Abstract
Multiresistant Acinetobacter baumannii, a common etiologic agent of severe nosocomial infections in compromised hosts, usually harbors aac(6′)-Ib. This gene specifies resistance to amikacin and other aminoglycosides, seriously limiting the effectiveness of these antibiotics. An antisense oligodeoxynucleotide (ODN4) that binds to a duplicated sequence on the aac(6′)-Ib mRNA, one of the copies overlapping the initiation codon, efficiently inhibited translation in vitro. An isosequential nuclease-resistant hybrid oligomer composed of 2′,4′-bridged nucleic acid-NC (BNANC) residues and deoxynucleotides (BNANC-DNA) conjugated to the permeabilizing peptide (RXR)4XB (“X” and “B” stand for 6-aminohexanoic acid and β-alanine, respectively) (CPPBD4) inhibited translation in vitro at the same levels observed in testing ODN4. Furthermore, CPPBD4 in combination with amikacin inhibited growth of a clinical A. baumannii strain harboring aac(6′)-Ib in liquid cultures, and when both compounds were used as combination therapy to treat infected Galleria mellonella organisms, survival was comparable to that seen with uninfected controls.
INTRODUCTION
Acinetobacter baumannii is an opportunistic human pathogen, mainly nosocomial, that causes bacteremia, meningitis, urinary tract infections, pneumonia, and necrotizing fasciitis among other infections (1–4). Multidrug-resistant A. baumannii strains are increasingly found in hospitals, complicating treatment of the infections they cause (4). Antisense technologies could be a path for designing new therapeutic strategies to overcome this problem. Options include the silencing of one or more essential genes (5–12) or the silencing of one or more resistance genes to induce phenotypic conversion to susceptibility (13–16). In the latter case, the antisense compound would be administered in combination with the appropriate antibiotic. However, in spite of important advances, silencing of bacterial genes by antisense oligomers is far from reaching its full potential (10). The main antisense mechanisms of gene silencing include degradation of the target mRNA by double-stranded RNA (dsRNA)-specific RNase, RNase H, or RNase P and steric hindrance of translation (interference with assembly of the ribosome or translation arrest) (10, 17). Practical application of any of these strategies requires that the antisense compounds resist the action of the ubiquitous nucleases and reach the cytosol to exert their action.
There are numerous nuclease-resistant nucleotide analogs available that are adequate for different antisense strategies (10, 18, 19). For example, hybrid molecules containing locked nucleic acid and deoxyribonucleotide residues (LNA-DNA) in different configurations have been successfully utilized in bacteria and eukaryotes (16, 20–23). New analogs related to LNAs, the 2′,4′-bridged nucleic acid-NC (BNANC) analogs (Fig. 1), that exhibit advantages such as higher binding affinity to a cRNA and excellent single-mismatch discriminating ability, have been recently introduced (24). Furthermore, tests carried out in mice showed that BNANC-based antisense molecules have minimal toxicity (25).
FIG 1.
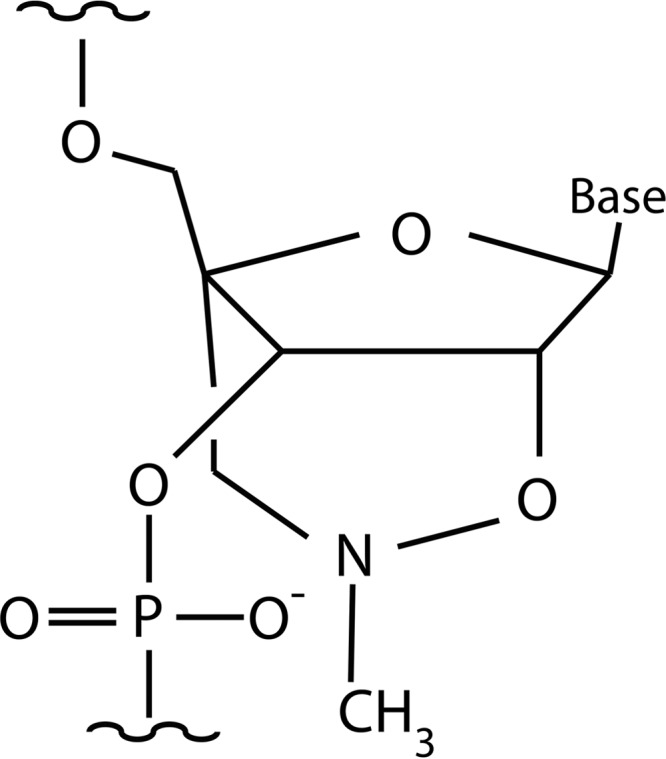
Chemical structure of a 2′,4′-BNANC residue.
