LETTER
The enteric microbiota of hospitalized patients serves as one reservoir for carbapenem-resistant Enterobacteriaceae (CRE) infections (1), although it has not been well characterized. To better understand this potential reservoir of nosocomial carbapenem-resistant organisms, we estimated the frequency of carriage of coliform bacteria harboring carbapenemase resistance genes in patient enteric flora.
We screened diarrheic stool samples submitted for Clostridium difficile testing from patients of The Ohio State University Wexner Medical Center (OSUWMC) to estimate the frequency of carriage of carbapenemase-producing enteric bacteria. Submissions (n = 692) received at the OSUWMC Clinical Diagnostic Laboratory between July and December 2013 were deidentified, aliquoted to a transport swab, and taken by courier to our laboratory. Initially, each sample swab was inoculated to MacConkey broth modified with 2 μg/ml cefotaxime, incubated overnight at 37°C, and subsequently inoculated to MacConkey agar supplemented with 2 μg/ml meropenem. Resulting lactose-positive Enterobacteriaceae isolates were tested for their ability to reduce carbapenem antimicrobials using modified Hodge and Carba NP tests. Identification to species level was performed via matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker, Billerica, MA), and carbapenemase carriage was confirmed by both PCR and whole-genome sequencing (Illumina MiSeq; San Diego, CA).
From our selective culture, 13 samples (1.9%) produced Enterobacteriaceae or Pseudomonas spp. resistant to meropenem from the 692 total stool submissions (Table 1). Of these, two Klebsiella pneumoniae isolates (0.3%) produced both positive modified Hodge and Carba NP test results. Standard PCR confirmed one K. pneumoniae isolate, CRE-185, to harbor blaKPC-3 while the second, CRE-626, carried blaNDM-1.
TABLE 1.
Enterobacteriaceae and Pseudomonas spp. resistant to carbapenems recovered from 692 patient diarrheic stool submissions received at the OSUWMC Clinical Diagnostic Laboratory for C. difficile culture between July and December 2013
| Enterobacteriaceae type and submission date (mo/day/yr) | Isolate identifier | Bacterial species | Resistance phenotypea | Cephalosporinase gene type |
|---|---|---|---|---|
| Carbapenemase producing | ||||
| 8/5/2013 | CRE-185 | Klebsiella pneumoniae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Cip Fis Gen Imi Mer Nal Pod P/T4 Xnl | KPC-3, LEN11 |
| 11/1/2013 | CRE-626 | Klebsiella pneumoniae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Fis Gen Imi Mer Nal Pod P/T4 Str Sxt Tet Xnl | NDM-1, CTX-M-15, TEM-1A, OKP-B-3 |
| Reduced susceptibility to carbapenems | ||||
| 7/8/2013 | CRE-4b | Not determined | Not determined | CTX-M group 1 |
| 7/15/2013 | CRE-53 | Klebsiella pneumoniae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Cip Fis Imi Mer Nal Pod P/T4 Str Sxt Tet Xnl | CTX-M group 1 |
| 7/24/2013 | CRE-123 | Klebsiella pneumoniae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Cip Fis Imi Mer Nal Pod P/T4 Str Sxt Tet Xnl | CTX-M group 1 |
| 7/25/2013 | CRE-126 | Enterobacter cloacae | Amp Aug2 Axo Cep Faz Fot Fox Caz Pod P/T4 Xnl | |
| 7/31/2013 | CRE-150 | Enterobacter cloacae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Imi Mer Pod P/T4 Xnl | |
| 8/7/2013 | CRE-209 | Enterobacter aerogenes | Amp Aug2 Axo Cep Faz Fot Fox Caz Imi Mer Pod P/T4 Xnl | |
| 8/8/2013 | CRE-222 | Enterobacter aerogenes | Amp Aug2 Axo Cep Faz Fot Fox Caz Pod P/T4 Xnl | |
| 8/19/2013 | CRE-266 | Enterobacter cloacae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Cip Fis Imi Mer Nal Pod P/T4 Tet Xnl | |
| 8/19/2013 | CRE-279 | Klebsiella pneumoniae | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Cip Fis Gen Mer Nal Pod P/T4 Str Sxt Tet Xnl | CMY |
| 9/16/2013 | CRE-438 | Pseudomonas aeruginosa | Amp Aug2 Axo Cep Faz Fep Fot Fox Caz Chl Fis Imi Mer Pod P/T4 Sxt Tet Xnl | CTX-M group 1 |
| 9/16/2013 | CRE-447 | Enterobacter cloacae | Amp Aug2 Axo Cep Faz Fot Fox Caz Chl Pod P/T4 Tet Xnl |
Abbreviations: Amp, ampicillin; Aug2, amoxicillin-clavulanic acid, 2:1 ratio; Axo, ceftriaxone; Cep, cephalothin; Faz, cefazolin; Fep, cefepime; Fot, cefotaxime; Fox, cefoxitin; Caz, ceftazidime; Chl, chloramphenicol; Cip, ciprofloxacin; Fis, sulfisoxazole; Gen, gentamicin; Imi, imipenem; Mer, meropenem; Nal, nalidixic acid; Pod, cefpodoxime; P/T4, piperacillin-tazobactam, constant 4; Str, streptomycin; Sxt, trimethoprim-sulfamethoxazole; Tet, tetracycline; Xnl, ceftiofur.
