibitor (Kunitz). Solvent accessibility in the folded conformation. Biochemistry. 1975 Jul 29;14(15):3413–3419. doi: 10.1021/bi00686a019. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Mayne L. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V., Shakhnovich E. I. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers. 1989 Oct;28(10):1681–1694. doi: 10.1002/bip.360281004. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Parak F., Young R. D. Conformational substates in proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- Kopelman R. Fractal reaction kinetics. Science. 1988 Sep 23;241(4873):1620–1626. doi: 10.1126/science.241.4873.1620. [DOI] [PubMed] [Google Scholar]
- Lewis M., Rees D. C. Fractal surfaces of proteins. Science. 1985 Dec 6;230(4730):1163–1165. doi: 10.1126/science.4071040. [DOI] [PubMed] [Google Scholar]
- Liebovitch L. S., Sullivan J. M. Fractal analysis of a voltage-dependent potassium channel from cultured mouse hippocampal neurons. Biophys J. 1987 Dec;52(6):979–988. doi: 10.1016/S0006-3495(87)83290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebovitch L. S. Testing fractal and Markov models of ion channel kinetics. Biophys J. 1989 Feb;55(2):373–377. doi: 10.1016/S0006-3495(89)82815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Wickett R. R., Ide G. J., Rosenberg A. A hydrogen-exchange study of lysozyme conformation changes induced by inhibitor binding. Biochemistry. 1974 Jul 30;13(16):3273–3277. doi: 10.1021/bi00713a015. [DOI] [PubMed] [Google Scholar]
- Woodward C., Simon I., Tüchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982 Oct 29;48(3):135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]


