Abstract
Ceftazidime-avibactam is active against most Enterobacteriaceae isolates with KPC carbapenemases. We investigated whether this activity could be compromised by mutation. Single-step and multistep selections were attempted using ceftazidime-avibactam (avibactam fixed at 1 or 4 μg/ml) versus two strains each of Enterobacter cloacae and Klebsiella pneumoniae, all with the KPC-3 enzyme. Mutant blaKPC alleles were sequenced, and their parentage was confirmed by typing. Ceftazidime-avibactam selected mutants at up to 16× MIC, with frequencies of ca. 10−9. This contrasted with previous experience for ceftaroline-avibactam, where mutant frequencies under similar conditions were <10−9. The MICs of ceftazidime with 1 μg/ml avibactam for the ceftazidime-avibactam-selected mutants rose from 1 to 8 μg/ml to 16 to >256 μg/ml and those of ceftazidime with 4 μg/ml avibactam from 0.25 to 1 μg/ml to 4 to 128 μg/ml; ceftaroline-avibactam MICs rose less, typically from 0.5 to 1 μg/ml to 1 to 8 μg/ml. The MICs of carbapenems and cephalosporins except ceftazidime and piperacillin-tazobactam were reduced for many mutants. Sequencing of blaKPC revealed point and insertion changes in 12/13 mutants investigated, representing all four parents; one mutant lacked blaKPC changes and possibly had reduced permeability. Amino acid changes commonly involved Ω loop alterations or 1 to 6 amino acid insertions immediately C-terminal to this loop. The most frequent change, seen in four mutants from three strains, was Asp179Tyr, replacing a residue that ordinarily forms a salt bridge to stabilize the Ω loop. Since ceftaroline-avibactam was less affected than ceftazidime-avibactam, we postulate that these mutations increase ceftazidimase specificity rather than conferring avibactam resistance. The clinical relevance remains uncertain.
INTRODUCTION
Avibactam is the first diazabicyclooctane β-lactamase inhibitor to reach advanced clinical development. It inhibits class A β-lactamases, including KPC types and AmpC types (1, 2). Activity against class D β-lactamases varies with the particular enzymes. Metallo (class B)-β-lactamases are unaffected. The combination of ceftazidime with avibactam was approved in February 2015 by the U.S. Food and Drug Administration for use in complicated intra-abdominal and urinary tract infections. Combinations with ceftaroline fosamil and aztreonam are in the earlier stages of development (ceftaroline fosamil is a prodrug of ceftaroline, to which it is rapidly converted following intravenous [i.v.] administration).
The ability of avibactam to inhibit KPC β-lactamases is of particular interest, given that these carbapenemases are expanding globally, with dramatic proliferation in, e.g., Italy, Greece, Brazil, the United States, and Israel (3). Enterobacteriaceae that produce KPC carbapenemases typically are resistant to all β-lactam antibiotics except temocillin, which retains strain-variable activity (4); most are also resistant to multiple other agents besides β-lactams (3). Although KPC enzymes are class A β-lactamases, strains that produce these enzymes are resistant to the available penicillin-clavulanate and penicillin-penicillanic acid sulfone combinations, apparently because the carbapenemases can inactivate these β-lactamase inhibitors (5).
It is important, however, to assess the vulnerabilities of new β-lactamase inhibitor combinations such as ceftazidime-avibactam to spontaneous mutational resistance. Amino acid substitutions in the TEM and SHV enzymes can reduce binding of clavulanate and penicillanic acid sulfones (6), although inhibitor-resistant TEM mutants appear not to have proliferated to the same extent as cephalosporin-hydrolyzing extended-spectrum β-lactamase (ESBL) mutants and are rarely selected in vivo (7). Much less is known as yet about the potential of diazabicyclooctane combinations to select resistance. Our previous work (8) with ceftaroline-avibactam found selection in vitro of (i) a CTX-M-15 β-lactamase mutant with a Lys237Gln substitution, (ii) mutants of AmpC-derepressed Enterobacter strains with large deletions (amino acids 213 to 226) in the AmpC Ω loop, and (iii) mutants of AmpC-derepressed Enterobacter strains with loss of OmpC/F porins, sometimes combined with substitutions to Asn346 in the C terminus of AmpC, a residue recently shown to be involved in the binding of avibactam (9). On the other hand, we failed to select stable resistance to ceftaroline-avibactam in Enterobacteriaceae with KPC carbapenemases, or in those with other ESBLs besides CTX-M-15; moreover, the ceftaroline-avibactam-selected CTX-M-15 mutant lost its ability to confer resistance to other cephalosporins besides ceftaroline. Breakthrough of variants with reduced susceptibility was not observed with ceftazidime-avibactam in neutropenic and immunocompetent mouse thigh and lung infection models performed with ceftazidime-resistant Enterobacteriaceae that carried various β-lactamase genes, including blaKPC-3 (10).
The present report describes the results of in vitro selection studies performed with ceftazidime-avibactam for Enterobacteriaceae with KPC-3 carbapenemase.
