Abstract
As part of the International Cooperative Biodiversity Groups (ICBG) Program, we were interested in identifying biologically active unfolded protein response (UPR) inducing compounds from marine microorganisms isolated from Costa Rican biota. With this aim in mind we have now generated more than 33,000 unique prefractionated natural product extracts from marine and terrestrial organisms that have been submitted to the Center of Chemical Genomics (CCG) at the University of Michigan for high throughput screening (HTS). An effective complementary cell-based assay to identify novel modulators of UPR signaling was used for screening extracts. Active fractions were iteratively subjected to reverse-phase HPLC chromatographic analysis, and together with lobophorin A, B, E, and F (1–4), three new lobophorin congeners, designated as CR1 (5), CR2 (6), and CR3 (7) were isolated. Herein, we report that secondary assays revealed that the new lobophorins induced UPR-associated gene expression, inhibited oral squamous cell carcinoma cell growth, and led to UPR-dependent cell death in murine embryonic fibroblast (MEF) cells.
Keywords: UPR, CHOP, ATF4, lobophorin, natural products, oral squamous cell carcinoma, oral cancer, ER stress, anticancer
In 2011 there were nearly 300,000 Americans living with oral or pharyngeal cancer and 40,000 new cases are expected this year.1 Late stage diagnosis and a paucity of new therapies have stagnated the five-year survival rate at less than 50%; and the serendipitous discovery of cisplatin almost 40 years ago provided one of the last major improvements in our ability to treat these patients.
Approximately 60% of the drugs currently on the market are derived from natural product sources.2 Since many bioactive metabolites initially isolated from invertebrates are biosynthesized by associated microorganisms, it is often not possible to obtain culturable species that produce sufficient biomass for compound isolation and identification. With only 0.001–0.1% of marine microorganisms identified, however, they represent a relatively unexplored source of potential bioactive compounds.3 The collection of natural products into screenable libraries and the development of reliable novel target assays for HTS is essential to find new active structures as well as to identify new activities for known compounds.
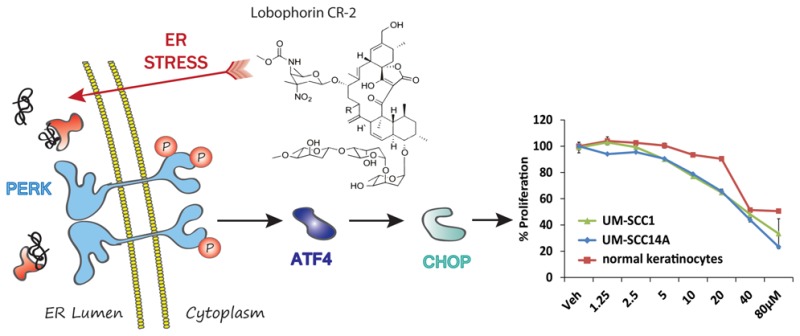
In our ongoing effort to identify novel compounds that activate the apoptotic arm of unfolded protein response (UPR) as a potential anticancer strategy (Figure 1), we screened a collection of natural product extracts at the University of Michigan Center for Chemical Genomics to identify those that activated UPR (luciferase) reporters.4 The UPR is a coordinated response to the accumulation of misfolded protein in the lumen of the endoplasmic reticulum (ER). Inositol-requiring enzyme 1 alpha (IRE1α), activating transcription factor 6 (ATF6), and protein kinase RNA-like endoplasmic reticulum kinase (PERK) are ER transmembrane sensors that monitor the productivity of protein folding in the lumen. When the ability of the ER to properly fold peptides is outpaced by the demand for new proteins, the UPR is initiated by genetically distinct pathways that attempt to enhance the dynamics of peptide processing (adaptive arm) or lead to cell death (apoptotic arm) if the challenge is prolonged.5 The adaptive response is characterized by the unconventional splicing of a 26 base intron from X-box binding protein (XBP1) mRNA by IRE1α in the cytoplasm. Spliced XBP1 encodes a multitarget transcription factor that drives the expression of chaperones, heat shock factors, and other enzymes that return to the ER to enhance folding. Inhibition of protein translation, a hallmark of ER stress, occurs via PERK mediated phosphorylation of eukaryotic initiation factor 2 alpha (eIF2α). eIF2α phosphorylation also leads to the accumulation of the proapoptotic transcription factors ATF4 and CHOP.6,7
Figure 1.
