Abstract
Bypass of the d,d-transpeptidase activity of penicillin-binding proteins by an l,d-transpeptidase (Ldtfm) results in resistance to ampicillin and glycopeptides in Enterococcus faecium M9, a mutant obtained by nine consecutive selection steps. Resistance requires activation of a cryptic locus for production of the essential tetrapeptide-containing substrate of Ldtfm and impaired activity of protein phosphatase StpA. Here, whole-genome sequencing revealed a high mutation rate for the entire selection procedure (79 mutations in 900 generations). Acquisition of a mutation in the mismatch repair gene mutL had little impact on the frequency of rifampin-resistant mutants although the mutation spectrum of M9 was typical of impaired MutL with high transversion to transition (40/11) and substitution to deletion (51/28) ratios. M9 did not mainly accumulate neutral mutations since base substitutions occurred more frequently in coding sequences than expected (χ2 = 5.0; P < 0.05) and silent mutations were underrepresented (χ2 = 5.72; P < 0.02). None of the mutations directly affected recognition of the tetrapeptide substrate of Ldtfm by peptidoglycan synthesis enzymes. Instead, mutations appear to remodel regulatory circuits involving two-component regulatory systems and sugar metabolism. The high number of mutations required for activation of the l,d-transpeptidase pathway may strongly limit emergence of cross-resistance to ampicillin and glycopeptides by this mechanism.
INTRODUCTION
Acquisition of resistance to antibiotics is mediated by four mechanisms, including drug detoxification, efflux and decreased cell wall permeability, decreased affinity of the target for the drug, and bypass of the target. For β-lactams, detoxification of the antibiotics by β-lactamases is widespread in nearly all bacterial phyla. In Gram-negative bacteria, β-lactamase production is frequently associated with reduced permeability of the outer membrane and efflux. In the Firmicutes, this permeability barrier does not exist, and resistance is often due to production of targets displaying a lower affinity for the drug following horizontal gene transfer or acquisition of mutations. The remaining mechanism, bypass of the target, has been identified for the first time in mutants of Enterococcus faecium selected for their resistance to ampicillin in laboratory conditions (1). In these mutants, the classical targets of β-lactams, the high-molecular-weight penicillin-binding proteins (PBPs) (2), are replaced by an l,d-transpeptidase (LDT) (3), which catalyzes the essential cross-linking step of peptidoglycan synthesis.
PBPs and LDTs are structurally unrelated and catalyze formation of peptidoglycan cross-links by distinct catalytic mechanisms involving active-site serine and cysteine residues, respectively (3–5). In the first step of the cross-linking reaction, PBPs react with an acyl donor containing a stem pentapeptide, which displays the sequence l-Ala1-γ-d-Gln2-l-Lys3(d-iAsx)-d-Ala4-d-Ala5 in E. faecium. The enzymes cleave the d-Ala4-d-Ala5 peptide bond, hence the d,d designation of the transpeptidases, and form an ester bond between the carbonyl of d-Ala4 and the active site serine. In the following step, the carbonyl of the resulting acyl enzyme is attacked by the nucleophilic α-amino group of d-iso-asparagine or D-iAsx in the side chain of the second substrate, the acyl acceptor. This results in the formation of 4→3 cross-links (d-Ala4→d-iAsx-l-Lys3) that connect the 4th and 3rd positions of stem peptides located on adjacent glycan chains. In contrast, LDTs use acyl donors containing a tetrapeptide stem (Fig. 1A). E. faecium produces a single l,d-transpeptidase (Ldtfm), which cleaves the l-Lys3-d-Ala4 peptide bond (l,d-designation) and forms 3→3 cross-links (l-Lys3→d-iAsx-l-Lys3). Since LDTs are not inactivated by ampicillin (6, 7), the l,d-transpeptidation pathway conveys high-level resistance to this antibiotic with a MIC of >1,000 μg/ml.
FIG 1.
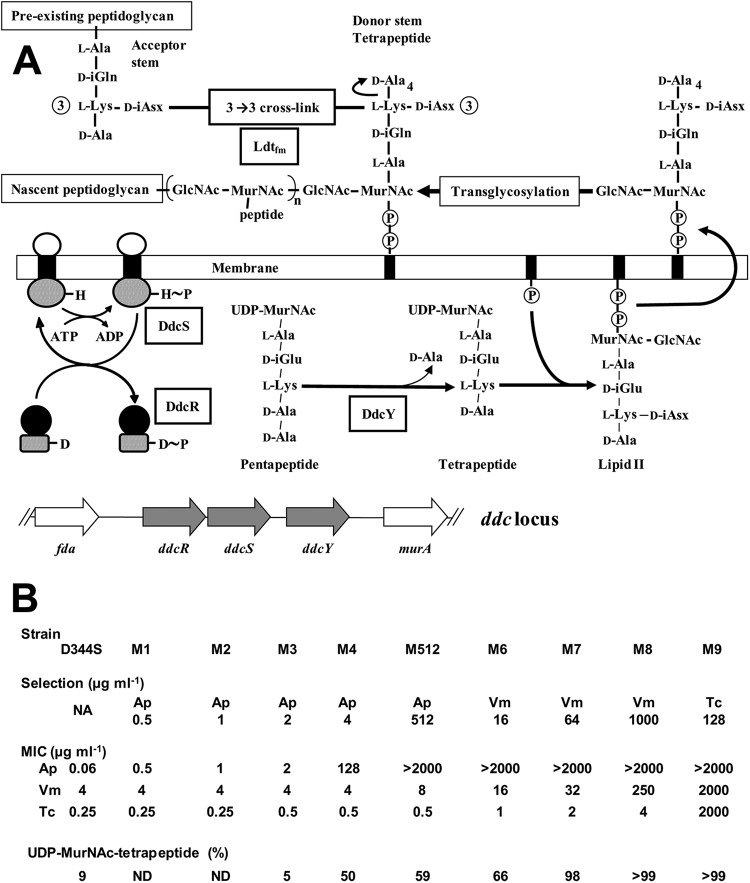
Activation of the l,d-transpeptidation pathway in response to ampicillin and glycopeptides in Enterococcus faecium. (A) Map of the ddc locus and role of the encoded proteins in peptidoglycan synthesis. The black boxes represent the undecaprenyl lipid transporter inserted into the cytoplasmic membrane. Phosphates are indicated by circled Ps. Transcriptional activation by DdcRS involves autophosphorylation of DdcS on a His residue (H∼P) and subsequent transfer of the phosphate group onto an Asp residue of DdcR (D∼P). Peptidoglycan cross-linking by l,d-transpeptidase Ldtfm involves cleavage of the l-Lys3-d-Ala4 peptide bond of a stem tetrapeptide (acyl donor) and formation of an amide bond between the carbonyl of l-Lys3 and the α-amino group of d-iAsn of the acyl acceptor. (B) Characteristics of the parental strain D344S and mutants M1 to M9. Serial selection was performed with ampicillin (Ap), vancomycin (Vm), or teicoplanin (Tc) at the indicated concentrations. The resulting increases in MICs for the three drugs are indicated. Selection for resistance resulted in increased UDP-MurNAc-tetrapeptide (shown as a percentage of the total cytoplasmic pool of precursors) to the detriment of UDP-MurNAc-pentapeptide following hydrolysis of d-Ala5 by d,d-carboxypeptidase DdcY. NA, not applicable.