MATERIALS AND METHODS
Test strains.
The test organisms comprised two clinical isolates each of Klebsiella pneumoniae and Enterobacter cloacae, all submitted to the Public Health England's (PHE) Antimicrobial Resistance and Healthcare Associated Infections Reference Unit by United Kingdom diagnostic laboratories between 2007 and 2011. Previous PCRs (11) had detected blaKPC genes in these isolates. The K. pneumoniae isolates were unique in terms of variable-number tandem repeat (VNTR) profiles (12), and the two E. cloacae differed in pulsed-field gel electrophoresis (PFGE) profiles. K. pneumoniae NCTC 13438 had a VNTR profile (3, 2, 2, 13, 2, 1, 3, —, 1), typical of the international sequence type (ST) 258 lineage (PHE, data on file). β-Lactamase gene profiles were reconfirmed with Check-MDR CT103XL DNA microarrays (Check Points, Wageningen, Netherlands). Strain NCTC 13438 (H073620453) was among the first isolates with a KPC carbapenemase to be recorded in the United Kingdom and was lodged with the National Collection of Type Cultures on that basis (13).
Antibiotics.
Avibactam was from AstraZeneca (Wilmington, DE, USA), as were ceftazidime and ceftaroline. The other antimicrobials were purchased from Sigma (Poole, United Kingdom), except for ertapenem (Merck, Hoddesdon, United Kingdom) and meropenem (AstraZeneca, Alderley Park, United Kingdom).
Single-step mutant selection.
Selection was undertaken as previously described (8). Approximately 109 CFU from overnight broth cultures were spread on Mueller-Hinton agar (Oxoid, Basingstoke, United Kingdom) with ceftazidime plus avibactam (fixed concentration, 1 or 4 μg/ml) at 2× to 16× the MICs found previously by the Clinical and Laboratory Standards Institute (CLSI) general agar dilution method (14). Colonies were counted after overnight incubation, with representatives retained for MIC determination. Two concentrations of avibactam were used to increase the possibility of mutant selection; although the 4 μg/ml is routinely used in susceptibility tests, it is evident that bacteria in infections and the gut flora are exposed to constantly changing rather than constant drug levels.
Multistep selection.
Inocula of 108 CFU were added to 10-ml volumes of nutrient broth containing ceftazidime-avibactam (1 or 4 μg/ml) at the ceftazidime-avibactam MICs found previously by CLSI agar dilution and incubated for 24 h (13). This procedure was repeated daily, each time doubling the ceftazidime concentration up to a maximum of 12 steps or 128 μg/ml ceftazidime, while keeping the avibactam concentration fixed at the initial 1 or 4 mg/L. Organisms from each step were retained for MIC determinations.
Determination of MICs.
MICs of ceftazidime-avibactam and other antibiotics were determined for each isolate by CLSI agar dilution (14). Where needed, avibactam was incorporated into the agar at 1 and 4 μg/ml; tazobactam was used at 4 μg/ml.
Typing.
K. pneumoniae parent strains and their mutants were compared by VNTR analysis (12). E. cloacae parents and mutants were compared by PFGE of XbaI-digested genomic DNA, as described previously (15).
Sequencing blaKPC.
A PCR template was prepared from single colonies, as described previously (16). Previously described primers were used to amplify blaKPC, namely, 6560U (5′-ACCCTTGCCATCCCGTGTGC-3′) and 8848L (5′-CGCCATCGTCAGTGCTCTAC-3′ (17). The PCR was set up using MyTaq Red Mix PCR Mastermix (Bioline, London, United Kingdom) in 25-μl volumes, with 0.1 mM primers and 2 μl of template and performed with an initial denaturation for 5 min at 95°C and then 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min, followed by a final extension at 72°C for 5 min. The PCR product was purified by polyethylene glycol (PEG) precipitation as previously described (18), with minor changes. Briefly, the PEG solution (20% [wt/vol] PEG-2.5 M sodium chloride) was added to the DNA, and the sample was mixed and incubated at room temperature for 30 min, followed by centrifugation for 45 min at 13,000 rpm and 4°C. The pellet was washed twice with ethanol and resuspended in molecular-grade water. Amplicons were sequenced using the primers 6560U and 8848L (above) as well as a primer designed in this work: 6560U_1A (5′-CGGACGCGAGGAAGCGAACC-3′). Sequencing was performed by Public Health England's Genomic Services Unit; sequence data were analyzed and assembled with BioNumerics software (version 6.1; Applied Maths, Belgium). The blaKPC gene open reading frame was determined by sequence homology searching with the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments of nucleotide and amino acid sequences were conducted with the Clustal Omega tool (http://www.ebi.ac.uk/Tools/msa/clustalo/).
RESULTS
β-Lactamase gene profiles.
