LETTER
IncL/M plasmids identified recently in clinical and environmental isolates are characterized by insertions of one or more complex multidrug resistance regions (MRRs) (1–3). Here, we analyzed the sequence of plasmid pIMP-HB623, which has a MRR with novel structural features and cocarries the carbapenemase blaIMP-34 and the fosfomycin resistance determinant fosC2. In 2011, we identified a multidrug-resistant Enterobacter cloacae strain, strain CRE623, in an isolate from a rectal swab of a 76-year-old man who was repatriated from Harbin, China, where he had been hospitalized for intracranial hemorrhage (see Table S1 in the supplemental material). PCR and sequencing using previously described primers showed that the strain was positive for blaIMP-34, fosC2, and aacC2 genes (4–6).
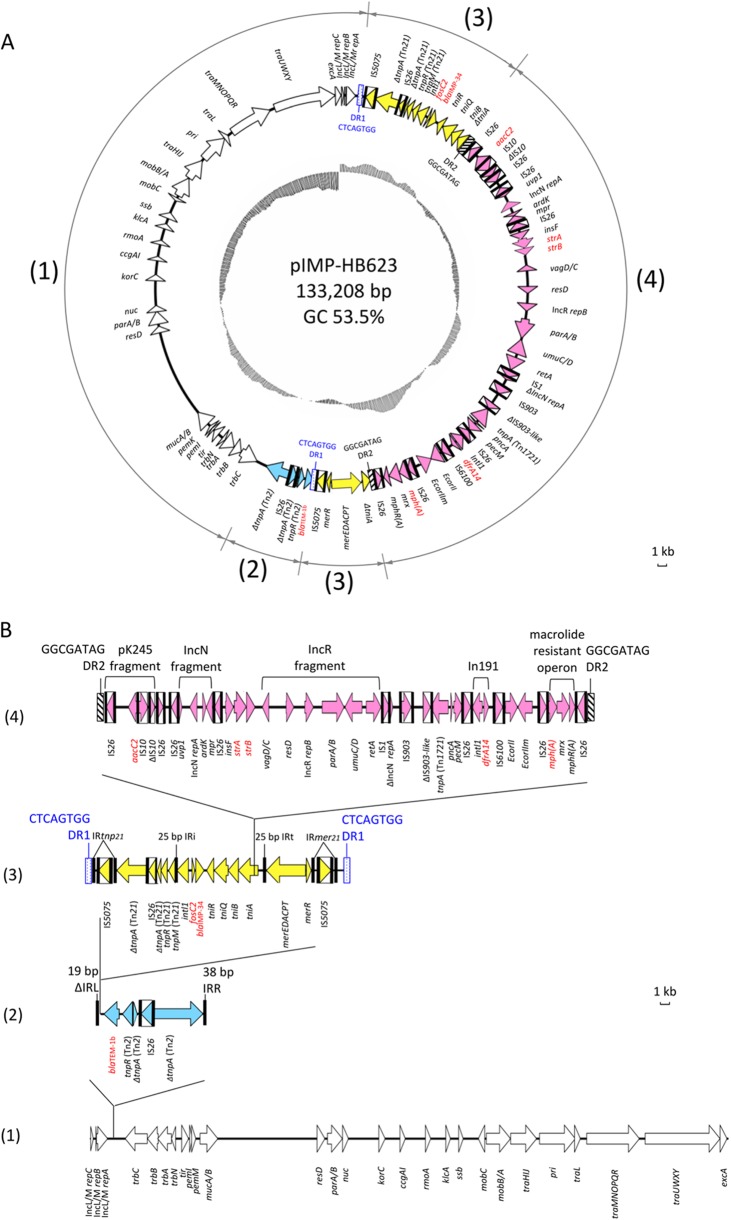
In filter mating (2), the blaIMP-34-carrying plasmid could be transferred to the J53 recipient at a frequency of 10−1 per donor cell. Plasmid DNA from the transconjugant was sequenced at approximately 150-fold coverage using a 454 GS-FLX system (4). The reads were assembled using a GS de novo Assembler (version 2.6) into 24 contigs, and the gaps were closed by additional PCR and Sanger sequencing (see Table S2 in the supplemental material). Plasmid pIMP-HB623 (GenBank accession no. KM877517) was 133,208 bp in size, comprising a 60,166-bp backbone and a 73,042-bp MRR, which was inserted into an integration hot spot between repA and trbC in IncL/M (Fig. 1). The plasmid backbone of pIMP-HB623 differs from those of pEL1573 (JX101693) and pCTX-M3 (AF550415) by only 3 and 56 nucleotides, respectively (see Table S3). These nucleotide variations were confirmed by Sanger sequencing.
FIG 1.