The most successful strategy to guide antisense oligomers inside cells is through conjugation to cell-penetrating peptides, which consist of a small number (no more than 30) of amino acids, are amphipathic, and have a net positive charge (13, 18, 26, 27). LNA-DNA co-oligomers have been conjugated to cell-penetrating peptides and successfully used to inhibit gene expression (23). In particular, antisense oligonucleotide analogs conjugated to the (RXR)4 or (RXR)4XB peptides (R, arginine; X, 6-aminohexanoic acid; B, β-alanine) were efficiently guided inside A. baumannii and showed biological activity (18, 28).
A. baumannii A155, a strain isolated from a urinary sample (29), harbors aac(6′)-Ib, the most common amikacin (AMK) resistance gene found in Gram-negative pathogens (30, 31). This gene is present in the chromosome as well as in integrons, transposons, plasmids, genomic islands, and other genetic structures, is broadly distributed among Gram-negative species, and is characterized as being highly heterogeneous at the N terminus (32). However, the aac(6′)-Ib allele present in the chromosome of A. baumannii A155 is found in numerous Gram-negative bacterial species. In this work, we show that a (RXR)4XB-conjugated BNANC-DNA antisense co-oligomer that targets a duplicated region in the aac(6′)-Ib mRNA, one that includes the start codon and another that encompasses the codons specifying amino acids 7 to 11, induced susceptibility to AMK.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and permeabilizing peptide-conjugated oligonucleotide analogs.
A. baumannii A155 is a multidrug-resistant clinical strain isolated from a urine sample (29, 33). Plasmid pFC9 (34), which carries the aac(6′)-Ib gene, was used as the template for generating a linear DNA fragment consisting of the T7 promoter followed by aac(6′)-Ib that was used to synthesize the mRNA in vitro as described before (16). The oligonucleotides used as primers to generate the DNA template were 5′-TTGTAATACGACTCACTATAGGGAGAAAGCGCGTTACGCCGTGGGTCGATG and 5′-GGGTTAGGCATCACTGCGTGT. Antisense oligodeoxynucleotides (ODNs) were purchased from IDT (Integrated DNA Technologies) and (RXR)4XB-Cys-SMCC-C6 amino-2′,4′-BNANC-DNA (R, arginine; X, 6-aminohexanoic acid; B, β-alanine) (CPPBD) from Bio-Synthesis Inc. (Table 1). The chemical structure of a 2′,4′-BNANC residue is shown in Fig. 1.
TABLE 1.
Oligodeoxynucleotides and analogsa
| Name | Sequence | Length (bp) |
|---|---|---|
| ODN1 | TTTTACTGCTGCGTAACATCGTTGCTG | 20 |
| ODN2 | GTAACATCGTTGCTG | 15 |
| ODN3 | GCGTAACATCGTTGC | 15 |
| ODN4 | CTGCTGCGTAACATC | 15 |
| ODNS | GATGTTACGCAGCAG | 15 |
| ODNAP | AGCGGTAAGGCATCT | 15 |
| CPPBD4 | (RXR)4XB-Cys-SMCC-C6 amino-C+TGCT+GCGT+AACA+TC | 15 |
| CPPBDAP | (RXR)4XB-Cys-SMCC-C6 amino-A+GCGG+TAAG+GCAT+CT | 15 |
ODNs are oligodeoxynucleotides; CPPBDs are permeabilizing peptide-conjugated 2′,4′-bridged nucleic acid residue and deoxynucleotide hybrid compounds. R, arginine; X, 6-aminohexanoic acid; B, β-alanine, +N, BNANC; SMCC, sulfosuccinimidyl-trans-4-(N-maleimidomethyl)cyclohexane-1-carboxylate.
General procedures.
The presence of the A. baumannii A155 aac(6′)-Ib allele in other Gram-negative bacteria was determined using BLAST (35). In vitro translation of AAC(6′)-Ib was carried out using an Escherichia coli S30 Cell-Free Extract System for Circular DNA kit (Promega). The reactions were performed as recommended by the supplier in the presence of 10 μCi (specific activity, 1,175 Ci/mmol) of [35S]methionine (Perkin-Elmer) and, when indicated, 6.6 μM ODN or CPPBD compounds. The products were analyzed using sodium dodecyl sulfate-18% polyacrylamide gel electrophoresis (36). Gels were treated with En3hance (PerkinElmer) for 20 min, immersed in a solution containing 3.3% glycerol and 3.3% polyethylene glycol for 20 min, and dried, and radioactivity was detected on a phosphorimager (Cyclone Storage Phosphor system; Packard). Growth inhibition assays in the presence of (RXR)4XB–BNANC-DNA oligonucleotide analogs were carried out by inoculating Mueller-Hinton broth (100 μl) with the additions indicated in the text in microtiter plates using a BioTek Synergy 5 microplate reader (37). Culture procedures were carried out at 37°C with shaking, and optical density at 600 nm (OD600) was recorded every 20 min.