DNA arrays confirmed the presence of blaKPC in all four parent strains, and sequencing indicated the KPC-3 variant in all cases. K. pneumoniae H105180643 additionally had an SHV ESBL with a Gly238Ser substitution, and both E. cloacae strains had genes encoding CTX-M-group 9 ESBLs. Although K. pneumoniae NCTC 13438 was an ST258 variant, it lacked evidence of the SHV ESBLs commonly found in this lineage, although classical blaSHV was detected. All four strains had blaTEM-1, with no ESBL mutations detected.
Mutant selection frequencies.
Mutants with increased ceftazidime-avibactam MICs were selected from all four strains, using avibactam at either 1 or 4 μg/ml (Table 1). At 2× MIC, frequencies were from 10−6 to 10−9. These frequencies diminished at higher MIC multiples, but remained around 10−9 for three of the four strains at 16× MIC for at least one of the avibactam concentrations. It may appear counterintuitive that mutant frequencies were sometimes higher with 4 μg/ml than 1 μg/ml avibactam, but it should be recognized that starting MICs of ceftazidime with 4 μg/ml avibactam were lower than those with 1 μg/ml, reducing the barrier to selection. Mutants with raised MICs were also selected in broth in a multistep procedure. To confirm that the mutants were derived from their parents and were not contaminants, we confirmed the consistency of the VNTR (K. pneumoniae) and PFGE profiles (E. cloacae) for representatives within each series, selected as showing diverse changes in carbapenem MICs (below) in parallel with raised ceftazidime-avibactam MICs. In all cases, the mutants' profiles were indistinguishable from those of their parents, confirming parentage.
TABLE 1.
Frequencies of mutants obtained in single-step selection on agar containing different MIC multiples of ceftazidime-avibactam, 1 or 4 μg/ml
| Strain | MIC for CAZ (μg/ml)a |
Mutant frequency × 10−9 with: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alone | +AVI, 1 μg/ml | + AVI, 4 μg/ml | CAZ-AVI, 1 μg/ml at 2× MIC | CAZ-AVI, 4 μg/ml at 2× MIC | CAZ-AVI, 1 μg/ml at 4× MIC | CAZ-AVI, 4 μg/ml at 4× MIC | CAZ-AVI, 1 μg/ml at 8× MIC | CAZ-AVI, 4 μg/ml at 8× MIC | CAZ-AVI, 1 μg/ml at 16× MIC | CAZ-AVI, 4 μg/ml at 16× MIC | |
| K. pneumoniae NCTC 13438 | 256 | 8 | 2 | >1,000 | 2,080 | 155 | 19.5 | 1.03 | 3.08 | 2.05 | <0.50 |
| K. pneumoniae H105180643 | 128 | 8 | 2 | 49.6 | 48.0 | 1.60 | 17.6 | 0.80 | 8.00 | <0.50 | 1.60 |
| E. cloacae H112260226 | 32 | 2 | 0.5 | 130 | 160 | 6.49 | 22.3 | 0.93 | 3.71 | 0.93 | 0.93 |
| E. cloacae H120900216 | 32 | 2 | 0.5 | 1.00 | 2.99 | 1.00 | <0.50 | <0.50 | 1.99 | <0.50 | <0.50 |
AVI, avibactam; CAZ, ceftazidime.
Phenotypes of mutants.