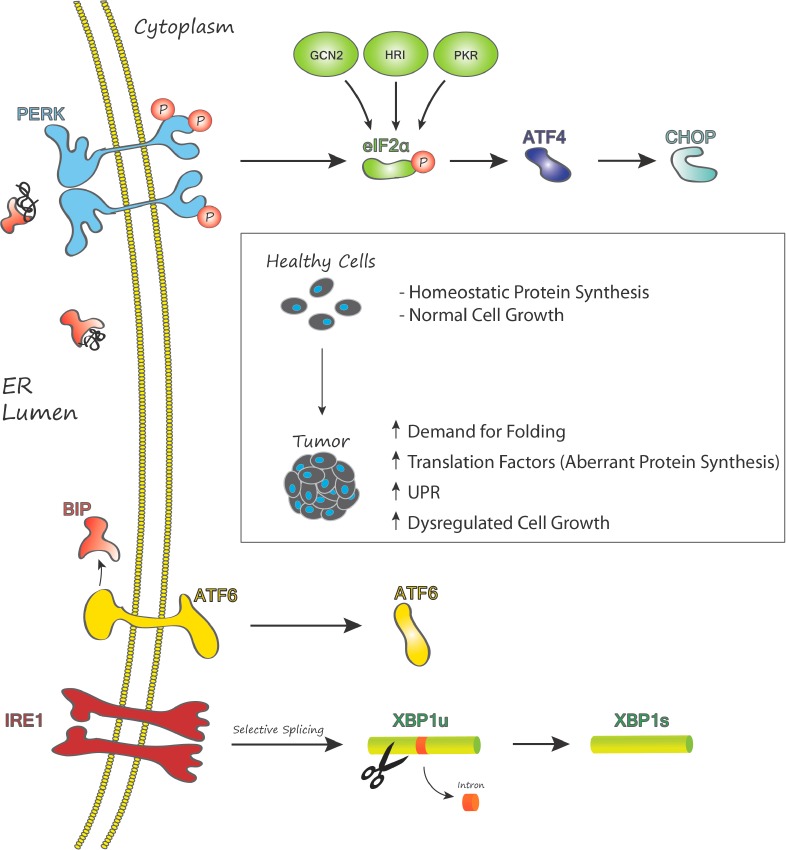
General model of ER stress and the UPR in cancer.
Given the highly secretory nature of many solid and hematological tumors it is not surprising that increased expression of translation factors and high basal levels of stress and UPR signaling characterize many human cancers. Recent studies have revealed that more than a dozen human cancers display increased expression of eukaryotic initiation factors including breast,8 pancreas, lymphoma, leukemia, and head and neck squamous cell carcinoma.9,10 As malignant cell populations begin to grow and invade host tissue, the extracellular tumor milieu becomes increasingly starved of oxygen, glucose, and other essential nutrients. Survival in these harsh conditions requires further UPR activation. We hypothesize that pharmacologic enforcement of the UPR might overwhelm the adaptive response in malignant cells and divert them toward apoptosis, while healthy adjacent cells might see the challenge, mount an effective adaptive UPR, and return to homeostasis.
A cell-based HTS was engineered using Chinese Hamster Ovary (CHO) cells stably transfected with luciferase reporters to specifically monitor XBP1 splicing or Chop promoter activation.11 With the goal of identifying new active structures from renewable marine microbial sources, a collection of 5036 extracts derived from cultivable marine microorganisms was prepared in collaboration with the National Institute for Biodiversity (INBio) in Costa Rica. Lobophorins E, F, and CR1–3 were identified in a DMSO extract that could significantly activate a Chop-luciferase reporter but not an XBP1-luciferase reporter (Figure S1), suggesting that one or more compounds could selectively activate the apoptotic arm of the UPR. Bioassay guided fractionation of extracts prepared from the active strain 7790_N4 allowed for the isolation of the spirotetronate macrolides lobophorin A, B, E, and F (1–4)12,13 and three new congeners designated CR1 (5), CR2 (6), and CR3 (7) (Figure 2).