Infection assays.
A. baumannii cells were collected by centrifugation and resuspended in phosphate-buffered saline (PBS) (7.2 pH) or in PBS (7.2 pH) with the indicated additions as described before (38). Bacterial inocula were estimated spectrophotometrically at 600 nm. The injection site was swabbed with ethanol immediately prior to injection using a syringe pump (New Era Pump Systems, Inc., Wantagh, NY) with a 26-gauge by half-inch needle to deliver 5-μl inocula containing 5 × 105 (± 0.5 log) A. baumannii A155 cells into the hemocoel at the last left proleg. Ten healthy randomly selected final-instar G. mellonella larvae (Grubco, Fairfield OH), weighing 250 mg to 350 mg, were used for each group (n = 30) in experiments performed in triplicate. If >2 deaths occurred in either the PBS-injected or the no-injection control groups, that trial was omitted. After injection, the larvae were incubated at 37°C in the dark. Survival was assessed at 24-h intervals over 120 h with removal of dead caterpillars at time of inspection. The survival curves were plotted using the Kaplan-Meier method. A P value of ≤0.05 was considered statistically significant for the log-rank test of survival curves (SAS Institute Inc., Cary, NC).
RESULTS
AAC(6′)-Ib is a ubiquitous aminoglycoside-modifying enzyme that confers multiresistance to aminoglycosides, including AMK, to the majority of AAC(6′)-I-producing Gram-negative clinical isolates (30, 39). The purpose of this work was to inhibit production of this enzyme in the A. baumannii A155 clinical strain using antisense BNANC-DNA co-oligomers complementary to the region of initiation of translation. We selected BNANC-DNA molecules as antisense molecules because of their high affinity of binding to the sense molecule, their specificity, which may lead to a highly efficient and selective inhibitory effect, and their low toxicity (24, 25).
Since numerous variants of aac(6′)-Ib have been identified, several of them differing at the N terminus (32, 40), we decided to find out if a potential antisense molecule that inhibits expression of the resistance gene could have applications beyond this isolate. For this, we determined if the aac(6′)-Ib allele found in A. baumannii A155 is present in other Gram-negative clinical isolates or is unique to this strain. A BLAST comparison of the nucleotides in the sequence of the complete gene plus 16 nucleotides upstream of the start codon showed 27 identical sequences and 39 that have 100% coverage of the sequence and 99% identity (see Table S1 in the supplemental material). This allele of the gene is found in chromosomes and plasmids in diverse strains of Gram-negative species, including Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, K. oxytoca, E. coli, E. fergusonii, Enterobacter cloacae, Pseudomonas aeruginosa, A. baumannii, Shigella flexneri, Aeromonas allosaccharophila, and Burkholderia cepacia. In addition, sequences targeted by some of the oligonucleotides tested are also present in other alleles of this gene such as those present in InV117, an integron found in a ca. 150-kbp Vibrio cholerae O1 biotype El Tor plasmid (34), and in In116, an integron described in Morganella morganii (41).
Inhibition of translation in vitro.
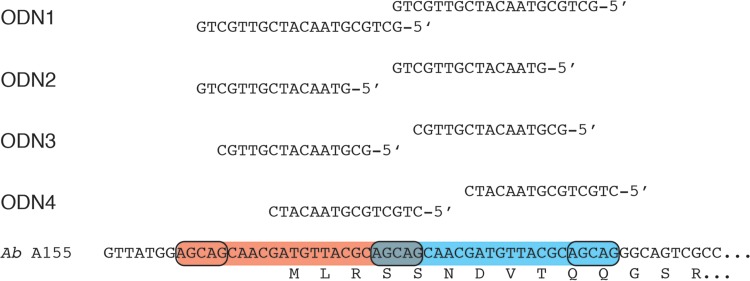
We first tested oligodeoxynucleotides (ODNs) complementary to a region encompassing the start codon where there is a 19-bp tandem duplication (Fig. 2). As a consequence, each antisense ODN was able to bind to two regions, one of which included the initiation codon (Fig. 2). Cell extracts were incubated in the presence of the mRNA with or without addition of the ODNs. The in vitro synthesis reaction produced two protein bands of ca. 20 and 25 kDa, respectively, and a smaller one of ca. 15 kDa (Fig. 3). Although we do not know why we systematically observed these products, we think that the ca. 25-kDa band corresponds to the full protein whereas the ca. 20- and 15-kDa proteins may be products of premature termination. Figure 3A shows that all four ODNs tested were robust inhibitors of translation of the aac(6′)-Ib mRNA but ODN1 and ODN4 showed the strongest activity. Since ODN4 was the shorter of the two ODNs, we selected this compound to carry out further assays.