Activation of the l,d-transpeptidation pathway has been obtained in a strain of E. faecium, D344S, deficient for production of low-affinity PBP5 following spontaneous deletion of the corresponding chromosomal gene (1, 8). Starting with this hypersusceptible strain (MIC of ampicillin, 0.06 μg/ml), five selection steps on increasing concentrations of ampicillin were required to obtain highly resistant mutant M512 (MIC, >1,000 μg/ml) (Fig. 1B) (8). The selection procedure did not result in any modification in the sequence or level of production of the E. faecium Ldtfm (9). The enzyme was active in the parental strain but only contributed to formation of 3% of the cross-links due to limited amounts of its essential tetrapeptide-containing donor substrate. Bypass of PBPs by Ldtfm required activation of a cryptic locus, ddc, which is only present in ca. 20% of E. faecium clinical isolates. The ddc locus encodes a metallo-d,d-carboxypeptidase, DdcY, related to VanY encoded by the vanA vancomycin resistance gene cluster of transposon Tn1546 (10). DdcY generates a stem tetrapeptide by hydrolysis of the d-Ala4-d-Ala5 peptide bond of the cytoplasmic peptidoglycan precursor UDP-N-acetylmuramoyl (MurNAc)-l-Ala1-γ-d-Glu2-l-Lys3-d-Ala4-d-Ala5 (UDP-MurNAc-pentapeptide) (9). The ddc locus also encodes a two-component regulatory system composed of a membrane-associated sensor kinase (DdcS) and a response regulator belonging to the OmpR-PhoB family (DdcR) (9).
Activation of the ddc locus in mutant M512 results from a mutation in gene ddcS, which impairs the phosphatase activity of the sensor kinase and leads to production of DdcR, DdcS, and DdcY (9). This mutation was detected at the 5th selection step that provided mutant M512 from mutant M4 (Fig. 1). A second mutation was detected in the protein phosphatase gene stpA of mutant M1 located upstream from the Ser/Thr protein kinase gene stk (11). The stpA mutation of M1 impairs the phosphatase activity of StpA, leading to hyperphosphorylation of Stk and several unidentified proteins (11). Genetic analyses showed that impaired phosphatase activities of DdcS and StpA are both required and sufficient for high-level ampicillin resistance (11).
The glycopeptide antibiotics vancomycin and teicoplanin bind to the peptidyl-d-Ala4-d-Ala5 extremity of peptidoglycan precursors and inhibit by steric hindrance both the elongation of glycan chains by glycosyltransferases and cross-linking of stem peptides by d,d-transpeptidases. LDTs use acyl donors containing a stem tetrapeptide ending in d-Ala4 that do not bind the drugs (12, 13). However, mutant M512 remained susceptible to glycopeptides in spite of production of tetrapeptide stems and bypass of PBPs by Ldtfm (12). Four additional selection steps on increasing concentrations of vancomycin or teicoplanin were required to obtain mutant M9 (12), which was coresistant to ampicillin and glycopeptides (MICs of all three drugs, >1,000) (Fig. 1B). Sequencing of the ddc locus revealed two mutations in ddcY that enhanced the d,d-carboxypeptidase activity of DdcY and affected the membrane anchor of the protein (9). These modifications led to full replacement of UDP-MurNAc-pentapeptide by UDP-MurNAc-tetrapeptide in the cytoplasm, and, consequently, to the exclusive formation of 3→3 cross-links by Ldtfm (9, 12) (Fig. 1B). Full elimination of precursors ending in d-Ala4-d-Ala5 is required to prevent inhibition of the transglycosylation reaction by glycopeptides (12, 14). In this study, whole-genome sequencing was performed to identify the complete set of mutations acquired by derivatives of E. faecium D344S in response to selection by ampicillin and glycopeptides.
MATERIALS AND METHODS
Estimate of the number of generations associated with selection of ampicillin- and glycopeptide-resistant derivatives of D344S.
A total of nine selection steps were required to obtain mutant M9 (Fig. 1B) (1, 12). For each selection step, we used one colony rather than a fraction of the population. Briefly, for the 1st subculture, bacteria were streaked for isolated colonies on brain heart infusion (BHI) agar containing approximately one-eighth of the drug concentration used for the previous selection step (27 generations). Specifically, ampicillin was used at concentrations of 0.0625, 0.125, 0.25, 1, and 32 μg/ml for M1, M2, M3, M4, and M512, respectively. Vancomycin was used at concentrations of 2, 4, and 64 μg/ml for M6, M7, and M8, respectively. Teicoplanin was used at 32 μg/ml for M9. For the 2nd subculture, one colony was inoculated in 10 ml of BHI broth containing the same drug concentration and grown to saturation (10 generations). In the 3rd subculture, bacteria were plated on 2-fold increasing inhibitory concentrations of the drugs (27 generations). In the 4th subculture, colonies of candidate mutants were streaked for isolated colonies on BHI agar containing one-eighth the drug concentration used for the selection (27 generations). In the 5th subculture, one colony was inoculated in 10 ml of BHI broth containing the same drug concentration and grown to saturation (10 generations). This culture was used to perform the next selection step and to conserve the mutants at −80°C. Thus, each selection step involved five subcultures and ca. 100 generations.
Frequency of mutants resistant to rifampin.
E. faecium D344S and mutants M1 to M9 were grown overnight in BHI broth, and appropriate dilutions were plated on BHI agar and on BHI agar containing 20 μg/ml of rifampin. CFU were enumerated after 72 h of incubation at 37°C, and the frequency of rifampin-resistant mutants was expressed as the ratio of the number of CFU detected on the rifampin-containing medium to the number of CFU detected on the antibiotic-free medium. The Mann-Whitney U test was used to compare frequencies with a value of P <0.05 for the threshold of significance. The same procedure was used to obtain the rifampin-resistant mutants that were characterized by genome sequencing except that plates were incubated for 1, 2, 3, or 6 days.
Whole-genome sequencing.
The Illumina single reads sequencing technology was used for sequencing. Illumina library preparation (genomic DNA sample prep kit v1) and sequencing followed standard protocols developed by the supplier. Briefly, genomic DNA was shared by nebulization, and sheared fragments were end repaired and phosphorylated. Blunt-end fragments were A tailed, and sequencing adapters were ligated to the fragments. Fragments with an insert size of around 200 bp were gel extracted and enriched with 14 cycles of PCR before library quantification and validation. Hybridization of the library to the flow cell and bridge amplification were performed to generate clusters. Single reads of 36 cycles were collected on a GAIIX (Illumina, San Diego, CA). After sequencing was complete, image analysis, base calling, and error estimation were performed using Illumina Analysis Pipeline v1.6.