Figure 2.
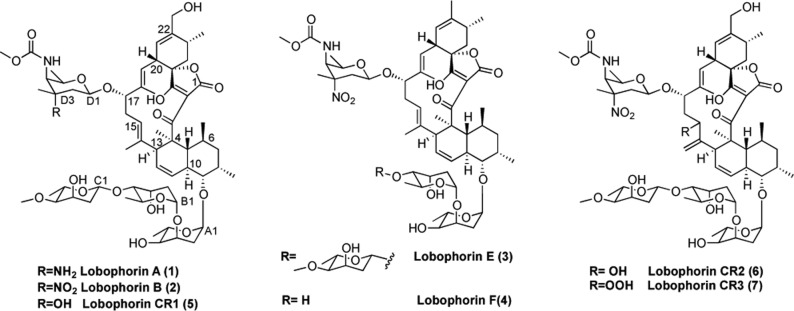
Structure of known lobophorins and novel congeners CR1–CR3.
Lobophorin CR1 (5) was isolated as a white powder. Its molecular formula was established by HREIMS as C61H92NO20 (m/z 1180.5993 [M + Na]+; calcd for C61H92NO20Na, 1180.6032), requiring 17 degrees of unsaturation and one more amu than compound 1 (C61H91N2O19).
Comparison of 1H NMR spectra of compound 5 with 1 did not reveal a clear difference. COSY, HSQC, and HMBC data correlated to (not with) the same signals represented by the aglycone and trisaccharide moieties that were present in 1. Differences between 1 and 5 were identified in a group of signals corresponding with the amino sugar. In this group, together with two methines (δH/δC 4.67/98.4, 3.25/57.4), one methylene (δH/δC 1.50,1.68/39.3), three methyls (δH/δC 1.07/16.1, 1.16/26.9, 3.63/51.4), and one quaternary carbon (δC 158.4) showing the same shifts as 1 appeared a new quaternary carbon at δC 71.5 instead of at δC 52.4 or 91.0 as in lobophorin A (1) and B (2), respectively. These data indicated that instead of a nitro or amino group at position D-3, there must be a hydroxyl group, maintaining the same configuration β as in 1 and 2. All of these data are consistent with the structure of compound 5 (Table S1 and Figures S2–S7).
The molecular formula of lobophorin CR2 (6), isolated as a white powder, was established as C61H90N2O22 by HREIMS (m/z 1247.5697 [M – H + 2Na]+; calcd for C61H89N2O22Na2, 1247.5702), indicating one less degree of unsaturation and 16 more amu than 2. NMR data showed the presence of the same sugars, three digitoxose and one kijanose, as lobophorin B. However, some interesting differences regarding the aglycone moiety were observed in the 1H and 13C NMR shifts when compared with the other congeners. Two new signals were present in the HSQC, an olefinic methylene (δH/δC 4.78, 5.10/114.0) and an oxymethine (δH/δC 4.06/67.9). These data together with the lack of the olefinic methine at C-15 (characteristic of lobophorin family) as well as the shift of methylene C-16 from δH 2.16, 2.49/32.3 in 2, to δH 1.80, 1.91 in 6, suggested the presence of an exocyclic double bond at C-14 and a hydroxyl group bonded to the methine C-15. COSY experiment allowed connecting the methine C-15 with the methylene C-16 through a H–H correlation between H-15 (δH 4.06) and H-16a (δH 1.80). Finally, the exocyclic methylene was placed at C-14 by HMBC correlations of H-30a (δH 4.78) with C-13 (δC 45.4) and C-15 (δC 67.9). These data enabled us to propose the structure of CR2 (see Figure 2), which was supported by a key ROESY cross peak between H-30a (δH 4.78) and H2-16 (δH16a 1.80 and δH16b 1.91) (Table S2 and Figures S8–S12). Insufficient CR2 was isolated to obtain reliable 13C NMR.