FIG 2.
Nucleotide sequences targeted by antisense ODNs. The nucleotide sequences around the initiation codon of the A. baumannii (Ab) A155 aac(6′)-Ib gene are shown at the bottom of the figure. The duplicated sequences are within red and blue boxes. The sequences of the ODNs tested and the locations of antisense regions are shown at the top.
FIG 3.
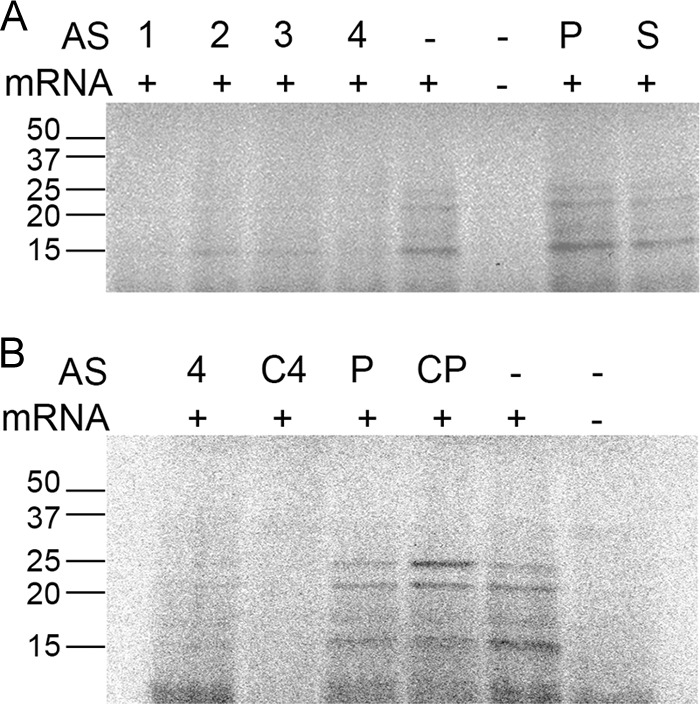
In vitro activities of ODNs. Cell-free translation reactions were carried out as described in Materials and Methods in the absence (−) or presence (+) of the indicated ODNs at 6.6 μM. The products were processed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the radioactivity was detected with a phosphorimager. A control lacking aac(6′)-Ib mRNA was included (−). (A) “AS” stands for “antisense,” and the numbers indicate the ODN numbers corresponding to those shown in Fig. 2. “P” and “S” stand for ODNs antisense to a phoA sequence and sense sequence, respectively. (B) “AS” stands for “antisense.” “C4” and “CP” stand for “CPPBD4” and “CPPBDAP,” respectively.
Since therapeutic use of oligonucleotides on bacterial systems requires the oligomers to be resistant to nucleases and to reach the cytoplasm, we designed a bridged nucleic acid-NC (BNANC) DNA (BNANC-DNA) hybrid oligomer with the permeabilizing peptide (RXR)4XB covalently bound to the 5′ end (CPPBD4; Table 1). A compound with the same BNANC-DNA configuration but with a nucleotide sequence antisense to a region of phoA was used as a negative control (CPPBDAP; Table 1). CPPBD4 was able to inhibit translation of the aac(6′)-Ib mRNA in the cell-free system with the same efficiency as that observed when the antisense molecule was DNA (Fig. 3B). As expected, addition of CPPBDAP to the reaction did not affect translation (Fig. 3B).
Inhibition of AMK resistance of A. baumannii A155 cells in culture.