Raw sequences files were filtered using programs developed by N. Joly (Biology IT Center, Institut Pasteur, Paris). Quality-filtered trimmed reads were mapped on the genome sequence of E. faecium D344SRF (GenBank accession no. ACZZ00000000), a derivative D344S resistant to rifampin and fusidic acid. Variant detection was done with CLC Genomics Workbench version 5 (CLC Bio, Denmark). Genome sequencing was performed on strain D344S issued from our laboratory collection, in addition to that of mutant M9, to eliminate differences between the reference genome of D344SRF and the parental strain D344S. The parameters used for the detection of variants are reported in Table 1. Assignment of proteins to functional categories was based on queries in the Pfam database.
TABLE 1.
Parameters for detection and mapping of variants
| Parameter | Strain |
|
|---|---|---|
| D344S | M9 | |
| No. of trimmed readsa | 9,202,931 | 8,576,759 |
| Unmapped reads (%)a | 7.06 | 3.89 |
| Mapped reads (%)a | 92.94% | 96.11 |
| Average coveragea | 76 | 71 |
| Minimum coverage | 10 | 10 |
| Maximum expected variantsb | 2 | 2 |
| Ignore quality scoresb | No | No |
| Ignore nonspecific matchesb | Yes | Yes |
| Variant probabilityb | 80.0 | 80.0 |
| Require presence in both forward and reverse readsb | No | No |
Mapping program clc_mapper version 4.10 (CLC Bio).
Detection of variants with CLC Genomics Workbench version 5. The genetic code 11 (bacterial and plant plastid) was used.
Phosphatase activity of StpA.
Recombinant StpA containing a C-terminal 6-histidine tag was produced in Escherichia coli BL21(DE3) and purified by nickel affinity and size exclusion chromatography methods, as previously described (11). Hydrolysis of para-nitrophenyl-phosphate (Sigma) by StpA was determined at 37°C in 50 mM Tris-HCl (pH 8.0). Reactions were initiated by the addition of MnCl2 at 50 μM or 2 mM, and the absorbance was monitored at 405 nm (ε of 12,500 M−1 cm−1).
Sequence accession numbers.
Sequence data have been deposited in the BioProject NCBI database under study accession number SRP058288 with BioSample accession numbers SAMN03654371 and SAMN03766829 for strain D344S and mutant M9, respectively.
RESULTS AND DISCUSSION
Sequencing of the genome of mutant M9.
The genome of mutant M9 differed from that of the parental strain E. faecium D344S by a total of 79 mutations. Sanger sequencing was performed to confirm the presence of the 79 mutations in M9 and to assign each of the mutations to one of the nine selection steps used to obtain mutant M9 (Table 2). The number of mutations acquired in individual selection steps ranged from 4 to 15 with an average of 8.8 (Fig. 2). Mutations were stably inherited with three exceptions involving the stpA and ddcR genes and a putative NADPH-dependent flavin mononucleotide (FMN) reductase gene (EDAG_01903), as described below. Mutant M1 acquired a mutation leading to a T101R substitution in StpA. However, the corresponding codon seemingly reverted to the wild-type sequence in the following step. Mutants M2 to M9 harbored another stpA mutation that led to an A35V substitution. Purification of StpA derivatives harboring T101R and A35V indicated that both substitutions impaired the phosphatase activity of the protein (Table 3). The impact of A35V, present in M2 to M9, was less than that of T101R, present in M1, suggesting that residual activity of StpA A35V might confer a selective advantage. For the ddc locus, mutant M4 acquired a mutation leading to an M55I substitution in DdcR. M512 harbored a wild-type codon at this position and acquired another mutation, leading to a T161A substitution in sensor kinase DdcS. This mutation was subsequently inherited by mutants M6 to M9. The ddc locus was activated to a similar extent in both mutants (9). Finally, the mutation in EDAG_01903 was detected in mutants M2, M512, M6, M7, M8, and M9 but not in M3 and M4. The exact sequence of events leading to reversion of the initial mutation and acquisition of a secondary mutation in these loci remain unknown. The simplest interpretation is that reversion of the initial mutation occurred, but it is not possible to exclude the possibility that the two alleles coexisted within a bacterial population or within a clone following transient gene duplication. However, such polymorphisms were not retrospectively detected in the subcultures conserved at −80°C (data not shown).
TABLE 2.
Mutations detected in mutants M1 to M9
| Mutant | EDAGa | Mutation | Predicted impact of the mutation |
|---|---|---|---|
| M1 | 00077 | C302G | T101R substitution in Ser Thr protein phosphatase StpA |
| 01693 | G5A | S2N substitution in ribosome maturation factor RimP | |
| 00369 | C623T | A208V substitution in signal recognition particle-docking protein FtsY | |
| 00670 | G979T | D327Y substitution in mannitol operon transcriptional antiterminator | |
| 01067 | C34T | R12C substitution in glyceraldehyde-3-phosphate dehydrogenase, type I | |
| 01287 | C486T | Silent mutation (member of the xanthine/uracil permease family) | |
| A61T | Intergenic, 61 bp upstream from EDAG_00488 | ||
| M2 | 02370 | C404A | A135E substitution in response regulator CroR |
| 00077 | C104T | A35V substitution in serine/threonine protein phosphatase StpA | |
| 00473 | C1616A | T539K substitution in PBPB | |
| 00961 | ΔA1726 | Truncation of DNA mismatch repair protein MutL | |
| 01903 | ΔA13 | Truncation of a putative NADPH-dependent FMN reductase | |
| ΔA360 | Intergenic, 360 bp downstream from EDAG_01362 | ||
| M3 | 00792 | A956G | Q319R substitution in a hypothetical protein |
| 02385 | A253G | T85A substitution in a hypothetical protein | |
| 01714 | A316G | N106D substitution in substrate-binding protein of a putative multiple sugar ABC transporter | |
| 00246 | iA158 | Truncation of a transcriptional regulator of the MarR family | |
| 00858 | ΔA19 | Truncation of glutamine-binding protein GlnH (ABC transporter) | |
| M4 | 00953 | C47T | P16L substitution in a putative phosphotyrosine protein phosphatase |
| 00202 | T542C | V181A substitution in 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase | |
| 01377 | G559A | D187N substitution in a putative DNA-directed DNA polymerase | |