Lobophorin CR3 (7) was obtained as a white powder, with the molecular formula C61H90N2O23 on the basis of its HREIMS (m/z 1241.5809 [M + Na]+; calcd for C61H90N2O23, 1241.5832), having the same number of degrees of unsaturation as 6 and 16 additional amu. After an exhaustive comparison of all 1H and 13C NMR shifts of 7 with those present in 6, only one difference was observed. Carbon C-15 shifted downfield from δC 67.9 to δC 81.0, which can only be explained by the presence of a hydroperoxyl group at C-15 instead of the hydroxyl (Figure 2 and Table S3 and Figures S13–S18).
The lobophorin family is related to kijanimicin,14,15 a spirotetronate antibiotic that is formed by a pentacyclic core bearing four l-digitoxoses and one nitrosugar. Lobophorins A (1), B (2), and E (3) have three l-digitoxose units instead of four, while lobophorin F (4) bears just two. Versipelostatin, also derived from Streptomyces sp., also has a structure very similar to the lobophorins and has been reported to activate the UPR.16,17 Although antimicrobial and antitumor activities have been reported for kijanimicin18 and lobophorins,13,19−23 herein we report the ability of lobophorins A, B, E, F, and CR1 (1–5) to activate the UPR and cell death in two human oral squamous cell carcinoma (OSCC) cell lines.
The floor of mouth OSCC cell lines UMSCC1 and UMSCC14A and nonmalignant primary human epidermal keratinocytes (nHEK) were treated with lobophorins A, B, E, F, and CR-1 (1.25–80 μM) for 24 and 48 h. Sufficient quantities of CR-2 and CR-3 were not obtained for inclusion in these analyses. Lobophorins B, E, F, and CR1 demonstrated the most potent effects on cell growth (Figure 3A and Table S4A,B). None of the lobophorins could reduce proliferation of the nonmalignant cells by 50% at 24 h and only lobophorins B and F could at 48 h. Next, attention was focused toward determining whether lobophorin F, one of the most potent congeners isolated, could induce markers of the UPR. UMSCC1, UMSCC14A, and UMSC23 cells were treated for 6 or 12 h. qRT-PCR analysis of cDNA pools generated from whole cell lysates revealed an accumulation of transcripts for CHOP, spliced XBP1 (XBP1s), GADD34, ATF3, and BiP/GRP78 (Figures 3B,C and S19). These data indicate that lobophorins activate the UPR and have significant antiproliferative selectively toward malignant cells.