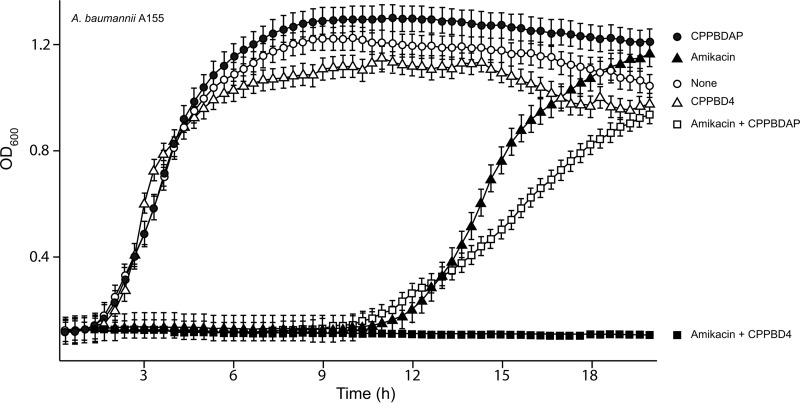
The ability of CPPBD4 to inhibit expression of resistance to AMK in A. baumannii A155 (MIC, 12 μg/ml) was assessed by performing growth curve experiments in the presence or absence of the antisense compound. Figure 4 shows that A. baumannii A155 cells cultured in the presence of AMK displayed a longer lag time than cells that were cultured in the absence of the antibiotic but reached the same OD600 at the stationary phase and displayed approximately the same doubling time. Figure 4 also shows that addition of CPPBD4 or CPPBDAP did not significantly affect the growth of A. baumannii A155 cells. However, when CPPBD4 was added to cultures containing AMK, there was complete growth inhibition for the duration of the experiment. Conversely, addition of the control CPPBDAP did not significantly modify the growth curve. These results strongly suggest that the peptide permeabilizer-conjugated BNANC-DNA compounds reach the cell cytosol and that CPPBD4 interferes with expression of resistance to AMK.
FIG 4.
Effect of CPPBD4 on resistance to AMK. A. baumannii A155 was cultured in 100 μl Mueller-Hinton broth in microtiter plates at 37°C, with the additions indicated in the figure, and the OD600 was determined every 20 min. CPPBD compounds were added at 0.5 μM and AMK at 4 μg/ml.
Effect of CPPBD4 in an experimental infection.
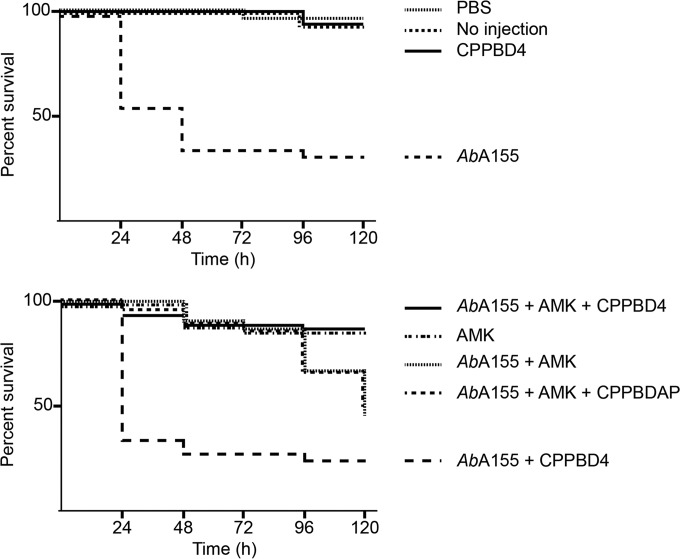
The potential capability of CPPBD4 combined with AMK for use as a therapeutic agent against A. baumannii A155 was examined using the G. mellonella virulence model, which has been extensively validated for the study of host-pathogen interactions and to determine the efficacy of antibiotic treatments against numerous pathogens, including A. baumannii (38, 42–44). Figure 5 shows that infection with A. baumannii A155 bacteria resulted in a significant increase in mortality compared with larvae that were not injected with the bacteria or were injected with sterile PBS. Similar virulence results were observed in those groups injected with CPPBD4. In the group treated with AMK, high mortality was also observed, although with a delay with respect to the negative control. This delay seems to correspond with the extension of the lag time observed in cultures when AMK was added to the media (see Fig. 4). The group treated with the combination of AMK plus CPPBD4 showed a significant increase in survival rates comparable to those seen with the controls injected with PBS or not injected. As expected, the group treated with AMK plus CPPBDAP showed high levels of mortality similar to those observed in the presence of AMK. The results described in this section show that the new hybrid analogs composed of BNANC and DNA, when conjugated to a permeabilizing peptide, can reach the A. baumannii cytosol and efficiently exert an antisense effect in tests using the G. mellonella model of infection.
FIG 5.
G. mellonella infection and treatment assays. Final-instar larva groups of 10 individuals were injected with the components shown in the figure, and a control group was not injected (“No injection”). The concentrations of the injected components were 10 mg AMK/kg of body weight and 0.5 μM CPPBD4 and CPPBDAP. The larvae were incubated at 37°C in the dark, and survival was recorded at 24-h intervals over 120 h.