(A) Circular map of pIMP-HB623. Open reading frames (ORFs) are colored coded, with the direction of transcription indicated by arrowheads. ORFs with unknown function are not shown. Insertion sequence (IS) elements are represented by boxes, with the arrows inside the boxes indicating the transposases. Black bars are used to indicate inverted repeats associated with IS elements (IS1, IS3, IS10, IS26, IS903, IS5075, and IS6100), Tn2 (ΔIRL, IRR), Tn21 family transposons (IRtmp21, IRmer21), and integrons (IRi and IRt). Duplication repeats are represented by rectangular boxes (DR1, blue and dotted boxes; DR2, black and hatched boxes). Resistance genes are indicated in red font. The numbers in the outer circle indicate the following regions: (1) IncL/M plasmid scaffold (white arrows), (2) Tn2-based module (blue arrows), (3) Tn5075 module (yellow arrows), and (4) IS26 transposon-like structure (pink arrows). The G+C plot is indicated in the inner circle (mean, 53.5%). (B) Linear map of pIMP-HB623 showing the putative insertion events that have contributed to the formation of the multidrug-resistant region. The modules are identified, and other features are represented as described for panel A. Scale bars show the sequence length for 1 kb. For the purpose of illustration, duplication repeats and inverted repeats are not drawn to scale.
The MRR had probably resulted from the integration of several mobile elements, giving rise to a complex structure with multiple resistance determinants. First, it has a Tn2 carrying blaTEM-1b (Fig. 1), as identified previously in several other IncL/M plasmids (pCTX-M360, pCTX-M3, pNDM-OM, pNDM-HK, and pEl1573). Second, there is a Tn21 backbone with copies of IS5075 inserted into inverted repeats (IRs) IRtnp21 and IRmer21 (7). The insertion of IS5075 should have occurred subsequent to Tn21 inserting into Tn2, as the interruption of the IR by IS5075 prevents further mobilization. A novel class 1 integron carrying fosC2 and blaIMP-34 (designated In1070 by INTEGRALL [http://integrall.bio.ua.pt/]) with a complete set of tni modules (tniR, tniQ, tniB, and tniA) and the mer operon were identified within Tn5075. This class 1 integron was bound within two Tn402 inverted repeats of 25 bp, IRi and IRt (as identified in GQ857074), while the tniA gene was split by the insertion of a 47.6-kb sequence with IS26 termini, flanked by 8-bp direct repeats (GGCGATAG). The 47.6-kb insertion includes multiple copies of IS26 and other mobile elements, suggesting that it was formed by multiple genetic events. Modular sequences that could be identified within this region include a 5.6-kb fragment carrying aacC2 (positions 15174 to 20828, with 100% identity to pK245) (DQ449578), a 3.3-kb fragment matching the IncN backbone (positions 22,402 to 25,699, with 100% identity to pHKU1) (KC960485), aminoglycoside resistance genes (strA and strB), a matching 12.8-kb fragment (positions 29621 to 42455, with 99.4% identity to pKP1780) (JX424614), a partial class 1 integron (In191) carrying dfrA14, and a macrolide resistance operon. In conclusion, this is the first report of the cocarriage of blaIMP and a fosfomycin resistance gene. It further highlights the importance of IncL/M plasmids as vectors for emerging resistance genes.
Nucleotide sequence accession number.
Sequence data from plasmid pIMP-HB623 have been deposited in GenBank under accession no. KM877517.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Health and Medical Research Fund of the Health and Food Bureau of the government of the Hong Kong Special Administrative Region (HKSAR).
We thank Chi-Ho Lin, Levina Lam, Ben Ho, and Wilson Chan of the Centre for Genomic Science, The University of Hong Kong, for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00375-15.
REFERENCES
- 1.Partridge SR, Ginn AN, Paulsen IT, Iredell JR. 2012. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho PL, Lo WU, Chan J, Cheung YY, Chow KH, Yam WC, Lin CH, Que TL. 2014. pIMP-PH114 carrying blaIMP-4 in a Klebsiella pneumoniae strain is closely related to other multidrug-resistant IncA/C2 plasmids. Curr Microbiol 68:227–232. doi: 10.1007/s00284-013-0471-x. [DOI] [PubMed] [Google Scholar]
- 5.Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, Chow KH. 2013. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol 62:1707–1713. doi: 10.1099/jmm.0.062653-0. [DOI] [PubMed] [Google Scholar]
- 6.Ho PL, Wong RC, Lo SW, Chow KH, Wong SS, Que TL. 2010. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J Med Microbiol 59:702–707. doi: 10.1099/jmm.0.015032-0. [DOI] [PubMed] [Google Scholar]
- 7.Partridge SR, Hall RM. 2003. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol 185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.