Figure 3.
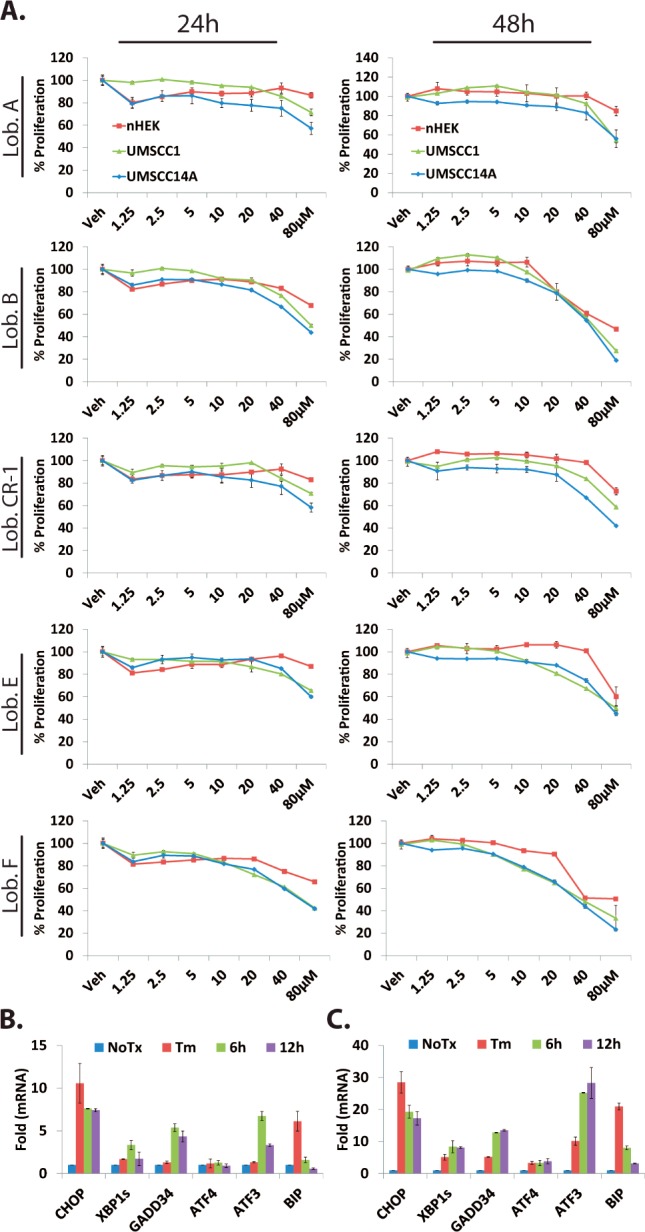
ATP-based luminescent proliferation assays with OSCC and normal keratinocytes (nHEK) treated with lobophorins. (A) Two-way ANOVA: OSCC vs nHEK, p < 0.0001 for dose and interaction (Table S4). qRT-PCR analysis of UPR transcripts in UMSCC14A (B) and UMSCC23 (C) treated with 40 μM lobophorin F 4 or 2.5 μg/mL tunicamycin (Tm).
Having observed lobophorin activated biomarkers of the UPR and inhibited OSCC growth, Atf4 −/–, Chop −/–, and eIF2-α (Ser51Ala) mutant murine embryonic fibroblasts (MEF) and wildtype littermate controls were utilized to determine at what level the UPR might regulate these effects. Atf4 null (Figure 4A) and Chop null (Figure 4B) cells were significantly and substantially protected in a dose response fashion, from lobophorin F after 16 h; Two-way ANOVA, p < 0.0001 for dose and interaction for wildtype vs null (Atf4 and Chop). Lobophorin B was not appreciably toxic in MEF cultures under any of the conditions examined (data not shown). These results indicate lobophorin F is more cytostatic than lobophorin B, and its ability to inhibit proliferation in MEF depends on intact Atf4 and Chop. Since the eIF2-α-Atf4-Chop axis is a well-characterized cornerstone of apoptotic UPR signaling, cDNA libraries generated from lobophorin F treated Atf4 null and eIF2-α mutant cells were interrogated for the accumulation of UPR and cell death transcripts. Lobophorin F-resistant Atf4-null cells failed to accumulate apoptotic mRNA transcripts for Noxa, Puma, and Trb3 after 6 h (Figure 4C). This is a strong indication that the observed reduction of proliferation in wild type MEF cells was the result of apoptosis. MEFs with an eIF2-α Ser51Ala knock-in mutation that cannot phosphorylate eIF2-α or activate Atf4 during ER stress were similarly unable to up-regulate Noxa, Puma, or Trb3 (Figure 4D). Similar results were obtained in identical experiments performed with lobophorin B (Figure S20A,B). Considered together, these data indicate that an intact eIF2-α-Atf4-Chop signaling is essential for lobophorin to induce apoptosis and inhibit MEF proliferation.
Figure 4.
