Abstract
Corynebacterium striatum BM4687 was resistant to gentamicin and tobramycin but susceptible to kanamycin A and amikacin, a phenotype distinct among Gram-positive bacteria. Analysis of the entire genome of this strain did not detect any genes for known aminoglycoside resistance enzymes. Yet, annotation of the coding sequences identified 12 putative acetyltransferases or GCN5-related N-acetyltransferases. A total of 11 of these coding sequences were also present in the genomes of other Corynebacterium spp. The 12th coding sequence had 55 to 60% amino acid identity with acetyltransferases in Actinomycetales. The gene was cloned in Escherichia coli, where it conferred resistance to aminoglycosides by acetylation. The protein was purified to homogeneity, and its steady-state kinetic parameters were determined for dibekacin and kanamycin B. The product of the turnover of dibekacin was purified, and its structure was elucidated by high-field nuclear magnetic resonance (NMR), indicating transfer of the acetyl group to the amine at the C-3 position. Due to the unique profile of the reaction, it was designated aminoglycoside 3-N-acetyltransferase type XI.
INTRODUCTION
Nondiphtherial corynebacteria are ubiquitous microorganisms capable of colonizing the skin and mucous membranes. Most strains are members of the human microbiota and are rarely involved in clinical infections. Among them, Corynebacterium striatum, which is routinely isolated from wound exudates and often discarded as a contaminant, has been associated with serious infections in both immunocompetent and immunocompromised patients (1). This microorganism has been reported from various diseases such as bacteremia associated with central venous catheter infections, arthritis, vertebral osteomyelitis, meningitis, endocarditis, breast abscess, pleuropulmonary infections, peritonitis, chorioamnionitis, and prosthetic joint infections (2). This species remains susceptible to vancomycin, but resistance to various antibiotic classes has been reported, although not documented at the genetic level in most instances (3).
The resistance profile of C. striatum strain BM4687 to aminoglycosides is of interest. The strain was resistant to gentamicin and tobramycin but remained susceptible to kanamycin A and amikacin. This phenotype cannot be accounted for by known mechanisms of resistance to aminoglycosides in Gram-positive bacteria, which involve chemical modification of the drugs by specific enzymes. These are aminoglycoside 3′-O-phosphotransferase type III [APH(3′)-III], conferring resistance to kanamycin A; 4′-O-adenylyltransferase [ANT(4′)-Ia], which confers resistance to tobramycin but not to gentamicin; and the broad-spectrum bifunctional 6′-N-acetyltransferase-2″-O-phosphotransferase [AAC(6′)-APH(2″)], conferring resistance to kanamycin A, tobramycin, and gentamicin. In the case of C. striatum, only the presence of APH(3′)-III has been reported to date (4).
We undertook to identify the determinant responsible for aminoglycoside resistance in C. striatum BM4687 by a multidisciplinary approach involving whole-genome sequencing, cloning of the putative resistance gene, and characterization of the purified enzyme. The collective data revealed that the strain produced a new aminoglycoside 3-N-acetyltransferase designated type XI. The distribution of the aac(3)-XI gene in clinical isolates of C. striatum was also explored and is reported here.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Corynebacterium striatum BM4687 was isolated from the heel biopsy specimen of an 80-year-old diabetic woman at the Hôpital Saint-Joseph in Paris, France, in 2013. Twenty C. striatum isolates of distinct origins from our strain collection were screened for the presence of the resistance gene. Strains were grown in brain heart infusion (BHI) or Luria-Bertani (LB) broth and agar at 37°C. Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton (MH) agar according to the standards of the Comité de l'Antibiogramme de la Société Française de Microbiologie. MICs were determined by Etest (bioMérieux) on BHI agar and by microdilution in BHI (5).
DNA preparation and recombinant DNA techniques.