| 00066 | iA511 | Truncation of a hypothetical protein | |
| 01651 | ΔA837 | Truncation of copper-translocating P-type ATPase | |
| 01550 | G165A | M55I substitution in response regulator DdcR | |
| M512 | 01551 | A481G | T161A substitution in sensor kinase DdcS |
| 01065 | T102C | Silent mutation in RNA polymerase sigma factor 54 gene | |
| 01143 | C97T | R33C substitution in ribosomal protein S21 | |
| 01730 | C98T | A33V substitution in a putative phage DNA packaging protein | |
| 00694 | C1498T | Truncation of a putative calcium-transporting ATPase | |
| 02113 | A1377G | Silent mutation (large subunit of a putative phage terminase) | |
| 01186 | A547G | S183G substitution in a putative sensor kinase | |
| 00615 | A778G | T260A substitution in RNase HIII | |
| 00746 | A549G | Silent mutation (putative sensor kinase) | |
| 01598 | C74A | P25H substitution in a phosphoesterase belonging to the DHH family | |
| 00511 | A530G | D177G substitution in glucose-1-phosphate thymidylyltransferase | |
| 00961 | ΔA661 | Truncation of DNA mismatch repair protein MutL | |
| 00469 | iG147 | Truncation of a putative lipoprotein protein | |
| 00089 | ΔA24 | Truncation of l-lactate dehydrogenase | |
| 00626 | ΔA1994 | Truncation of a putative V-type ATPase | |
| M6 | 00718 | ΔA178 | Truncation of a hypothetical protein |
| 01553 | G379A | E127K substitution in the catalytic domain of DdcY | |
| 00728 | C1556T | A519V substitution in ATP-dependent DNA helicase IV | |
| 01753 | T623C | L208S substitution in a hypothetical protein | |
| 01577 | C2248T | P750S substitution in valyl-tRNA synthetase | |
| 02299 | A29G | H10R substitution in a hypothetical protein | |
| 02038 | T159C | Silent mutation (hypothetical ATP/GTP-binding protein) | |
| 01180 | ΔA463 | Truncation of a putative member of the YitT family | |
| 00623 | ΔA518 | Truncation of fructose-1,6-bisphosphatase | |
| 02047 | ΔA1033 | Truncation of a hypothetical PTS system transcriptional activator | |
| 01858 | ΔT20 | Truncation of a hypothetical protein | |
| M7 | 02391 | C70T | Truncation of a putative sodium/dicarboxylate symporter |
| 00183 | C482T | P161L substitution in a hypothetical protein | |
| 01207 | A517G | T173A substitution in a hypothetical protein | |
| 01042 | G901A | A301T substitution in a sensor histidine kinase | |
| 01754 | T86C | V29A substitution in membrane-bound protein LytR | |
| 00781 | ΔA482 | Truncation of a hypothetical protein | |
| 01454 | ΔA234 | Truncation of a member of a Spx/MgsR transcriptional regulator family | |
| ΔA32 | Intergenic, 32 bp downstream from EDAG_01142 | ||
| ΔT101 | Intergenic, 101 bp downstream from EDAG_05560 | ||
| ΔA147 | Intergenic, 147 bp downstream from EDAG_00488 | ||
| M8 | 00248 | G139C | D47H substitution in acyl carrier protein |
| 00719 | G1461A | Truncation of glycerol kinase | |
| 02004 | ΔA114 | Truncation of ferrous iron transporter B | |
| 02466 | ΔA9 | Truncation of d,d-carboxypeptidase DdcP | |
| M9 | 01136 | G169A | G57R substitution in a putative alpha-galactosidase |
| 02125 | A278T | H93L substitution in a hypothetical protein | |
| 00476 | G592A | A198T substitution in transferase MurG | |
| 00308 | C342T | Silent mutation in permease OpuCB gene | |
| 00549 | C419T | T140I substitution in DNA topoisomerase III | |
| 02412 | T476A | I159N substitution in phosphate-binding protein PstS | |
| 02124 | ΔT1893 | Truncation of phage tail tape measure protein | |
| 01553 | T41A | I14N substitution in the membrane anchor of DdcY | |
| 00606 | ΔA297 | Truncation of a hypothetical protein | |
| 01532 | ΔA117 | Truncation of a hypothetical RNA-binding | |
| 01554 | C614T | A205V substitution in MurAB | |
| 01561 | ΔA290 | Truncation of beta-glucoside specific transporter | |
| 00718 | A414T | Silent mutation (conserved hypothetical protein) | |
| C122T | Intergenic, 122 bp downstream from EDAG_02398 | ||
| ΔT262 | Intergenic, 262 bp downstream from EDAG_00339 |
EDAG number as appearing in the D344SRF genome database at http://www.broadinstitute.org/annotation/genome/enterococcus_rice/MultiHome.html.
FIG 2.
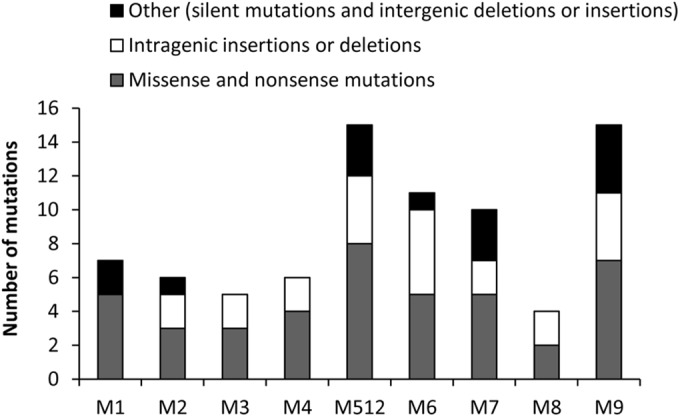
Number and spectrum of mutations acquired by mutants M1 to M9.
TABLE 3.
Catalytic constants for para-nitrophenyl-phosphate hydrolysis by StpA from parental strain D344S and ampicillin resistant mutants M1 and M2
| Substitution in StpA (strain) | Catalytic constants determined in the presence of Mn2+ ata: |
|||||
|---|---|---|---|---|---|---|
| 2 mM |
50 μM |
|||||
| Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) | |
| None (D344S) | 0.66 ± 0.09 | 1,200 ± 100 | 1,800 ± 300 | 0.69 ± 0.17 | 12 ± 1 | 17 ± 5 |
| T101R (M1) | 1.2 ± 0.2 | 0.83 ± 0.04 | 0.69 ± 0.12 | NDb | ND | ND |
| A35V (M2) | 1.0 ± 0.4 | 350 ± 40 | 350 ± 140 | 0.67 ± 0.28 | 0.22 ± 0.02 | 0.33 ± 0.14 |
Values ± SE of regression were obtained by fitting experimental data to the Michaelis-Menten equation, V = kcat ES/(Km + S), where V is the initial velocity and E and S are the initial enzyme and substrate concentrations, respectively.
ND, not determined.
Mutation rate associated with activation of the l,d-transpeptidase pathway under the selective pressure of ampicillin and glycopeptides.
In Escherichia coli, a rate of 2.2 × 10−10 mutations per nucleotide per generation has recently been reported for neutral mutations based on whole-genome sequencing of 59 bacterial lines evolved for a total of 2.5 × 105 generations (15). Estimates of the mutation rate obtained by scoring for resistance to rifampin or nalidixic acid in classical fluctuation tests were lower (0.33 and 0.21 × 10−10 mutations per nucleotide per generation, respectively). Since ca. 900 generations were required to obtain mutant M9 (see Materials and Methods), the presence of 79 mutations implies a mutation rate of 2.9 × 10−8 mutations per nucleotide per generation. Thus, the observed mutation rate in our selection procedure is 130-fold higher than the mutation rate reported for neutral mutations in E. coli (15). It is also higher than the mutation rate reported for whole-genome sequence analyses of E. coli and Staphylococcus aureus populations evolved under the selective pressure of antibiotics (8- to 22-fold) (16–18).
