Abstract
We developed a two-step PCR-based strategy to detect genes encoding OqxAB, allowing a specific assignment of Tn6010-associated oqxAB in Enterobacteriaceae. Chromosomal location in this setup was confirmed by hybridization with I-CeuI-restricted genomes. This approach led us to find that Klebsiella sp. and Raoultella sp. reference strains chromosomally carried oqxAB.
TEXT
Four plasmid-mediated quinolone resistance (PMQR) mechanisms have been described in Enterobacteriaceae so far: Qnr, AAC(6′)-Ib-cr, QepA, and OqxAB (1–4). OqxAB belongs to the resistance-nodulation-cell-division (RND) family multidrug efflux pump, and OqxA and OqxB have shown high homology with the AcrAB pump from Escherichia coli (4). It has been shown that OqxAB-encoding genes may be located in Enterobacteriaceae on the conjugative plasmid pOLA52, within a composite transposon Tn6010 flanked by IS26 (5), but also natively on the chromosome in Klebsiella pneumoniae without association to a transposon (6, 7).
As for other PMQR, OqxAB alone confers low-level resistance to fluoroquinolones (8). Although the increased MIC of ciprofloxacin remains below the critical concentration, categorizing the enterobacterial isolate as susceptible, it has been shown that PMQR pose a challenge to antimicrobial treatments (9). Moreover, differentiation between plasmid-borne and chromosomal locations of these genes has a stake in preventing the spread of such resistance markers (10). All together, these data underscore the importance of setting up molecular detection assays. The aim of this study was to develop a reliable PCR-based method to detect not only oqxA and oqxB but also any associations with IS26, allowing the detection of the transposon-associated oqxAB (Fig. 1A). The mobile version of oqxAB, which is embedded in Tn6010, has been almost exclusively reported on plasmids (10–13). To our best knowledge, only two Salmonella isolates carrying a chromosomally encoded OqxAB associated with IS26 have been reported so far (14). It is noteworthy that the transposon-borne version is associated with boosted expression because of the lack of rarA that normally downregulates oqxR and the presence of several plasmid copies of the plasmid-borne transposon version (7).
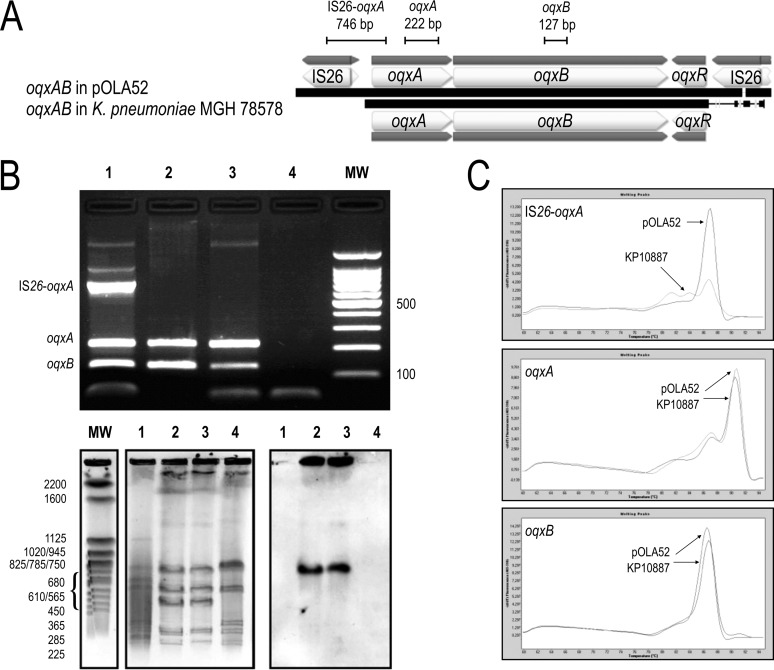
FIG 1.
(A) Schematic representation of oqxAB carried in pOLA52 and K. pneumoniae strain MGH 78578 (GenBank accession no. NC_009648.1). Lines indicate the size of the PCR fragments. (B) Top, agarose gel electrophoresis (2%) used for the separation of multiplex PCR products. Lane 1, E. coli CSH26/pOLA52; lanes 2 and 3, K. pneumoniae KP10887; lane 4, E. coli CIP 54.8T; MW, molecular weight marker (100-bp ladder; Invitrogen, Cergy-Pontoise, France). Bottom, PFGE migration profiles of I-CeuI restricted whole-cell DNAs. Lane 1, E. coli CSH26/pOLA52; lanes 2 and 3, K. pneumoniae KP10887; lane 4, E. coli CIP 54.8T; MW, Saccharomyces cerevisiae chromosomes (Bio-Rad Laboratories, Hercules, CA, USA) used as a molecular weight marker. The set on the right shows hybridization performed with a specific probe for the oqxB gene. (C) Derivative fluorescence melting curve (melting peak) for IS26-oqxA, oqxA, and oqxB in SYBR green real-time PCR with LC480. The melting temperatures (Tm) were 86.86°C, 90.79°C, and 86.16°C for IS26-oqxA, oqxA, and oqxB, respectively.