DISCUSSION
Although the most obvious consequence of the increase in the number of multiresistant bacterial species is the complication of treatment of infectious diseases, the problem also threatens medical procedures such as surgery, cancer treatment, transplants, prosthetic replacements, care for premature infants, and some dentistry procedures (45). Part of the management of this problem could be the extension of the useful life of existing antibiotics by finding inhibitors of the resistance mechanisms or their expression (30, 46, 47). Here we tested one of the latest oligonucleotide analogs, a hybrid oligomer composed of 2′,4′-bridged nucleic acid-NC residues and deoxynucleotides conjugated to the permeabilizing peptide (RXR)4XB, as an antisense inhibitor of resistance to AMK mediated by AAC(6′)-Ib, one of the most widespread aminoglycoside-modifying enzymes (30, 39). We used the clinical A. baumannii A155 strain (29, 37), which carries an aac(6′)-Ib allele that has been characterized as possessing a sequence duplication encompassing the initiation codon. As determined by BLAST analysis, this is a quite common variant present in most Gram-negative species. We took advantage of the duplication to design antisense oligonucleotides that target this sequence and therefore can bind simultaneously to two regions in the same mRNA molecule. ODNs antisense to this region were robust inhibitors of expression of the gene in vitro. On the basis of these results, we speculate that the mechanism of inhibition of resistance to AMK is interference with ribosome assembly and/or steric hindrance of translation. Although we cannot discount a level of contribution of RNase H-mediated mRNA degradation, previous results obtained testing LNA-DNA co-oligomers with various configurations that showed that only gapmers with at least 6 contiguous deoxynucleotides induce significant RNase H activity (48–50) would discourage interpretation of this as a significant mechanism.
Since it has been shown that a successful solution to the problem of cellular uptake of oligonucleotide analogs, such as peptide nucleic acids or phosphorodiamidate morpholino oligomers, was their conjugation to permeabilizing peptides (13, 51), a BNANC-DNA hybrid co-oligomer with the most active sequence, that of ODN4, was conjugated to the (RXR)4XB peptide. This compound, CPPBD4, was then tested for its ability to reduce the levels of resistance to AMK of A. baumannii A155 cells. In combination with AMK, CPPBD4 showed sequence-specific inhibition of growth of A. baumannii A155 cells in culture and a reduction of their virulence in tests in the G. mellonella infection model. These results, taken together with those indicating that BNA-based compounds exhibit low toxicity for the host (25), suggest that hybrid oligomers composed of 2′,4′-bridged nucleic acid-NC residues and deoxynucleotides conjugated to permeabilizing peptides can be a viable option to develop antisense therapeutics. Previous studies showed that there are several variables that affect the activity of antisense compounds. For example, the efficiency of LNA-DNA hybrid compounds as antisense molecules is dependent on the configuration of the residues (16) and the efficiency of antisense peptide-phosphorodiamidate morpholino oligomers has been shown to be dependent on the composition of the permeabilizing peptide (52). Therefore, our future experiments will include examining a variety of permeabilizing peptides and oligomer configurations that will permit identification of the best compounds to treat different Gram-negative species.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant 2R15AI047115-04 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to M.E.T.), and by Miami University research funds.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01304-15.
REFERENCES
- 1.Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. 2012. The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartstein AI, Rashad AL, Liebler JM, Actis LA, Freeman J, Rourke JW Jr, Stibolt TB, Tolmasky ME, Ellis GR, Crosa JH. 1988. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilator circuits and resuscitation bags. Am J Med 85:624–631. doi: 10.1016/S0002-9343(88)80233-X. [DOI] [PubMed] [Google Scholar]
- 3.Charnot-Katsikas A, Dorafshar AH, Aycock JK, David MZ, Weber SG, Frank KM. 2009. Two cases of necrotizing fasciitis due to Acinetobacter baumannii. J Clin Microbiol 47:258–263. doi: 10.1128/JCM.01250-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai H, You Y, Yan H, Meng J, Xue X, Hou Z, Zhou Y, Ma X, Sang G, Luo X. 2012. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials 33:659–667. doi: 10.1016/j.biomaterials.2011.09.075. [DOI] [PubMed] [Google Scholar]
- 6.Dryselius R, Aswasti SK, Rajarao GK, Nielsen PE, Good L. 2003. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides 13:427–433. doi: 10.1089/154545703322860753. [DOI] [PubMed] [Google Scholar]