MIC distributions for 101 mutants (7 to 33 per parent strain) selected in single-step (SS) and multistep procedures are shown in Table 2. There was little or no relationship between the selective MIC multiple and the shifts in ceftazidime-avibactam MICs observed, justifying pooling of data for mutants selected under different conditions.
TABLE 2.
MIC distributions for ceftazidime-avibactam-selected mutants of K. pneumoniae and E. cloacae with KPC carbapenemases
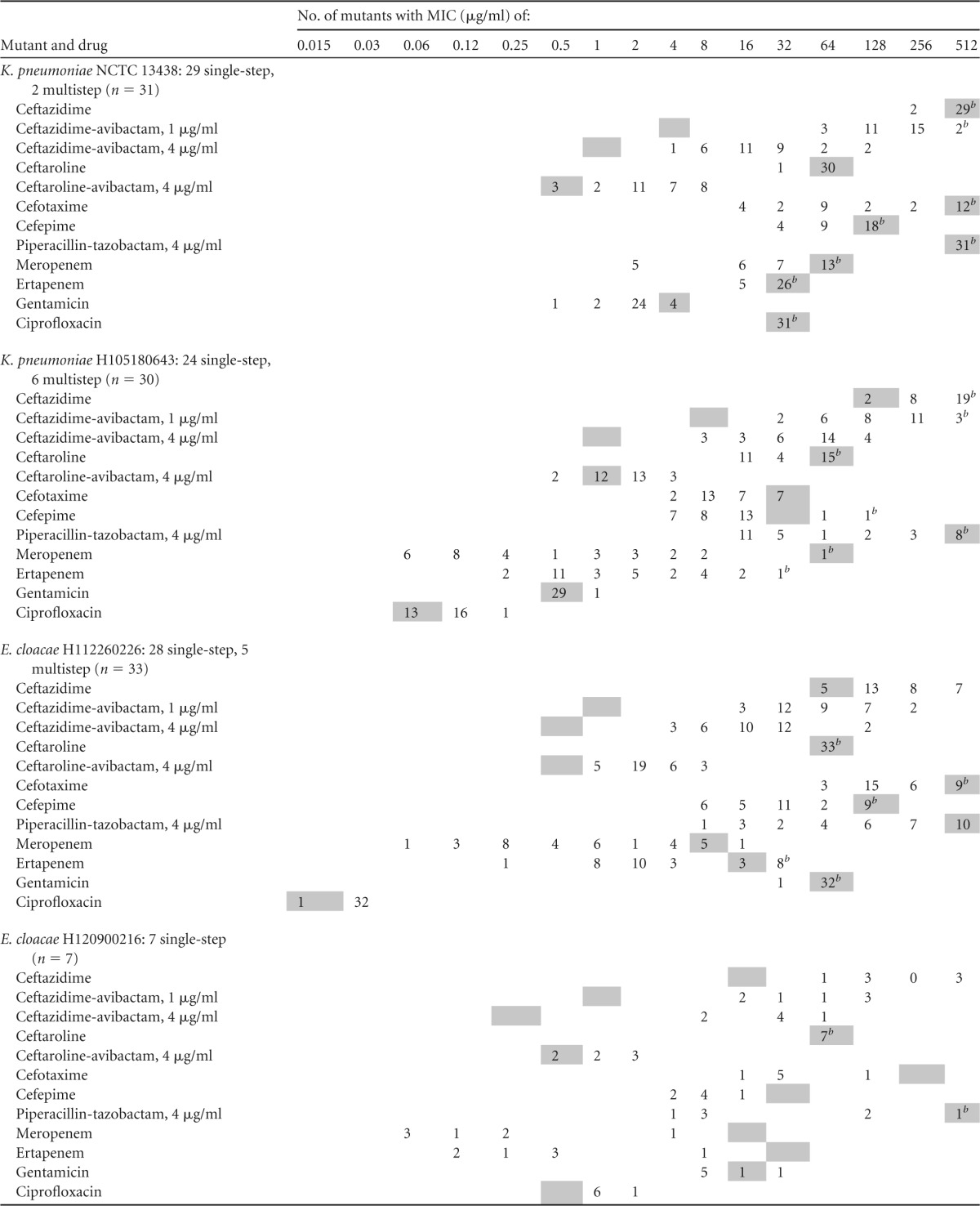
A gray square indicates the MIC for the parent strain.
MIC of more than the indicated value, which was the highest dilution tested.
The MICs of ceftazidime with avibactam at 1 μg/ml rose from 1 to 8 μg/ml for the parent strains to 16 to >256 μg/ml for their mutants; those of ceftazidime with avibactam at 4 μg/ml rose from 0.25 to 1 μg/ml to 4 to 128 μg/ml (generally 8 to 64 μg/ml). The MICs of ceftaroline-avibactam were much less affected, being raised from 0.5 to 1 μg/ml to 1 to 8 μg/ml. The MICs of the other β-lactams, particularly carbapenems, were often reduced. Thus, among the 31 ceftazidime-avibactam-selected mutants derived from K. pneumoniae NCTC 13438 (with a VNTR type corresponding to ST258), five showed reductions in meropenem MICs, from >32 μg/ml to 2 μg/ml, and, among 30 mutants derived from K. pneumoniae H105180643, only one retained the parental level of resistance to ertapenem and meropenem (MICs of >16 to 32 μg/ml), whereas 21 became susceptible to meropenem at ≤1 μg/ml and 13 to ertapenem at 0.5 μg/ml. In the case of E. cloacae H112260226, 10/33 mutants showed no change in the meropenem MIC (±1 doubling dilution) from the parental value of 8 μg/ml, whereas 22 became susceptible at ≤1 μg/ml. For ertapenem, 11/33 continued to exhibit parental MICs of around 16 μg/ml, whereas the MICs for the remainder fell to 1 to 4 μg/ml and in one case to 0.25 μg/ml. Last, 6/7 mutants of E. cloacae H120900216 showed dramatic MIC reductions for meropenem and ertapenem, from ≥16 μg/ml to ≤0.5 μg/ml, whereas the MICs for the remaining mutant remained at 4 μg/ml (meropenem) and 8 μg/ml (ertapenem). MIC shifts were consistent throughout between the two carbapenems, i.e., a mutant that showed a marked MIC reduction to meropenem showed a reduction also for ertapenem.
Many of the mutants also showed reductions in the MICs of piperacillin-tazobactam and those of cephalosporins other than ceftazidime and ceftaroline. Unlike for carbapenems, however, the analysis here is complicated by the fact that, except for K. pneumoniae NCTC 13438, the test strains had secondary SHV or CTX-M ESBLs, which would continue to contribute to cephalosporin resistance in their mutants irrespective of whether the KPC enzyme lost activity. The ciprofloxacin and gentamicin MICs remained consistent within each mutant series.