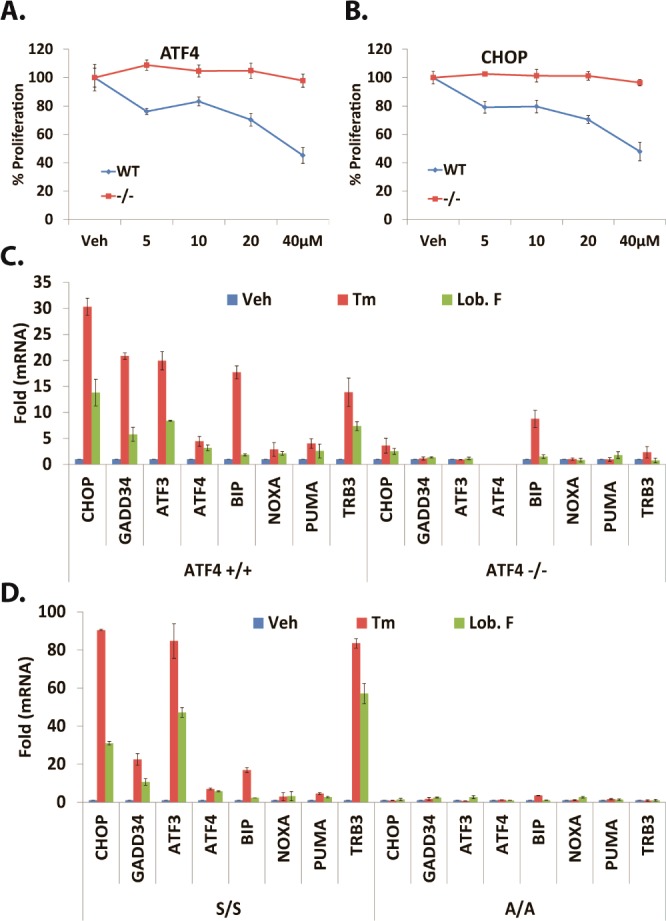
ATP-based luminescent proliferation assays with Atf4 wildtype (wt) and knockout (−/−) (A) and Chop wt and −/– (B) MEF at 16 h. qRT-PCR for UPR and apoptotic transcripts in Atf4 wt and −/– (C) or eIF2α wt (S/S) and Ser51Ala mutant (A/A) (D) in MEF 40 μM lobophorin F 4 or 2.5 μg/mL Tm 6 h.
Herein we have discussed the discovery of five congeners of lobophorin in bacterial cultures from marine sediments collected in Costa Rica. While our studies were underway two of these lobophorins were reported elsewhere and named lobophorin E and F. The three new lobophorins have been named CR 1–3 to denote they were discovered in Costa Rica. Lobophorins are structurally similar to the kijanimicin, trierixin, and versipelostatin. The ability of lobophorin, kijanimicin, and versipelostatin to inhibit cancer cell growth is known, and versipelostatin has been reported to activate the UPR. Trierixin, however, was demonstrated to inhibit markers of the UPR, and its effect on cancer cell growth is unknown.
Lobophorins E, F, and CR 1–3 were identified in a DMSO extract that could significantly activate a Chop-luciferase reporter but not an XBP1-luciferase reporter. Purified lobophorins recapitulated this finding by potently inducing CHOP gene expression and only modestly splicing XBP1. Though relatively high concentrations of lobophorins are required to activate the UPR and interfere with cancer cell growth it is encouraging that these compounds preferentially activated the apoptotic arm of the UPR. Studies currently underway are focused to increase the cultivatable lobophorin in culture and to elucidate the mechanism by which active congeners activate the UPR.
Acknowledgments
The authors are grateful to Ms. Danielle Garshott for statistical analysis of various datasets, and for assistance with the production of figures and the graphical abstract.
Glossary
ABBREVIATIONS
- ATF4
activating transcription factor 4
- CCG
Center for Chemical Genomics
- CHO
Chinese hamster ovary
- CHOP
C/EBP homologous protein
- eIF2α
eukaryotic initiation factor two-alpha
- HTS
high throughput screening
- INBIO
National Institute for Biodiversity
- ER
endoplasmic reticulum
- MEF
murine embryonic fibroblasts
- Noxa
phorbol-12-myristate-13-acetate-induced protein 1
- Puma
p53 upregulated modulator of apoptosis
- Trb3
tribbles homologue three
- UPR
unfolded protein response
- XBP1
X-box binding protein 1
Supporting Information Available
Experimental procedures for cell culture and reagents, proliferation, and polymerase chain reaction, 13C and 1H NMR data for lobophorins CR1–CR3, 1H NMR spectra of lobophorins CR1–CR3, 13C NMR spectra of lobophorins CR1–CR3, COSY spectra of lobophorins CR1–CR3, HSQC spectra of lobophorins CR1–CR3, HMBC spectra of lobophorins CR1–CR3, and ROESY spectra of lobophorins CR1–CR3. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.5b00127.