Mutations affecting the mismatch repair protein MutL.
Two mutations were detected in gene mutL encoding the putative 702-residue MutL ortholog of E. faecium. Deletion of A1726 in M2 and, additionally, of A661 in M512 led to frameshift mutations in codons 576 and 221 of mutL (Table 2). To evaluate the impact of these mutations, we compared the frequency of spontaneous mutants resistant to rifampin (Fig. 3). This antibiotic was chosen since single amino acid substitutions in the RpoB subunit of RNA polymerase are known to be sufficient for high-level resistance. The frequency of rifampin-resistant mutants was ca. 4-fold higher in mutant M2 than in the preceding mutant (M1) or in the parental strain (D344S). This difference was statistically significant for both comparisons (P < 0.05, Mann-Whitney U test). The 4-fold difference appears rather modest since a 138-fold increase in the frequency of mutations per generation has been reported for a mutL null mutation in E. coli (15). The high mutation rate observed in M2 was not found in the downstream mutants M3 to M9 and the second mutation detected in mutL of M512 had no additional impact on the frequency of rifampin-resistant mutants. We therefore looked for compensatory mutations in proteins involved in DNA metabolism. A mutation leading to substitution D187N in a putative DNA polymerase was detected in mutant M4. In E. coli, the closest homologue is the translesion error-prone DNA polymerase V subunit, which causes increased mutagenesis by promoting translesion synthesis of DNA damaged by UV or chemicals (19). Impaired activity is predicted to lead to a reduction in the mutation rate and might therefore act as a suppressor of the mutL mutation detected in M2.
FIG 3.

Frequency of rifampin-resistant mutants in derivatives of E. faecium D344S. Bars indicate the median values of a minimum of four experiments. The frequency is higher in M2 than in the remaining strains (P < 0.05, Mann-Whitney U test).
In conclusion, impaired activity of MutL may have contributed to the high incidence of mutations observed in our selection procedure. However, the impact of the mutL mutation on the frequency of rifampin-resistant mutants was moderate (4-fold) and only detected in one mutant (M2) (Fig. 3). Of note, the number of mutations obtained at each selection step on media containing ampicillin fluctuates with 4 to 7 mutations for M1, M2, M3, M4, and M8 versus 10 to 15 mutations for M512, M6, M7, and M9 (Fig. 2). These differences did not correlate with the frequency of mutants detected on media containing rifampin. As previously reported, the mutation rate is elevated in hospital-adapted strains of E. faecium (4.9 ± 0.3 × 10−5 substitutions per nucleotide per year) (20). In the latter study, the frequency of fosfomycin resistance determined in laboratory conditions for these strains (ca. 5 × 10−7) was similar to the value of 1 × 10−6 obtained for D344S (Fig. 3), indicating that this strain is not atypical with respect to the mutation rate.
Mutation spectrum of ampicillin- and glycopeptide-resistant derivatives of D344S.
The molecular spectrum of the 79 mutations detected in mutant M9 is depicted in Table 4. Transitions were predominant (n = 40) among single nucleotide substitutions (total of 51), and these mutations were mainly missense mutations (n = 39). Single base deletions (n = 24) were more frequent than insertions (n = 4) and all 24 deletions involved As or Ts. In the coding strand of intragenic sequences, deletions more frequently involved As (n = 18) than Ts (n = 2) (data not shown). To evaluate the relative contributions of neutral and selective mutations to the large panel of mutations detected in M9, we compared the number of single base substitutions in coding (n = 49) and intergenic sequences (n = 2) as previously described (15). The ratio deduced (49/2 = 24.5) was higher than the ratio of 5.58 deduced from the length of coding (2,506,356 bp) and noncoding (448,938 bp) sequences (χ2 = 5.0; P < 0.05). Thus, single base substitutions occurred more frequently in coding sequences than expected. In contrast, the observed (23/5 = 4.60) and expected (5.58) ratios were similar for deletions and insertions (P = 0.69). To further evaluate whether mutations in M9 were neutral, we also compared the occurrence of nonsynonymous (n = 42) and synonymous (n = 7) mutations. The deduced ratio (42/7 = 6.00) was higher than the value of 2.34 deduced from the relative frequencies of transitions and transversions (Table 4) and from the codon usage in the 835,452 codons encoding the entire proteome (2,860 proteins) (χ2 = 5.72; P < 0.02). Thus, the mutation spectrum of mutant M9 (Table 4) indicates that the high mutation rate observed in our selection procedure did not mainly result from the accumulation of neutral mutations.
TABLE 4.
Characteristics of mutations detected in mutants M9
| Type of mutation | Intragenic | Intergenic |
|---|---|---|
| Single base substitution (n = 51) | 49 | 2 |
| Transitions (n = 40) | ||
| A:T→G:C | 16 | 0 |
| G:C→A:T | 23 | 1 |
| Transversions (n = 11) | ||
| A:T→T:A | 4 | 1 |
| A:T→C:G | 0 | 0 |
| G:C→T:A | 4 | 0 |
| G:C→C:G | 2 | 0 |
| A:T sites (n = 21) | 20 | 1 |
| G:C sites (n = 30) | 29 | 1 |
| Consequence | ||
| Synonymous | 7 | NAa |
| Missense | 39 | NAa |
| Nonsense | 3 | NAa |
| Insertion and deletion (n = 28) | 23 | 5 |
| Deletions (n = 24) | ||
| A or T | 20 | 4 |
| G or C | 0 | 0 |
| Insertions (n = 4) | ||
| A or T | 2 | 1 |
| G or C | 1 | 0 |
| Total (n = 79) | 72 | 7 |
NA, not applicable.
Mutation spectrum of rifampin-resistant derivatives of D344S.
Since long incubations (3 to 6 days) on inhibitory concentrations of ampicillin and glycopeptides were required to obtain mutants M1 to M9, we evaluated whether the number of mutations increased in aging plates. For this purpose, whole-genome sequencing was performed in four rifampin-resistant mutants (D1, D2, D3, and D6) that were detected after 1, 2, 3, and 6 days of incubation, respectively, on rifampin-containing plates. Each mutant harbored one mutation in the rpoB gene among a total of 8, 3, 5, and 4 mutations for D1, D2, D3, and D6, respectively. Thus, the number of mutations did not increase upon aging of the plates.