Following PCR confirmation of the carriage of a CTX-M group 1 gene and subsequent preservation on a nutrient slant, isolate CRE-4 became nonviable and was not available for additional characterization.
Whole-genome sequencing identified CRE-185 as multilocus sequence type (MLST) ST258 and CRE-626 as ST1602. K. pneumoniae ST258 harboring blaKPC-3 is a prominent CRE strain in the United States and worldwide (2). K. pneumoniae ST1602 isolates carrying blaNDM-1 have not been previously reported.
K. pneumoniae isolates harboring blaKPC have been previously recovered from patients of the OSUWMC by the Clinical Diagnostic Laboratory. As illustrated in the dendrogram (Fig. 1), BioNumerics (Applied Maths, Kortrijk, Belgium) analysis of pulsed-field gel electrophoresis (PFGE) banding patterns of CRE-185 and five blaKPC K. pneumoniae isolates recovered from OSUWMC patient clinical diagnostic submissions during the 2 months prior to our recovery of CRE-185 revealed highly dissimilar strains, suggesting a diverse reservoir.
FIG 1.
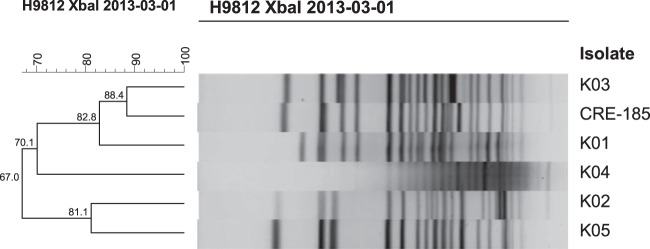
Dendrographic percent relatedness of K. pneumoniae CRE-185 recovered from an OSUWMC patient stool sample and 5 blaKPC-bearing K. pneumoniae isolates recovered from OSUWMC patient clinical diagnostic submissions during the same month. After electrophoresis, banding patterns were compared and levels of similarity were assigned using generally accepted criteria (7). blaKPC-bearing K. pneumoniae isolates were assessed using the Dice coefficient similarity index and the unweighted pair-group method with arithmetic averages with clustering settings of 1.00% optimization and 1.00% band position tolerance via BioNumerics software (Applied Maths, Kortrijk, Belgium).
In addition to blaKPC-3, K. pneumoniae CRE-185 carried a second β-lactam resistance gene, blaLEN-11. Multiple aminoglycoside and quinolone resistance genes and single fosfomycin, phenicol, and sulfonamide resistance genes were also detected. Four plasmid replicon types were identified—FIB(K), FII(K), R, and ColE. K. pneumoniae ST258 isolates with similar plasmid content and harboring blaKPC-3 have been previously recovered in Italy (3).
Sequencing of CRE-626 harboring blaNDM-1 identified 3 additional β-lactam resistance genes—blaCTX-M-15, blaTEM-1A, and blaOKP-B-3. Multiple aminoglycoside and sulfonamide resistance genes were detected as well as individual genes conveying resistance to quinolones, tetracycline, and trimethoprim. CRE-626 carried the NDM-MAR plasmid, which represents incompatibility groups FIB(Mar) and HI1B. This plasmid with a highly similar resistance genotype was originally identified in K. pneumoniae recovered from hospitalized patients in Morocco and has been subsequently fully sequenced (4). Additionally, CRE-626 carried incompatibility group FIA(HI1), FII(K), and ColE plasmids.
We did not detect carbapenemase production by the remaining 11 meropenem-resistant isolates. This subset included 4 Enterobacter cloacae, 3 K. pneumoniae, 2 Enterobacter aerogenes, and 1 Pseudomonas aeruginosa isolate and a single isolate not identified to species level that could not be recovered from storage. Carbapenem resistance in these isolates may be due to several factors, including increased expression of efflux systems, reduced porin expression, increased chromosomal cephalosporinase activity, or some combination of these (5).
Although we know very little about the deidentified patient population from which these samples were obtained, it is reasonable to assume that these carbapenem-resistant isolates were recovered from antimicrobial-associated diarrhea cases because it is standard practice at the OSUWMC to test stool specimens by PCR for opportunistic C. difficile infection in patients who develop diarrhea while receiving antibiotics. Our results indicate that while the prevalence of CRE is very low in patient fecal flora, even in this high-risk population, the threat of nosocomial CRE infections disseminated from an enteric flora reservoir exists in health care settings.
blaNDM-1 has been detected in a clinical diagnostic submission to the OSUWMC Clinical Diagnostic Laboratory only once subsequent to our study (K. pneumoniae, December 2014), from a patient referred to the OSUWMC who likely acquired the infection prior to admission. Our detection of carbapenemase-producing enteric bacteria in this population emphasizes the need for CRE surveillance and patient risk assessment in order to prevent the nosocomial dissemination of this important resistance genotype (6).
ACKNOWLEDGMENT
This work was supported in part by the USDA National Institute of Food and Agriculture, AFRI project 1000839.
REFERENCES
- 1.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 2.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A. 2012. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother 67:1645–1650. doi: 10.1093/jac/dks114. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]