Isolation of total DNA, small- and large-scale preparation of plasmid DNA, digestion with restriction endonucleases, ligation with T4 DNA ligase, and transformation with recombinant plasmid DNA were carried out by standard methods. DNA amplification was performed in a GeneAmp PCR 9700 system (PerkinElmer Cetus) with Phusion high-fidelity DNA polymerase (Thermo Scientific). Plasmid DNA or PCR product sequencing was performed with a CEQ 8000 DNA analysis system automatic sequencer (Beckman Instruments).
Whole-genome sequencing and analysis.
The entire genome of C. striatum BM4687 was sequenced on an Illumina instrument after construction of a pair-end-indexed library producing approximately 900 Mb (∼320-fold coverage). The data were assembled using the Velvet assembler (https://www.ebi.ac.uk), leading to 56 scaffolds with a cumulative size of 2.9 Mb. To reduce the number of undetermined bases, GapCloser (http://soap.genomics.org.cn) was used. The annotation of coding sequences (CDS) and prediction of gene functions were performed using the MicroScope platform (http://www.genoscope.cns.fr/agc/microscope/home/index.php) (6).
The deduced acetyltransferases inferred from the sequences were compared with aminoglycoside acetyltransferases by the BLAST tool of the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov). Multiple sequence alignments and determination of the amino acid identities of the acetyltransferases were carried out by the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/) (7). The unrooted phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.0 aLRT) with the WAG matrix and a gamma correction for variable evolutionary rates (8).
Cloning of the aac(3)-XI gene.
The gene for the putative aminoglycoside acetyltransferase was amplified by PCR using genomic DNA from C. striatum BM4687 as a template and primers EcoRI AacF and BamHI AacR (Table 1), cloned in pCRBlunt, and sequenced. The 473-bp EcoRI-BamHI fragment was ligated in pUC18. To improve protein expression, a similar approach, using primers NdeI AacF and XhoI AacR (Table 1), was carried out for cloning the aac gene in expression vector pET28/16 (9).
TABLE 1.
Primers used for amplification, reverse transcription, and sequencing
| Locus | Primer name | Sequence (5′→3′)a | Positionb |
|---|---|---|---|
| aac(3) | EcoRI AacF | GGAATTCCGTTAAGGAGGCGGCATATGACTACAACCAAC | 988–1020 |
| BamHI AacR | GGGATCCGGCCTAAAGCTCCCGGATGTAGAGCAGCATG | 1458–1434 | |
| aac(3) | NdeI AacF | GGGCATATGACTACAACCAACGAGGATC | 1003–1026 |
| XhoI AacR | CCCTCGAGCTACTAAAGCTCCCGGATGTAGAG | 1452–1432 | |
| aac(3) | AacF | ATGACTACAACCAACGAGATC | 1006–1026 |
| aac(3) | AacR | CTAAAGCTCCCGGATGTAGAG | 1452–1432 |
| aac(3) | Aac intR | ACGTAGGCGAAACCAATATCGCC | 1193–1171 |
| tnpCx | TnpF | ATGTCATCGTCGATACCATCTGC | 387–409 |
| tnpCx | TnpR | AAGCTTTGGCATCGGCGTGCGTT | 865–843 |
| Kinase | KinR | GTTGCAACCTGGACCGAACCATTC | 1616–1593 |
Restriction sites are in bold; the ribosome-binding site is underlined; start and stop codons are in italics.
The coordinates refer to the 1,800-bp sequence presented in Fig. 2.
Assay for aminoglycoside-modifying enzyme.
The activity of aminoglycoside-N-acetyltransferase in bacterial extracts of Escherichia coli TOP10/pUC18Ωaac(3)-XI was detected by the phosphocellulose paper-binding method with [1-14C]acetyl coenzyme A (CoA) as the second substrate. The reaction was allowed to proceed for 30 min at 37°C (10).
RNA isolation and cDNA synthesis.