Molecular profiling of mutants.
We sequenced blaKPC from 13 mutants (Table 3) and their corresponding parents. These mutants were selected as showing a diversity of shifts in the MICs of comparator antibiotics and represented all four parents. VNTR and PFGE profiles were consistent between parents and mutants within each series. All four parents were reconfirmed to have classical blaKPC-3. The nucleotide sequence analysis of blaKPC from the mutants revealed point mutations or insertion changes in all but one case. In 10 of the mutants, the alteration occurred within or immediately C-terminal to the Ω loop, which extends from Arg164 (standard numbering for class A β-lactamases [19], corresponding to Arg163 in the actual sequence) to Asp179. The most frequent change seen, observed in four mutants from three parents, was Asp179Tyr, whereas other changes in the loop or immediately adjacent to it included (i) Asp163Gly, (ii) Pro174Leu, (iii) insertion of serine between amino acids 180 and 181, (iv) insertion of two serine residues between amino acids 181 and 182, and (v) insertion of Arg-Ala-Thr-Thr-Ser-Ser-Pro between positions 183 and 184. Two mutants lacked modifications around the Ω loop but had more remote changes, namely, Thr243Pro and insertion of Ala-Arg between positions 265 and 266. Only mutant 4 (SS) of K. pneumoniae NCTC 13438 lacked detected changes to the sequence of blaKPC. Uniquely among the mutants studied in detail, this variant showed near-identical (8- to 16-fold) MIC rises for both ceftazidime-avibactam and ceftaroline-avibactam. Moreover, and unlike most other mutants, it showed no reductions in resistance to other β-lactams. It is plausible that its behavior reflected a generalized reduction in permeability.
TABLE 3.
Amino acid changes to KPC enzymes among ceftazidime-avibactam-selected mutants
| Mutanta | Changes to KPC-3 enzyme, based on sequencing blaKPCb | MIC (μg/ml)c |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ + AVI1 | CAZ + AVI4 | CPT | CPT + AVI | CTX | CPM | PTZ | MEM | ERP | GEN | CIP | ||
| K. pneumoniae NCTC 13438d | >256 | 8 | 1 | >32 | 0.5 | >256 | >64 | >256 | >32 | >16 | 4 | >16 | |
| Mut 4 (SS) | Unchanged | >256 | 128 | 8 | >32 | 8 | >256 | >64 | >256 | >32 | >16 | 2 | >16 |
| Mut 14 (SS) | Asp179Tyr | >256 | 256 | 8 | >32 | 0.5 | 16 | 32 | >256 | 2 | 16 | 2 | >16 |
| Mut 22 (SS) | Asp179Tyr | >256 | 256 | 64 | >32 | 4 | 16 | 32 | >256 | 2 | 16 | 4 | >16 |
| K. pneumoniae H105180643 | 128 | 8 | 1 | >32 | 1 | 32 | 32 | >256 | 32 | >16 | 0.5 | 0.06 | |
| Mut 13 (SS) | Asp163Gly | 128 | 64 | 32 | >32 | 2 | 128 | >64 | >256 | >32 | >16 | 0.5 | 0.06 |
| Mut 19 (SS) | Ser inserted between 180 and 181 | >256 | 256 | 64 | 16 | 1 | 8 | 8 | 16 | 0.06 | 0.25 | 0.5 | 0.06 |
| Mut 26 (MS) | Asp179Tyr | >256 | 256 | 64 | 16 | 1 | 8 | 4 | 16 | 0.06 | 0.5 | 0.5 | 0.06 |
| E. cloacae H120900216 | 16 | 1 | 0.25 | >32 | 0.5 | 256 | 32 | >256 | 16 | >16 | 16 | 0.5 | |
| Mut 2 (SS) | Ser-Ser inserted between 181 and 182 | 256 | 128 | 32 | >32 | 2 | 32 | 8 | 8 | 0.12 | 0.5 | 8 | 1 |
| Mut 3 (SS) | Pro174Leu | 64 | 16 | 8 | >32 | 0.5 | 128 | 16 | >256 | 4 | 8 | 16 | 1 |
| Mut 7 (SS) | Thr243Pro | 64 | 32 | 32 | >32 | 2 | 32 | 4 | 128 | 0.25 | 0.25 | 8 | 1 |
| E. cloacae H112260226 | 64 | 1 | 0.5 | >32 | 0.5 | >256 | 64 | >256 | 8 | 16 | >32 | 0.015 | |
| Mut 1 (SS) | Pro174Leu | 128 | 32 | 16 | >32 | 2 | >256 | >64 | >256 | 8 | >16 | >32 | 0.03 |
| Mut 2 (SS) | Ala-Arg inserted between 265 and 266 | 64 | 32 | 16 | >32 | 2 | 128 | 16 | 128 | 0.5 | 1 | >32 | 0.03 |
| Mut 12 (SS) | Arg-Ala-Val-Thr-Thr-Ser-Ser-Pro inserted between 183 and 184 | >256 | 256 | 128 | >32 | 8 | 128 | 32 | 16 | 0.125 | 2 | >32 | 0.03 |
| Mut 19 (SS) | Asp179Tyr | 256 | 64 | 16 | >32 | 2 | 128 | 8 | 8 | 0.06 | 0.25 | >32 | 0.03 |
Mut, mutant; SS, single step; MS, multistep.