The average number of mutations in rifampin-resistant mutants D1, D2, D3, and D6 was 5.0 (20 mutations in 4 mutants). This observation suggests that an average of 4 mutations were acquired by the mutants in addition to the mutation required for rifampin resistance. Since single modifications of RpoB are expected to be sufficient for resistance, these non-rpoB mutations may be neutral. The corresponding mutation rate, 1.4 10−8 per generation and per nucleotide is 64-fold higher than the value of 2.2 × 10−10 reported for E. coli (15). Transitions greatly exceeded transversions in M9 (40/11 = 3.64) (Table 4) but not in rifampin-resistant mutants (6/13 = 0.46; P = 0.00024) (Table 5). Insertions and deletions were less abundant than single-nucleotide substitutions in M9 (28/51 = 0.55), but the difference was greater in rifampin-resistant mutants (1/19 = 0.052; P = 0.0075). Of note, 4 transitions and 3 transversions (4/3 = 1.33) but no deletion were detected in mutant M1, which harbored a wild-type allele of mutL. These observations indicate that the mutL mutation of M2 shifted the spectrum of mutations in favor of both transitions and deletions. In E. coli, transitions and frameshift mutations also predominate in mutL mutants (Table 6). For example, transition to transversion ratios of 43 (1,588/37) and 1.3 (131/102) were reported for a mutL strain and its isogenic wild-type parental strain (15). The ratio of single-nucleotide insertions and deletions to single-nucleotide substitutions was also increased in a mutL null mutant (299/1,625 = 0.18) in comparison to that of the wild type (19/233 = 0.08). Together, these observations indicate that the mutL mutation detected in M2 modified the mutation spectrum (Table 6), in addition to the moderate increase in the frequency of rifampin-resistant mutants reported above.
TABLE 5.
Characteristics of mutations detected in rifampin-resistant mutants D1, D2, D3, and D6
| Type of mutation | Intragenic | Intergenic |
|---|---|---|
| Single base substitution (n = 19) | 16 | 3 |
| Transitions (n = 6) | 1 | 1 |
| A:T→G:C | ||
| G:C→A:T | 4 | 0 |
| A:T→C:G | 1 | 0 |
| G:C→T:A | 5 | 2 |
| G:C→C:G | 0 | 0 |
| A:T sites (n = 8) | 7 | 1 |
| G:C sites (n = 11) | 9 | 2 |
| Consequence | ||
| Synonymous | 2 | NAb |
| Missense | 14a | NAb |
| Nonsense | 0 | NAb |
| Insertion and deletion (n = 1) | 1 | 0 |
| Deletions (n = 0) | ||
| A or T | 0 | 0 |
| G or C | 0 | 0 |
| Insertions (n = 1) | ||
| A or T | 1 | 0 |
| G or C | 0 | 0 |
| Total (n = 20) | 17 | 3 |
NA, not applicable.
Including mutations leading to amino acid substitutions P27S, Q473K, Q473K, and Q473L in RpoB of mutants D1, D2, D3, and D6, respectively.
TABLE 6.
Comparison of mutations in wild-type and mutL E. faecium and E. coli
| Type of mutation (expected) |
E. faeciuma |
E. coli |
||
|---|---|---|---|---|
| MutL (Apr and Vmr) | Wild type (Rifr) | MutL (neutral) | Wild type (neutral) | |
| Mutation rate | 2.9 × 10−8 | 1.4 × 10−8 | 3.0 × 10−8 | 2.2 × 10−10 |
| % nonsynonymous (70) | 85 | 88 | 40 | 41 |
| % missense (85) | 96 | 84 | 47 | 43 |
| % transitions | 78 | 32 | 98 | 57 |
| % insertions and deletions | 35 | 5 | 15 | 7 |
Apr, ampicillin resistance; Vmr, vancomycin resistance; Rifr, rifampin resistance.
Peptidoglycan synthesis enzymes affected by mutations detected in M1 to M9.
A mutation was detected in a class B PBP gene encoding a putative orthologue of Streptococcus pneumoniae PBP2x, designated pbpB (21). The mutation led to a T539K substitution (Table 2) located immediately downstream from the conserved KTG motif at positions 536 to 538. In clinical isolates of streptococci, the L to V substitution in the residue immediately preceding this conserved motif is a common contributor to acquired penicillin resistance (22–24). A T to A substitution in the residue following the conserved KTG motif is rarely observed in clinical isolates although it is responsible for acquisition of low-level resistance to cephalosporins in mutants selected in laboratory conditions (22–24). These observations suggest that decreased affinity of PBPB for ampicillin was selected along with activation of the l,d-transpeptidation pathway. This may appear counterintuitive since PBPs are fully bypassed by l,d-transpeptidase Ldtfm in mutant M512 (1, 6). However, the T539K substitution was observed in mutant M2, which is only moderately resistant to β-lactams (Fig. 1). In this mutant, the proportion of l-Lys3→d-iAsx-l-Lys3 cross-links is increased in comparison to that in the parental strain D344S, but d,d-transpeptidases retain an essential role in cross-link formation (8). Thus, the T539K substitution in PBPB may have facilitated acquisition of ampicillin resistance in early selection steps although it does not play a direct role in the l,d-transpeptidation pathway. Alternatively, acylation of PBPB by ampicillin may inhibit peptidoglycan polymerization complexes even though this PBP is not required for peptidoglycan cross-linking. Such a dominant effect has been previously observed for inhibition of bifunctional class A PBPs by moenomycin (21).
Mutant M8 harbored a null mutation in gene ddcP, which encodes the major d,d-carboxypeptidase in E. faecium (25). As described in the Introduction, stepwise activation of the l,d-transpeptidation pathway in mutants M1 to M9 requires synthesis of DdcY and optimization of its d,d-carboxypeptidase activity for full conversion of soluble cytoplasmic precursors into the UDP-MurNAc-tetrapeptide (9, 12). DdcP and DdcY catalyzed the same reaction, potentially on different substrates depending on the specificity and location of the enzymes (9, 25). These substrates may include UDP-MurNAc-pentapeptide, as demonstrated for DdcY, lipid intermediates I and II, and stem peptides in nascent peptidoglycan (9, 25). Selection of a null mutation in gene ddcP of M8 shows that the d,d-carboxypeptidase activity of DdcP is not essential to the l,d-transpeptidation pathway. The role of the ddcP mutation in resistance, if any, remains to be established, but it is tempting to speculate that DdcY and DdcP compete for the same localization or cofactor. Of note, deletion of ddcP in a wild-type E. faecium background has been reported to decrease the MIC of ampicillin from 43 to 8 μg/ml (24).
An A205V substitution was detected in MurAB, one of the two UDP-N-acetylglucosamine 1-carboxyvinyltransferases that catalyze the first committed step on peptidoglycan synthesis in E. faecium. In Enterococcus faecalis, inactivation of gene murAA, but not of paralogue murAB, was reported to decrease cephalosporin resistance (26). Since the respective roles of these two enzymes, which are both functional for UDP-MurNAc synthesis, remain enigmatic (26), we cannot propose a mechanism for the possible contribution of the MurAB substitution to glycopeptide resistance of M9.
Likewise, there is no obvious link between the A198T substitution in GlcNAc transferase MurG and activation of the l,d-transpeptidation pathway since this enzyme, which forms lipid intermediate II, displays little specificity for the peptide stems of peptidoglycan precursors (27). Thus, MurG is not expected to be affected by the replacement of pentapeptide by tetrapeptide.