Author Present Address
# Pharma Mar, Avda. de los Reyes, 1 P.I. La Mina-Norte, Colmenar Viejo, Madrid 28770, Spain.
Author Contributions
⊥ These authors contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This research was supported by DE019678 and the Children’s Research Foundation of Michigan (to A.M.F.), Hyundai Hope on Wheels and the Detroit Country Day Men’s Lacrosse Team (to A.M.F. and M.U.C), the International Cooperative Biodiversity Groups initiative (U01 TW007404) at the Fogarty International Center (to D.H.S. and G.T.-C.), and the Hans W. Vahlteich Professorship (to D.H.S.). We thank the Technical Office, CONAGEBIO, Ministry of the Environment and Telecommunications, Costa Rica for providing sample collection permits. Portions of this work were supported by NIH grants DK042394, DK088227, and HL052173 (to R.J.K.).
The authors declare no competing financial interest.
Supplementary Material
References
- Surveillance, Epidemiology, and End Results Program. SEER Stat Fact Sheets: Oral Cavity and Pharynx, 2014.
- Molinari G. Natural products in drug discovery: present status and perspectives. Adv. Exp. Med. Biol. 2009, 655, 13–27. 10.1007/978-1-4419-1132-2_2. [DOI] [PubMed] [Google Scholar]
- Kennedy J.; Marchesi J. R.; Dobson A. D. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Fact. 2008, 7, 27. 10.1186/1475-2859-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribley A. M.; Cruz P. G.; Miller J. R.; Callaghan M. U.; Cai P.; Narula N.; Neubig R. R.; Showalter H. D.; Larsen S. D.; Kirchhoff P. D.; Larsen M. J.; Burr D. A.; Schultz P. J.; Jacobs R. R.; Tamayo-Castillo G.; Ron D.; Sherman D. H.; Kaufman R. J. Complementary cell-based high-throughput screens identify novel modulators of the unfolded protein response. J. Biomol. Screening 2011, 16, 825–835. 10.1177/1087057111414893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T.; Arnold S. M.; Miller C. N.; Wu J.; Li J.; Gunnison K. M.; Mori K.; Sadighi Akha A. A.; Raden D.; Kaufman R. J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006, 4, e374. 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.; Back S. H.; Hur J.; Lin Y. H.; Gildersleeve R.; Shan J.; Yuan C. L.; Krokowski D.; Wang S.; Hatzoglou M.; Kilberg M. S.; Sartor M. A.; Kaufman R. J. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner B. M.; Pincus D.; Gotthardt K.; Gallagher C. M.; Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harbor Perspect. Biol. 2013, 5, a013169. 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerekatte V.; Smiley K.; Hu B.; Smith A.; Gelder F.; Benedetti A. D. The proto-oncogene/translation factor eIF4E: A survey of its expression in breast carcinomas. Int. J. Cancer 1995, 64, 27–31. 10.1002/ijc.2910640107. [DOI] [PubMed] [Google Scholar]
- Franklin S.; Pho T.; Abreo F. W.; Nassar R.; Benedetti A. D.; Stucker F. J.; Nathan C.-A. O. Detection of the Proto-oncogene eIF4E in Larynx and Hypopharynx Cancers. Arch. Otolaryngol., Head Neck Surg. 1999, 125, 177. 10.1001/archotol.125.2.177. [DOI] [PubMed] [Google Scholar]
- Nathan C.-A. O.; Liu L.; Li B. D.; Abreo F. W.; Nandy I.; Benedetti A. D. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene 1997, 15, 579–584. 10.1038/sj.onc.1201216. [DOI] [PubMed] [Google Scholar]