Amino acid abbreviations follow the standard 3-letter code.
CAZ, ceftazidime; AVI1, avibactam, 1 μg/ml; AVI4, avibactam, 4 μg/ml; CPT, ceftaroline; CTX, cefotaxime; CPM, cefepime; PTZ, piperacillin-tazobactam, 4 μg/ml; MEM, meropenem; ERP, ertapenem; GEN, gentamicin; CIP, ciprofloxacin.
All K. pneumoniae NCTC 13438 mutants shared the parental VNTR profile 3, 2, 2, 13, 2, 1, 3, —, 1 and all K. pneumoniae H105180643 mutants shared its profile 5, 5, 3, 2, —, 2, 2, 4. An explanation of VNTR profiles and their numbering is given in, e.g., Turton et al. (12), but in the present context, the numbers should be seen only as a fingerprint, confirming mutant parentage; similarly, PFGE profiles of E. cloacae parents and mutants were consistent within each strain.
DISCUSSION
Based on previous experience with ceftaroline-avibactam (8), we initiated these studies with little expectation of selecting mutants other, perhaps, than those with permeability lesions. The results confounded these expectations. While mutant frequencies remained in the 10−9 range at MIC multiples of 4- to 16-fold, it was easier to select mutants of KPC-3-producing Enterobacteriaceae that were resistant to ceftazidime-avibactam than it had been to select mutants that were resistant to ceftaroline-avibactam, where mutation frequencies for strains with KPC carbapenemases were consistently below the detection limit of 10−9 despite ceftaroline being more vulnerable to β-lactamases than ceftazidime in general (20).
Most of the mutants selected here showed smaller MIC rises to ceftaroline-avibactam than to ceftazidime-avibactam, consistent with the observation that direct selection with ceftaroline-avibactam was more difficult. Also striking is that fact that the MICs of other β-lactams, particularly those of meropenem and ertapenem, were reduced for many of the mutants. A lack of generalized resistance to avibactam combinations suggests a ceftazidime-related effect, rather than evolution of avibactam resistance per se by the KPC enzymes. Plausible mechanisms would be an increase in enzymatic affinity for ceftazidime, resulting in a reduced Km, or a change in the balance of ceftazidime acylation versus deacylation. Either of these mechanisms would effectively lead to ceftazidime protecting the enzyme from avibactam. Amino acid substitutions that change β-lactamase interactions with ceftazidime are well known: both TEM and CTX-M ESBLs can undergo mutations that specifically increase “ceftazidimase” activity, as with CTX-M-15 β-lactamase versus CTX-M-3 (21, 22). Moreover, mutations that increase acylation relative to deacylation for ceftazidime have been described for KPC carbapenemases (23) as well as for TEM ESBLs (24). Crucially, Levitt et al. (23) described several substitutions to Arg164 in the Ω loop of KPC β-lactamase, generated by site-directed mutagenesis, that increased resistance to ceftazidime by changing the balance between acylation and deacylation. Exactly as with many of our “natural,” mutants, these showed reduced resistance to imipenem, cefotaxime, and cefepime. Levitt et al. (23) did not test avibactam combinations against their mutants. While none of our mutants had changes to Arg164, the majority had substitutions within the Ω loop or insertions immediately C-terminal to it. The most common single mutation seen was Asp179Tyr, replacing the residue that ordinarily forms a salt bridge with Arg164 to stabilize the Ω loop. It is clearly plausible that changes to this site therefore might exert effects similar to those at Arg164, and this view is supported by the results of Winkler et al. (25), who found that multiple substitutions to Asp179 degraded the ability of the KPC-2 enzyme to confer resistance to ampicillin, aztreonam, and imipenem while generally retaining the ability to confer ceftazidime resistance. Once again, the activities of avibactam combinations were not reported.
Other factors may have contributed to the resistance of some of the present mutants and potentially explain, for example, (i) mutant 4 (SS) of K. pneumoniae NCTC 13438, which lacked changes to blaKPC, and (ii) the variation in the MICs of ceftazidime-avibactam and other agents among different mutants of the same strain with the same KPC-3 Asp179Tyr mutation (e.g., K. pneumoniae NCTC 13438 mutants 14 [SS] and 22 [SS]). Porin mutations seem particularly likely in the K. pneumoniae NCTC 13438 mutant 4 (SS), which showed broad increases in resistance without the reductions in resistance to some comparator agents.