First, we carried out a multiplex conventional PCR to detect oqxA, oqxB, and an amplification product covering IS26 and oqxA. DNA extractions were all performed using the NucliSens easyMAG (bioMérieux, Marcy l'Etoile, France), according to the manufacturer's recommendations. Multiplex PCRs were run using the MyCycler thermocycler (Bio-Rad, Marnes-la-Coquette, France). Amplicons of 222 bp, 127 bp, and 746 bp targeting oqxA, oqxB, and the region spanning the IS26 right end and oqxA 5′ terminus, respectively (Fig. 1A), were generated in a 50-μl PCR that contained 5 μl of target DNA, 5 μl of PCR buffer, 0.5 μl of MgCl2 (25 mM), 0.5 μl of deoxynucleoside triphosphate (dNTP) (25 mM), 25 or 100 pmol of each primer (Table 1), 0.5 μl of Taq polymerase (5 U/μl), and 26.5 μl of sterile water. The PCRs were run using the following temperature-time profile: 95°C for 3 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. PCR products of the expected size were verified by electrophoresis in a 2% agarose gel. The control strains used for setting up the multiplex PCR were E. coli CSH26/pOLA52 (4), a clinical isolate of K. pneumoniae (KP10887) that we previously detected as positive for oqxAB (personal data), and E. coli CIP 54.8T as a negative control. An IS26-oqxA fragment specific to Tn6010 was amplified only in the plasmid-borne control E. coli CSH26/pOLA52 (Fig. 1B). The hybridization of I-CeuI pulsed-field gel electrophoresis (PFGE) profiles using an oqxB probe showed a chromosomal location only for K. pneumoniae KP10887 (15).
TABLE 1.
Primers used in this study
| Target | Primer | Primer sequence (5′ to 3′)a | Tm (°C) | Concn (pmol/reaction) | Product size (bp) |
|---|---|---|---|---|---|
| IS26-oqxA | IS26-oqxA_Fm2 | GTTTTTCCATTTCAGGCGCATA | 60 | 100b | 746 |
| IS26-oqxA_Rm | TTCACTTTATCAATGTATCCCGAGAC | 60 | 100b | ||
| oqxA | oqxA_Fm5 | GCCAAAACRCAGGCCAGYCT | 60 | 25 | 222 |
| oqxA_Rm3 | TCARCGCSCGGCTGGCGCG | 60 | 25 | ||
| oqxB | oqxB_Fm3 | GGCTGGATTTTCCGTCCGTT | 60 | 25 | 127 |
| oqxB_Rm4 | GCGGCRCARAGCAGCAG | 60 | 25 |
Degenerate residues are underlined. All primers were designed using Geneious 7.1 (Biomatters Ltd., Auckland, New Zealand).
These primer concentrations were used for the multiplex PCR, while all primers were used at 25 pmol/reaction in the simplex real-time PCR.
Second, we succeeded in setting up a simplex real-time PCR targeting each amplicon described above. These assays were carried out with the LightCycler 480 (Roche Molecular Diagnostics, Germany) using the same primers as described above (Table 1), with a PCR mix as follows: 2 μl of target DNA, 10 μl of 1× Kapa SYBR Fast quantitative PCR (qPCR) master mix (Kapa Biosystems, Wilmington, MA, USA), 0.4 μl of each primer, and 7.2 μl of sterile water. The PCR thermocycling program was as follows: 95°C for 7 min, followed by 40 cycles of 95°C for 10 s, 52°C for 30 s, and then melting resolution from 60°C to 98°C. As shown in Fig. 1C, distinct peaks were clearly distinguished for each amplicon except IS26-oqxA for K. pneumoniae KP10887, demonstrating the absence of a neighboring IS26.
According to the prevalence of oqxAB (6, 16, 17), we proposed to combine our assays into a two-step strategy, with screening of oqxAB by detecting oqxB in real-time PCR and then determination of oqxAB location only on strains that were positive by multiplex PCR. This method was applied to (i) 25 E. coli, 25 K. pneumoniae, and 3 Raoultella ornithinolytica strains prospectively isolated from urine samples from patients hospitalized in our emergency room in December 2014 and April 2015, (ii) 38 E. coli and 35 K. pneumoniae strains whose epidemiological links were studied previously (18), and (iii) the reference strains of the related genera Klebsiella and Raoultella (19) (Klebsiella oxytoca CIP 103434T, K. pneumoniae subsp. ozaenae CIP 52.211T, K. pneumoniae subsp. pneumoniae CIP 82.91T, K. pneumoniae subsp. rhinoscleromatis CIP 52.210T, Klebsiella michiganensis CIP 110787T, Klebsiella singaporensis CIP 108642T, Klebsiella variicola CIP108585T, R. ornithinolytica CIP 103364T, Raoultella planticola CIP 100751T, and Raoultella terrigena CIP 80.7T). For all the strains with no detection of IS26-oqxA using our method, we confirmed the chromosomal location of oqxAB by amplifying a 1.3 kb-long fragment overlapping the upstream region of oqxA that matches with the chromosome and oqxA.