- Fribley A. M.; Miller J. R.; Reist T. E.; Callaghan M. U.; Kaufman R. J. Large-scale analysis of UPR-mediated apoptosis in human cells. Methods Enzymol. 2011, 491, 57–71. 10.1016/B978-0-12-385928-0.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z.-D.; Jensen P. R.; Fenical W. Lobophorins A and B, new antiinflammatory macrolides produced by a tropical marine bacterium. Bioorg. Med. Chem. Lett. 1999, 9, 2003–2006. 10.1016/S0960-894X(99)00337-6. [DOI] [PubMed] [Google Scholar]
- Niu S.; Li S.; Chen Y.; Tian X.; Zhang H.; Zhang G.; Zhang W.; Yang X.; Zhang S.; Ju J.; Zhang C. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, 64, 711–716. 10.1038/ja.2011.78. [DOI] [PubMed] [Google Scholar]
- Mallams A. K.; Puar M. S.; Rossman R. R. Kijanimicin. 1. Structures of the individual sugar components. J. Am. Chem. Soc. 1981, 103, 3938–3940. 10.1021/ja00403a062. [DOI] [Google Scholar]
- Mallams A. K.; Puar M. S.; Rossman R. R.; McPhail A. T.; Macfarlane R. D. Kijanimicin. 2. Structure and absolute stereochemistry of kijanimicin. J. Am. Chem. Soc. 1981, 103, 3940–3943. 10.1021/ja00403a063. [DOI] [Google Scholar]
- Zhao P.; Ueda J. Y.; Kozone I.; Chijiwa S.; Takagi M.; Kudo F.; Nishiyama M.; Shin-ya K.; Kuzuyama T. New glycosylated derivatives of versipelostatin, the GRP78/Bip molecular chaperone down-regulator, from Streptomyces versipellis 4083-SVS6. Org. Biomol. Chem. 2009, 7, 1454–1460. 10.1039/b817312e. [DOI] [PubMed] [Google Scholar]
- Park H. R.; Chijiwa S.; Furihata K.; Hayakawa Y.; Shin-Ya K. Relative and absolute configuration of versipelostatin, a down-regulator of molecular chaperone GRP78 expression. Org. Lett. 2007, 9, 1457–1460. 10.1021/ol070042t. [DOI] [PubMed] [Google Scholar]
- Bradner W. T.; Claridge C. A.; Huftalen J. B. Antitumor activity of kijanimicin. J. Antibiot. 1983, 36, 1078–1079. 10.7164/antibiotics.36.1078. [DOI] [PubMed] [Google Scholar]
- Wei R.-B.; Xi T.; Li J.; Wang P.; Li F.-C.; Lin Y.-C.; Qin S. Lobophorin C and D, New Kijanimicin Derivatives from a Marine sponge-associated Actinomycetal strain AZS17. Mar. Drugs 2011, 9, 359–368. 10.3390/md9030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Wang J.; Guo H.; Hou W.; Yang N.; Ren B.; Liu M.; Dai H.; Liu X.; Song F.; Zhang L. Three antimycobacterial metabolites identified from a marine-derived Streptomyces sp. MS100061. Appl. Microbiol. Biotechnol. 2013, 97, 3885–3892. 10.1007/s00253-012-4681-0. [DOI] [PubMed] [Google Scholar]
- Pan H.-Q.; Zhang S.-Y.; Wang N.; Li Z.-L.; Hua H.-M.; Hu J.-C.; Wang S.-J. New Spirotetronate Antibiotics, Lobophorins H and I, from a South China Sea-Derived Streptomyces sp. 12A35. Mar. Drugs 2013, 11, 3891–3901. 10.3390/md11103891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z.; Koch M.; Pond C. D.; Mabeza G.; Seronay R. A.; Concepcion G. P.; Barrows L. R.; Olivera B. M.; Schmidt E. W. Structure and activity of lobophorins from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121–126. 10.1038/ja.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.; Zhang Q.; Zhu Y.; Li S.; Zhang G.; Zhang H.; Saurav K.; Zhang C. Characterization of the sugar-O-methyltransferase LobS1 in lobophorin biosynthesis. Appl. Microbiol. Biotechnol. 2013, 97, 9043. 10.1007/s00253-013-5083-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