We have not excluded possible changes to the SHV ESBL of K. pneumoniae H105180643 or to the CTX-M-group 9 enzymes of the two Enterobacter species. Even unchanged, these enzymes no doubt contributed to resistance to unprotected cephalosporins. Nevertheless, it is implausible that changes to the coproduced ESBLs were the primary determinant of the present findings since (i) there was a clear association between the mutations of blaKPC and the ceftazidime-avibactam resistance, (ii) the mutations selected here had similar effects, in reducing resistance to multiple comparator β-lactams, to analogous site-directed mutations introduced to blaKPC in the laboratory, (iii) putative changes to the coproduced ESBLs evidently cannot explain the reductions in carbapenem resistance, and (iv) similar results were obtained with K. pneumoniae NCTC 13438, which lacked secondary ESBLs, and the other strains, which had these enzymes.
Our data show the potential for the emergence of ceftazidime-avibactam resistance via mutations in blaKPC. Most often these remodeled the enzyme's Ω loop. Only time and experience will reveal whether such mutants are selected at infection sites or in the gut flora or are a laboratory curiosity. Selection of resistance has not been reported in animal infection models nor in phase II clinical trials with ceftazidime-avibactam (26, 27), although these only examined the infection sites and not the gut flora. Moreover, although single point mutations to blaTEM and blaSHV can lead to production of enzyme variants resistant to clavulanate and penicillanic acid sulfones, this seems to be a rare event in the clinic (28). Furthermore, it is plausible (but unproven) that the reductions in resistance engendered in other β-lactams may counterselect these mutations at a hospital level in milieux where a diversity of antibiotics are used. What is more, the selection experiments were done using fixed avibactam concentrations, whereas, at the infection site or in the gut, bacteria with KPC carbapenemases are exposed to changing avibactam concentrations and to changing ceftazidime/avibactam concentrations ratios, and these differences may modulate selectivity.
The observations should, however, be a consideration, along with spectrum and pharmacokinetic compatibility, in the development of β-lactamase inhibitor combinations, indicating that the choice of partner agent can affect the potential for mutational resistance.
ACKNOWLEDGMENTS
AstraZeneca provided financial support.
We are grateful to Public Health England's Genomic Services Unit for sequencing blaKPC genes.
Formatting of references was provided by Faye Gould of Prime Medica Ltd., Knutsford, Cheshire, and was funded by AstraZeneca.
Conflicts of interest: D.M.L. has served on advisory boards or performed ad hoc consultancy for Achaogen, Adenium, Alere, Allecra, Astellas, AstraZeneca, Basilea, Bayer, BioVersys, Cubist, Curetis, Cycle, Discuva, Forest, GSK, Meiji, Pfizer, Roche, Shionogi, Tetraphase, VenatoRx, and Wockhardt; has been a paid lecturer for AOP Orphan, Astellas, AstraZeneca, Bruker, Curetis, Merck, Pfizer, and Leo; and has shareholdings in Dechra, GSK, Merck, PerkinElmer, and Pfizer amounting to <10% of portfolio value. N.W. has no personal interests to declare. PHE's AMRHAI Reference Unit has received financial support for conference attendance, lectures, and research projects or contracted evaluations from numerous sources, including Achaogen Inc., Allecra Antiinfectives GmbH, Amplex, AstraZeneca, Becton Dickinson Diagnostics, The British Society for Antimicrobial Chemotherapy (BSAC), Cepheid, Check-Points B.V., Cubist Pharmaceuticals, Department of Health, Food Standards Agency, Glaxo SmithKline Services Ltd., Henry Stewart Talks, IHMA Ltd., Merck Sharpe & Dohme Corp., Meiji Seika Kiasya Ltd., Momentum Biosciences Ltd., Nordic Pharma Ltd., Norgine Pharmaceuticals, Rempex Pharmaceuticals Ltd., Rokitan Ltd., Smith & Nephew UK Ltd., Trius Therapeutics, VenatoRx, and Wockhardt Ltd. W.W.N. is an employee of AstraZeneca. The other authors declare no conflicts of interest.