- 7.Kurupati P, Tan KS, Kumarasinghe G, Poh CL. 2007. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob Agents Chemother 51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang S, He Y, Xia Y, Wang H, Wang L, Gao R, Zhang M. 2015. Inhibiting the growth of methicillin-resistant Staphylococcus aureus in vitro with antisense peptide nucleic acid conjugates targeting the ftsZ gene. Int J Infect Dis 30:1–6. doi: 10.1016/j.ijid.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Panchal RG, Geller BL, Mellbye B, Lane D, Iversen PL, Bavari S. 2012. Peptide conjugated phosphorodiamidate morpholino oligomers increase survival of mice challenged with Ames Bacillus anthracis. Nucleic Acid Ther 22:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen LC, Sperling-Petersen HU, Mortensen KK. 2007. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb Cell Fact 6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawyer AJ, Wesolowski D, Gandotra N, Stojadinovic A, Izadjoo M, Altman S, Kyriakides TR. 2013. A peptide-morpholino oligomer conjugate targeting Staphylococcus aureus gyrA mRNA improves healing in an infected mouse cutaneous wound model. Int J Pharm 453:651–655. doi: 10.1016/j.ijpharm.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sala CD, Soler-Bistue AJ, Korprapun L, Zorreguieta A, Tolmasky ME. 2012. Inhibition of cell division induced by external guide sequences (EGS Technology) targeting ftsZ. PLoS One 7:e47690. doi: 10.1371/journal.pone.0047690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. 2001. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol 19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C, Salavati R, Altman S. 1997. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci U S A 94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarno R, Ha H, Weinsetel N, Tolmasky ME. 2003. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob Agents Chemother 47:3296–3304. doi: 10.1128/AAC.47.10.3296-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soler Bistué AJ, Martin FA, Vozza N, Ha H, Joaquin JC, Zorreguieta A, Tolmasky ME. 2009. Inhibition of aac(6′)-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc Natl Acad Sci U S A 106:13230–13235. doi: 10.1073/pnas.0906529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies-Sala C, Soler-Bistue A, Bonomo RA, Zorreguieta A, Tolmasky ME. 2015. External guide sequence technology: a path to development of novel antimicrobial therapeutics. Ann N Y Acad Sci doi: 10.1111/nyas.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai H, Luo X. 2012. Antisense antibacterials: from proof-of-concept to therapeutic perspectives, p 319–344. In Bobbarala V. (ed), A search for antibacterial agents InTech, Rijeka, Croatia. [Google Scholar]
- 19.Kurreck J. 2003. Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem 270:1628–1644. [DOI] [PubMed] [Google Scholar]
- 20.Mutso M, Nikonov A, Pihlak A, Zusinaite E, Viru L, Selyutina A, Reintamm T, Kelve M, Saarma M, Karelson M, Merits A. 2015. RNA interference-guided targeting of hepatitis C virus replication with antisense locked nucleic acid-based oligonucleotides containing 8-oxo-dG modifications. PLoS One 10:e0128686. doi: 10.1371/journal.pone.0128686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgado E, Okabe H, Preziosi M, Russell JO, Alvarado TF, Oertel M, Nejak-Bowen KN, Zhang Y, Monga SP. 2015. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J Hepatol 62:380–387. doi: 10.1016/j.jhep.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Martino MT, Gulla A, Gallo Cantafio ME, Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D, Rossi M, Frandsen NM, Tagliaferri P, Tassone P. 2014. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One 9:e89659. doi: 10.1371/journal.pone.0089659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng J, Da F, Ma X, Wang N, Wang Y, Zhang H, Li M, Zhou Y, Xue X, Hou Z, Jia M, Luo X. 2015. Antisense growth inhibition of methicillin-resistant Staphylococcus aureus by locked nucleic acid conjugated with cell-penetrating peptide as a novel FtsZ inhibitor. Antimicrob Agents Chemother 59:914–922. doi: 10.1128/AAC.03781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman SM, Seki S, Utsuki K, Obika S, Miyashita K, Imanishi T. 2007. 2′,4′-BNA(NC): a novel bridged nucleic acid analogue with excellent hybridizing and nuclease resistance profiles. Nucleosides Nucleotides Nucleic Acids 26:1625–1628. doi: 10.1080/15257770701548980. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto T, Harada-Shiba M, Nakatani M, Wada S, Yasuhara H, Narukawa K, Sasaki K, Shibata MA, Torigoe H, Yamaoka T, Imanishi T, Obika S. 2012. Cholesterol-lowering action of BNA-based antisense oligonucleotides targeting PCSK9 in atherogenic diet-induced hypercholesterolemic mice. Mol Ther Nucleic Acids 1:e22. doi: 10.1038/mtna.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Järver P, Coursindel T, Andaloussi SE, Godfrey C, Wood MJ, Gait MJ. 2012. Peptide-mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Mol Ther Nucleic Acids 1:e27. doi: 10.1038/mtna.