In conclusion, 4 of the 20 enzymes committed to peptidoglycan biosynthesis were affected by amino substitutions (PBPB, DdcP, MurAB, and MurG). This proportion is higher than the incidence of nonsynonymous mutations at the scale of the entire proteome (20% versus 1.6%, respectively). None of the substitutions had any obvious role in the activation of the l,d-transpeptidation pathway. In particular, the substitutions did not involve enzymes that recognize the peptide stems of peptidoglycan precursors and, for this reason, might be affected by replacement of pentapeptide by tetrapeptide stems in the peptidoglycan precursors of the mutants. Such enzymes include d-aspartate ligase, MraY transferase, and d-Glu and d-Asp amidases that were not affected by substitutions in M9. Thus, replacement of pentapeptide by tetrapeptide appears to be well tolerated by enzymes involved in the late steps of peptidoglycan precursor assembly.
Functional classes of proteins affected by mutations in mutants M1 to M9.
Among the 79 mutations detected in M9, 65 were nonsynonymous mutations (Tables 2 and 4). Assignment of the corresponding proteins in functional classes (Fig. 4) revealed sequence alterations in eight proteins involved in transcription regulation, including CroR, a response regulator of a two-component regulatory system that contributes to intrinsic β-lactam resistance in the enterococci by an unknown mechanism (28–30). As described above for amino acid substitution T539K in PBPB, this modification may not be directly related to the activation of the l,d-transpeptidation pathway since it was acquired in an early step of the selection procedure (2nd) by a mutant (M2) which still partially relies on PBPs for peptidoglycan cross-linking. Nonsynonymous mutations also affected two sensor kinases, suggesting that regulatory circuits involving two-component regulatory systems are affected in response to the acquisition of ampicillin and glycopeptide resistance. Since certain response regulators of two-component regulatory systems are substrates of Ser/Thr protein kinases (31), the sensor kinase genes might be targets for compensatory mutations in response to the pleiotropic effects induced by impaired StpA phosphatase activity (11). Ten of the 65 proteins affected by nonsynonymous mutations were involved in the transport of various molecules through the membrane, including sugars, amino acids, phosphate, proteins, and metals. Five proteins were involved in the assimilation of carbon and generation of energy from carbon sources, including glyceraldehyde-3-phosphate dehydrogenase, l-lactate dehydrogenase, fructose-1,6-bisphosphatase, and glycerol kinase. These results suggest that the fluxes through central metabolic pathways, including glycolysis, might be profoundly remodeled in mutant M9. Of note, a connection between CroR and the phosphotransferase system (PTS) system has recently been reported in E. faecalis mutants hypersusceptible to β-lactam antibiotics (32).
FIG 4.
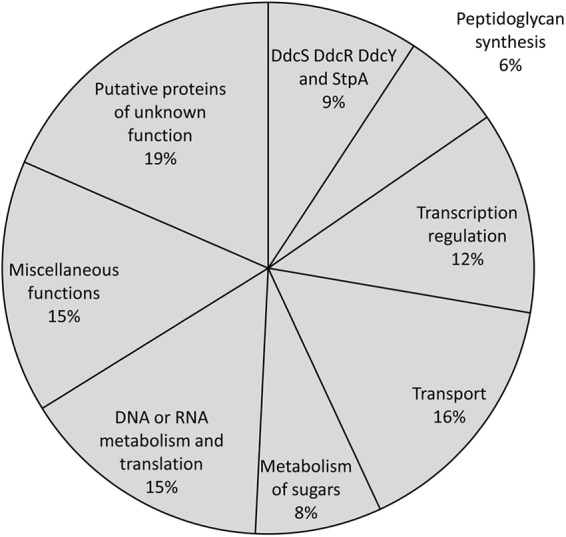
Functional classes of proteins affected by the 65 nonsynonymous mutations detected in mutant M9.
In conclusion, several lines of evidence indicate that multiple mutations are required for acquisition of high-level resistance to ampicillin and glycopeptides mediated by activation of the l,d-transpeptidation pathway. First, it is worth noting that nearly all E. faecium isolates naturally produce a low-affinity PBP (PBP5) that confers moderate levels of resistance to ampicillin and high-level resistance to this drug following mutational alteration of the corresponding gene. However, the l,d-transpeptidation pathway conveys even higher levels of resistance and, additionally, resistance to glycopeptides. Second, nine consecutive selection steps were required for full conversion of pentapeptide to tetrapeptide and exclusive formation of 3→3 cross-links in the peptidoglycan (Fig. 1). Third, mutants with increased resistance to ampicillin and glycopeptides were obtained at frequencies ranging from 10−9 to 10−10 in each of the nine selection steps. The frequency of mutants resistant to rifampin was at least 1,000-fold higher (Fig. 3). These observations suggest that more than one mutation was required to significantly increase the level of resistance to ampicillin and glycopeptides in each of the nine selection steps. Acquisition of a null mutation in the gene encoding mismatch repair protein MutL may have facilitated emergence of resistance by extending the repertoire of spontaneous mutations in M2 and subsequent mutants. An increase in the mutation rate per se was not involved since the frequency of mutants resistant to rifampin was only increased in one of the nine mutants (M2), and this increase was modest (4-fold) (Fig. 3). MutL appears functional in the parental strain D344S since the mutL null mutation resulted in an increase in the frequency of transversions and deletions, which is a signature of impaired MutL activity (Table 6) (15). Thus, the contribution of the mutL mutation to successful selection of resistance to ampicillin and glycopeptides may have mainly involved an increase in the frequency of frameshift mutations. This would imply that loss of function has played a key role in the activation of the l,d-transpeptidation pathway. Accordingly, two mutations that impaired the phosphatase activity of DdcS and StpA were experimentally shown to be required for ampicillin resistance (11). The impact of other mutations on the expression of ampicillin and glycopeptide resistance remains to be determined. However, the number and function of the proteins affected by mutations in M9 (Fig. 4) suggest that activation of the l,d-transpeptidation pathway involves profound alterations in the use of carbon sources and in transcriptional regulatory circuits. This may limit emergence of this resistance mechanism, which has never been detected in any clinical isolates, in natural conditions although a modest contribution of an l,d-transpeptidase distantly related to Ldtfm to intrinsic cephalosporin resistance has been reported in E. faecium (25). Bypass of d,d-transpeptidases by l,d-transpeptidases has been a viable evolutionary option for mycobacteria since the peptidoglycan of these organisms mainly (75 to 80%) contains 3→3 cross-links (33–35). In this case, β-lactam resistance might not have been the main selective pressure for the emergence and maintenance of the 3→3 mode of peptidoglycan cross-linking since these bacteria produce broad spectrum β-lactamases (36–38) and the l,d-transpeptidation pathway is inhibited by β-lactams that block production of tetrapeptide stems by d,d-carboxypeptidases belonging to the PBP family (35, 39).
ACKNOWLEDGMENTS
We thank L. Ma for technical assistance in genome sequencing.
This work was supported by the National Institute of Allergy and Infectious Diseases (Grants R01 307 AI046626).