REFERENCES
- 1.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodford N, Pike R, Meunier D, Loy R, Hill R, Hopkins KL. 2014. In vitro activity of temocillin against multidrug-resistant clinical isolates of Escherichia coli, Klebsiella spp. and Enterobacter spp, and evaluation of high-level temocillin resistance as a diagnostic marker for OXA-48 carbapenemase. J Antimicrob Chemother 69:564–567. doi: 10.1093/jac/dkt383. [DOI] [PubMed] [Google Scholar]
- 5.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. 2010. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob Agents Chemother 54:890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canton R, Morosini MI, de la Maza OM, de la Pedrosa EG. 2008. IRT and CMT β-lactamases and inhibitor resistance. Clin Microbiol Infect 14(Suppl 1):S53–S62. [DOI] [PubMed] [Google Scholar]
- 7.Martín O, Valverde A, Morosini MI, Rodriguez-Dominguez M, Rodriguez-Banos M, Coque TM, Canton R, del Campo R. 2010. Population analysis and epidemiological features of inhibitor-resistant-TEM-β-lactamase-producing Escherichia coli isolates from both community and hospital settings in Madrid, Spain. J Clin Microbiol 48:2368–2372. doi: 10.1128/JCM.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore DM, Mushtaq S, Barker K, Hope R, Warner M, Woodford N. 2012. Characterization of β-lactamase and porin mutants of Enterobacteriaceae selected with ceftaroline + avibactam (NXL104). J Antimicrob Chemother 67:1354–1358. doi: 10.1093/jac/dks079. [DOI] [PubMed] [Google Scholar]
- 9.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacVane SH, Crandon JL, Nichols WW, Nicolau DP. 2014. In vivo efficacy of humanized exposures of ceftazidime-avibactam in comparison with ceftazidime against contemporary Enterobacteriaceae isolates. Antimicrob Agents Chemother 58:6913–6919. doi: 10.1128/AAC.03267-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turton JF, Perry C, Elgohari S, Hampton CV. 2010. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol 59:541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 13.Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, Macdonald A, Brudney D, Sompolinsky D, Navon-Venezia S, Livermore DM. 2008. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother 62:1261–1264. doi: 10.1093/jac/dkn396. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A9. Clinical and Laboratory Standards Institute, Wayne PA. [Google Scholar]
- 15.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 16.Wimalarathna HM, Richardson JF, Lawson AJ, Elson R, Meldrum R, Little CL, Maiden MC, McCarthy ND, Sheppard SK. 2013. Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol 13:160. doi: 10.1186/1471-2180-13-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas T, Cuzon G, Truong HV, Nordmann P. 2012. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenthal A, Coutelle O, Craxton M. 1993. Large-scale production of DNA sequencing templates by microtitre format PCR. Nucleic Acids Res 21:173–174. doi: 10.1093/nar/21.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambler RP, Coulson AF, Frere JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A β-lactamases. Biochem J 276:269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mushtaq S, Warner M, Ge Y, Kaniga K, Livermore DM. 2007. In vitro activity of ceftaroline (PPI-0903M, T-91825) against bacteria with defined resistance mechanisms and phenotypes. J Antimicrob Chemother 60:300–311. doi: 10.1093/jac/dkm150. [DOI] [PubMed] [Google Scholar]
- 21.Baraniak A, Fiett J, Hryniewicz W, Nordmann P, Gniadkowski M. 2002. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J Antimicrob Chemother 50:393–396. doi: 10.1093/jac/dkf151. [DOI] [PubMed] [Google Scholar]
- 22.Karisik E, Ellington MJ, Pike R, Livermore DM, Woodford N. 2006. Development of high-level ceftazidime resistance via single-base substitutions of blaCTX-M-3 in hyper-mutable Escherichia coli. Clin Microbiol Infect 12:803–806. doi: 10.1111/j.1469-0691.2006.01423.x. [DOI] [PubMed] [Google Scholar]
- 23.Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the Ω-loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem 287:31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antunes NT, Frase H, Toth M, Mobashery S, Vakulenko SB. 2011. Resistance to the third-generation cephalosporin ceftazidime by a deacylation-deficient mutant of the TEM β-lactamase by the uncommon covalent-trapping mechanism. Biochemistry 50:6387–6395. doi: 10.1021/bi200403e. [DOI] [PubMed] [Google Scholar]
- 25.Winkler ML, Taracila MA, Papp-Wallace KM, Page RA, Bonomo RA. 2014. Two sides to every story: exploring the functional role of D179 in the Ω-loop of KPC-2 β-lactamase, abstr C-153 Abstr 45th Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC Abstract C-153. [Google Scholar]
- 26.Lucasti C, Popescu I, Ramesh MK, Lipka J, Sable C. 2013. Comparative study of the efficacy and safety of ceftazidime/avibactam plus metronidazole versus meropenem in the treatment of complicated intra-abdominal infections in hospitalized adults: results of a randomized, double-blind, phase II trial. J Antimicrob Chemother 68:1183–1192. doi: 10.1093/jac/dks523. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez JA, Gonzalez Patzan LD, Stricklin D, Duttaroy DD, Kreidly Z, Lipka J, Sable C. 2012. Efficacy and safety of ceftazidime-avibactam versus imipenem-cilastatin in the treatment of complicated urinary tract infections, including acute pyelonephritis, in hospitalized adults: results of a prospective, investigator-blinded, randomized study. Curr Med Res Opin 28:1921–1931. doi: 10.1185/03007995.2012.748653. [DOI] [PubMed] [Google Scholar]
- 28.Jacquier H, Marcade G, Raffoux E, Dombret H, Woerther PL, Donay JL, Arlet G, Cambau E. 2013. In vivo selection of a complex mutant TEM (CMT) from an inhibitor-resistant TEM (IRT) during ceftazidime therapy. J Antimicrob Chemother 68:2792–2796. doi: 10.1093/jac/dkt278. [DOI] [PubMed] [Google Scholar]