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tilley LD, Hine OS, Kellogg JA, Hassinger JN, Weller DD, Iversen PL, Geller BL. 2006. Gene-specific effects of antisense phosphorodiamidate morpholino oligomer-peptide conjugates on Escherichia coli and Salmonella enterica serovar Typhimurium in pure culture and in tissue culture. Antimicrob Agents Chemother 50:2789–2796. doi: 10.1128/AAC.01286-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geller BL, Marshall-Batty K, Schnell FJ, McKnight MM, Iversen PL, Greenberg DE. 2013. Gene-silencing antisense oligomers inhibit Acinetobacter growth in vitro and in vivo. J Infect Dis 208:1553–1560. doi: 10.1093/infdis/jit460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arivett BA, Fiester SE, Ream D, Centrón D, Ramirez MS, Tolmasky ME, Actis LA. 2015. Draft genome of the multidrug-resistant Acinetobacter baumannii A155 clinical isolate. Genome Announc 3:e00212-15. doi: 10.1128/genomeA.00212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez MS, Nikolaidis N, Tolmasky ME. 2013. Rise and dissemination of aminoglycoside resistance: the aac(6′)-Ib paradigm. Front Microbiol 4:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramírez MS, Vilacoba E, Stietz MS, Merkier AK, Jeric P, Limansky AS, Marquez C, Bello H, Catalano M, Centron D. 2013. Spreading of AbaR-type genomic islands in multidrug resistance Acinetobacter baumannii strains belonging to different clonal complexes. Curr Microbiol 67:9–14. doi: 10.1007/s00284-013-0326-5. [DOI] [PubMed] [Google Scholar]
- 34.Soler Bistué AJ, Martin FA, Petroni A, Faccone D, Galas M, Tolmasky ME, Zorreguieta A. 2006. Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob Agents Chemother 50:1903–1907. doi: 10.1128/AAC.50.5.1903-1907.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Lin DL, Tran T, Alam JY, Herron SR, Ramirez MS, Tolmasky ME. 2014. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib by zinc: reversal of amikacin resistance in Acinetobacter baumannii and Escherichia coli by a zinc ionophore. Antimicrob Agents Chemother 58:4238–4241. doi: 10.1128/AAC.00129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiester SE, Nwugo CC, Penwell WF, Neary JM, Beckett AC, Arivett BA, Schmidt RE, Geiger SC, Connerly PL, Menke SM, Tomaras AP, Actis LA. 2015. Role of the carboxyl terminus of SecA in iron acquisition, protein translocation and virulence of the bacterial pathogen Acinetobacter baumannii. Infect Immun 83:1354–1365. doi: 10.1128/IAI.02925-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casin I, Bordon F, Bertin P, Coutrot A, Podglajen I, Brasseur R, Collatz E. 1998. Aminoglycoside 6′-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob Agents Chemother 42:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Power P, Galleni M, Di Conza J, Ayala JA, Gutkind G. 2005. Description of In116, the first blaCTX-M-2-containing complex class 1 integron found in Morganella morganii isolates from Buenos Aires, Argentina. J Antimicrob Chemother 55:461–465. doi: 10.1093/jac/dkh556. [DOI] [PubMed] [Google Scholar]
- 42.Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. 2012. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182:3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Infectious Diseases Society of America. 2010. The 10 × '20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis 50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 46.Labby KJ, Garneau-Tsodikova S. 2013. Strategies to overcome the action of aminoglycoside-modifying enzymes for treating resistant bacterial infections. Future Med Chem 5:1285–1309. doi: 10.4155/fmc.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi K, Caldwell SJ, Fong DH, Berghuis AM. 2013. Prospects for circumventing aminoglycoside kinase mediated antibiotic resistance. Front Cell Infect Microbiol 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kauppinen S, Vester B, Wengel J. 2005. Locked nucleic acid (LNA): high affinity targeting of RNA for diagnostics and therapeutics. Drug Discov Today Technol 2:287–290. doi: 10.1016/j.ddtec.2005.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sørensen MD, Kvaernø L, Bryld T, Håkansson AE, Verbeure B, Gaubert G, Herdewijn P, Wengel J. 2002. Alpha-L-ribo-configured locked nucleic acid (alpha-L-LNA): synthesis and properties. J Am Chem Soc 124:2164–2176. doi: 10.1021/ja0168763. [DOI] [PubMed] [Google Scholar]
- 50.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J. 2000. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A 97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geller BL, Deere JD, Stein DA, Kroeker AD, Moulton HM, Iversen PL. 2003. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrob Agents Chemother 47:3233–3239. doi: 10.1128/AAC.47.10.3233-3239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mellbye BL, Puckett SE, Tilley LD, Iversen PL, Geller BL. 2009. Variations in amino acid composition of antisense peptide-phosphorodiamidate morpholino oligomer affect potency against Escherichia coli in vitro and in vivo. Antimicrob Agents Chemother 53:525–530. doi: 10.1128/AAC.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.