Among the E. coli clinical strains, we did not detect any oqxAB genes. However, all the clinical K. pneumoniae isolates (61/61) carried oqxAB without detection of Tn6010. Interestingly, all the Klebsiella sp. and Raoultella sp. reference strains carried oqxAB in their chromosome, indicating normal housekeeping functions in these specimens.
Norman et al. (5) have proposed that K. pneumoniae may be a reservoir for oqxAB and that an oqxAB-carrying plasmid, such as pOLA52, may have arisen by capture from the chromosome of K. pneumoniae recruited by IS26. Nonetheless, some studies failed to detect oqxAB in all the K. pneumoniae isolates studied (6, 16, 17). This raised the question of the chromosomal location of oqxAB in K. pneumoniae, which might be particular not only for capture from the chromosome but also for its ability to lose oqxAB. However, it is noteworthy that a lack of detection in previous studies might have resulted from inadequate primers.
The method we describe here led us to show that (i) all Klebsiella spp. (clinical, n = 61; reference, n = 7) and Raoultella spp. (reference, n = 3) strains harbored oqxAB as part of their native chromosome, and (ii) the degenerate primers used in the present study allowed the detection of oqxAB in all cases. Moreover, our findings highlight the hypothesis stated by Norman et al. (5) and also demonstrate the importance of discriminating native and Tn6010-associated oqxAB regarding the overexpression of the mobile version.
ACKNOWLEDGMENTS
We thank Janick Madoux from CHU Reims for her excellent technical assistance. We also thank Hélène Moret and Lydie Morcrette for their helpful discussion.
This work was supported by an annual grant from Université de Reims Champagne-Ardenne (EA 4687).
REFERENCES
- 1.Martínez-Martínez L, Pascual A, Jacoby GA. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 2.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 3.Perichon B, Courvalin P, Galimand M. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51:2464–2469. doi: 10.1128/AAC.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother 48:3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norman A, Hansen LH, She Q, Sørensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53:3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70:81–88. doi: 10.1093/jac/dku340. [DOI] [PubMed] [Google Scholar]
- 8.Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 9.Guillard T, Cambau E, Chau F, Massias L, de Champs C, Fantin B. 2013. Ciprofloxacin treatment failure in a murine model of pyelonephritis due to an AAC(6′)-Ib-cr-producing Escherichia coli strain susceptible to ciprofloxacin in vitro. Antimicrob Agents Chemother 57:5830–5835. doi: 10.1128/AAC.01489-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen LH, Sørensen SJ, Jørgensen HS, Jensen LB. 2005. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb Drug Resist 11:378–382. doi: 10.1089/mdr.2005.11.378. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y, Wang M. 2012. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother 67:1655–1659. doi: 10.1093/jac/dks086. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Chen Z, Chen S, Deng Y, Liu Y, Tian W, Huang X, Wu C, Sun Y, Sun Y, Zeng Z, Liu JH. 2010. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother 54:4219–4224. doi: 10.1128/AAC.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong MHY, Chan EW, Liu LZ, Chen S. 2014. PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella Typhimurium. Front Microbiol 5:521. doi: 10.3389/fmicb.2014.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong MHY, Chen S. 2013. First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob Agents Chemother 57:658–660. doi: 10.1128/AAC.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillard T, Cambau E, Neuwirth C, Nenninger T, Mbadi A, Brasme L, Vernet-Garnier V, Bajolet O, de Champs C. 2012. Description of a 2,683-base-pair plasmid containing qnrD in two Providencia rettgeri isolates. Antimicrob Agents Chemother 56:565–568. doi: 10.1128/AAC.00081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Luo Y, Li J, Ma Y, Hu C, Jin S, Ye L, Cui S. 2010. Characterization of clinical Escherichia coli isolates from China containing transferable quinolone resistance determinants. J Antimicrob Chemother 65:453–459. doi: 10.1093/jac/dkp478. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez-Martínez JM, Díaz-de-Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F, Rodríguez Baño J, Martínez-Martínez L, Pascual Á. 2013. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother 68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]
- 18.Guillard T, Bertrand X, de Champs C, Cholley P, Bajolet O, Gbaguidi-Haore H. 2014. aac(6′)-Ib-cr is the major plasmid-mediated quinolone resistance determinant in extended-spectrum-β-lactamase-producing Escherichia coli in eastern France. J Glob Antimicrob Resist 2:111–113. doi: 10.1016/j.jgar.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Drancourt M, Bollet C, Carta A, Rousselier P. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol 51:925–932. doi: 10.1099/00207713-51-3-925. [DOI] [PubMed] [Google Scholar]