REFERENCES
- 1.Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. 2000. Novel mechanism of beta-lactam resistance due to bypass of dd-transpeptidation in Enterococcus faecium. J Biol Chem 275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 2.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant transpeptidation pathway. J Biol Chem 280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 4.Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C. 2006. Crystal structure of a novel beta-lactam-insensitive peptidoglycan transpeptidase. J Mol Biol 359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32:386–408. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- 6.Mainardi JL, Hugonnet JE, Rusconi F, Fourgeaud M, Dubost L, Moumi AN, Delfosse V, Mayer C, Gutmann L, Rice LB, Arthur M. 2007. Unexpected inhibition of peptidoglycan l,d-transpeptidase from Enterococcus faecium by the beta-lactam imipenem. J Biol Chem 282:30414–30422. doi: 10.1074/jbc.M704286200. [DOI] [PubMed] [Google Scholar]
- 7.Triboulet S, Dubée V, Lecoq L, Bougault C, Mainardi JL, Rice LB, Ethève-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet JE, Simorre JP, Arthur M. 2013. Kinetic features of l,d-transpeptidase inactivation critical for β-lactam antibacterial activity. PLoS One 8:e67831. doi: 10.1371/journal.pone.0067831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mainardi JL, Morel V, Fourgeaud M, Cremniter J, Blanot D, Legrand R, Frehel C, Arthur M, Van Heijenoort J, Gutmann L. 2002. Balance between two transpeptidation mechanisms determines the expression of beta-lactam resistance in Enterococcus faecium. J Biol Chem 277:35801–35807. doi: 10.1074/jbc.M204319200. [DOI] [PubMed] [Google Scholar]
- 9.Sacco E, Hugonnet JE, Josseaume N, Cremniter J, Dubost L, Marie A, Patin D, Blanot D, Rice LB, Mainardi JL, Arthur M. 2010. Activation of the l,d-transpeptidation peptidoglycan cross-linking pathway by a metallo-d,d-carboxypeptidase in Enterococcus faecium. Mol Microbiol 75:874–885. doi: 10.1111/j.1365-2958.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 10.Arthur M, Depardieu F, Cabanie L, Reynolds P, Courvalin P. 1998. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol Microbiol 30:819–830. doi: 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 11.Sacco E, Cortes M, Josseaume N, Rice LB, Mainardi JL, Arthur M. 2014. Serine/threonine protein phosphatase-mediated control of the peptidoglycan cross-linking l,d-transpeptidase pathway in Enterococcus faecium. mBio 5:e01446-01414. doi: 10.1128/mBio.01446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cremniter J, Mainardi JL, Josseaume N, Quincampoix JC, Dubost L, Hugonnet JE, Marie A, Gutmann L, Rice LB, Arthur M. 2006. Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J Biol Chem 281:32254–32262. doi: 10.1074/jbc.M606920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nieto M, Perkins HR. 1971. Modifications of the acyl-d-alanyl-d-alanine terminus affecting complex-formation with vancomycin. Biochem J 123:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arthur M, Depardieu F, Reynolds P, Courvalin P. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol 21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Popodi E, Tang H, Foster PL. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A 109:E2774–E2283. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, Ozan VB, Senturk GH, Cokol M, Yeh P, Toprak E. 2014. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol Biol Evol 31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. 2012. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo M, Cui L, Kim J, Hiramatsu K. 2013. Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains derived from hVISA clinical strain Mu3. Antimicrob Agents Chemother 57:5843–5853. doi: 10.1128/AAC.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaisman A, McDonald JP, Woodgate R. 19 March 2012. Translesion DNA synthesis. Ecosal Plus doi: 10.1128/ecosalplus.7.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arbeloa A, Segal H, Hugonnet JE, Josseaume N, Dubost L, Brouard JP, Gutmann L, Mengin-Lecreulx D, Arthur M. 2004. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J Bacteriol 186:1221–1228. doi: 10.1128/JB.186.5.1221-1228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granger D, Boily-Larouche G, Turgeon P, Weiss K, Roger M. 2005. Genetic analysis of pbp2x in clinical Streptococcus pneumoniae isolates in Quebec, Canada. J Antimicrob Chemother 55:832–839. doi: 10.1093/jac/dki118. [DOI] [PubMed] [Google Scholar]
- 23.Grebe T, Hakenbeck R. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob Agents Chemother 40:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sifaoui F, Kitzis MD, Gutmann L. 1996. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral beta-lactam antibiotics is associated with alterations of PBP2x. Antimicrob Agents Chemother 40:152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Paganelli FL, Bierschenk D, Kuipers A, Bonten MJ, Willems RJ, van Schaik W. 2012. Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium. PLoS Genet 8:e1002804. doi: 10.1371/journal.pgen.1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vesić D, Kristich CJ. 2012. MurAA is required for intrinsic cephalosporin resistance of Enterococcus faecalis. Antimicrob Agents Chemother 56:2443–2451. doi: 10.1128/AAC.05984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev 32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 28.Comenge Y, Quintiliani R Jr, Li L, Dubost L, Brouard JP, Hugonnet JE, Arthur M. 2003. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J Bacteriol 185:7184–7192. doi: 10.1128/JB.185.24.7184-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Breton Y, Muller C, Auffray Y, Rince A. 2007. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl Environ Microbiol 73:3738–3741. doi: 10.1128/AEM.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller C, Le Breton Y, Morin T, Benachour A, Auffray Y, Rince A. 2006. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J Bacteriol 188:2636–2645. doi: 10.1128/JB.188.7.2636-2645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright DP, Ulijasz AT. 2014. Regulation of transcription by eukaryotic-like serine-threonine kinases and phosphatases in Gram-positive bacterial pathogens. Virulence 5:863–885. doi: 10.4161/21505594.2014.983404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder H, Kellogg SL, Skarda LM, Little JL, Kristich CJ. 2014. Nutritional control of antibiotic resistance via an interface between the phosphotransferase system and a two-component signaling system. Antimicrob Agents Chemother 58:957–965. doi: 10.1128/AAC.01919-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J Bacteriol 193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HI, Barry CE 3rd. 2012. Meropenem inhibits d,d-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol 86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hugonnet JE, Blanchard JS. 2007. Irreversible inhibition of the Mycobacterium tuberculosis beta-lactamase by clavulanate. Biochemistry 46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE III, Blanchard JS. 2009. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soroka D, Dubee V, Soulier-Escrihuela O, Cuinet G, Hugonnet JE, Gutmann L, Mainardi JL, Arthur M. 2014. Characterization of broad-spectrum Mycobacterium abscessus class A beta-lactamase. J Antimicrob Chemother 69:691–696. doi: 10.1093/jac/dkt410. [DOI] [PubMed] [Google Scholar]
- 39.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Riegel P, Gutmann L, Mainardi JL. 2009. The beta-lactam-sensitive d,d-carboxypeptidase activity of Pbp4 controls the l,d and d,d transpeptidation pathways in Corynebacterium jeikeium. Mol Microbiol 74:650–661. doi: 10.1111/j.1365-2958.2009.06887.x. [DOI] [PubMed] [Google Scholar]