Total RNA from C. striatum BM4687 was extracted from exponentially grown bacteria (optical density at 600 nm [OD600] of ≈0.9) using the Fast RNA Pro Blue kit (Qbiogene). RNA samples were treated with the DNA-free kit (Ambion) to remove any genomic DNA carryover. The cDNA synthesis was carried out using either a random hexamer (Thermo Scientific RevertAid first-strand cDNA synthesis kit) or AacR- or TnpR-specific primers (Table 1).
Purification of aminoglycoside acetyltransferase.
Escherichia coli BL21 Star(DE3) was transformed with plasmid pET28/16Ωaac(3)-XI DNA. Bacteria were grown in LB medium supplemented with ampicillin (50 mg/liter) at 37°C to an OD600 of 0.7, protein expression was induced with 0.6 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), and bacteria were allowed to grow overnight at 20°C. Cells were harvested at 3,200 × g at 4°C for 20 min, resuspended in lysis buffer containing 25 mM HEPES (pH 7.0) and 200 mM NaCl supplemented with Complete EDTA-free protease inhibitor (Roche), and disrupted by sonication. Cellular debris was removed by centrifugation at 14,000 × g for 1 h at 4°C. The supernatant was incubated with 5 ml nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Qiagen) equilibrated in lysis buffer for 1 h at 4°C with gentle agitation. The resin-lysate slurry was poured into a column, and the flowthrough containing unbound proteins was discarded. The column was then washed with 50 ml buffer A (25 mM HEPES [pH 7.0], 200 mM NaCl, and 20 mM imidazole), and the enzyme was eluted with a 100-ml linear gradient from 0 to 100% buffer B (25 mM HEPES [pH 7.0], 200 mM NaCl, 250 mM imidazole) at 1 ml/min. Samples were analyzed by 15% SDS-PAGE, the fractions containing the enzyme were pooled and dialyzed against buffer B without imidazole, and the solution was concentrated by centrifugal filtration (3,000-molecular-weight cutoff [MWCO]). The protein was quantified using a bicinchoninic acid (BCA) assay (Thermo), stored at 4°C, and characterized within a week of purification.
Enzymatic modification of dibekacin and purification of the acetylated product.
Dibekacin was acetylated according to the methodology reported for other aminoglycoside acetyltransferases (11). The reaction was monitored using thin-layer chromatography (TLC) plates containing silica gel resolved in methanol-ammonia (5:2) and visualized with p-anisaldehyde. The enzyme was removed from the reaction mixture by centrifugal concentration (3,000 MWCO), and the filtrate was concentrated by rotary evaporation. The residue was suspended in 5 ml water and loaded on a silica gel in a 30-ml sintered-glass funnel. The sample was washed with 100 ml methanol-water (1:1) to remove acetyl coenzyme A (acetyl-CoA) and coenzyme A (CoASH). The modified aminoglycoside was eluted from a silica gel with 30 ml of methanol-water-ammonia (1:1:0.5). The eluent was concentrated by rotary evaporation and applied to a 20-ml weak cation-exchange column, Amberlite CG-50 (NH4+ form; Mallinckrodt). The column was washed with 150 ml water, and acetylated dibekacin was eluted with a stepwise gradient of ammonium hydroxide (0.06%, 0.125%, 0.25%, 0.5%, 0.75%, 1%, 1.5%, 2%, 5%). Fractions were analyzed by TLC using a solvent system of chloroform-water-17% ammonium hydroxide (2:2:1) and visualized by ninhydrin. Fractions containing acetylated dibekacin (1.5% NH4OH) were concentrated by rotary evaporation, lyophilized to dryness, and reconstituted in D2O.
NMR analysis of the structurally modified dibekacin.
All nuclear magnetic resonance (NMR) spectra, except a one-dimensional (1D) 13C{1H} spectrum, were recorded at 25°C on a Bruker Avance II 800 spectrometer equipped with a 5-mm triple resonance TCI (1H, 13C, 15N) cryoprobe and operating at a proton resonance frequency of 800.18 MHz. The 1D 13C{1H} spectrum was measured at 25°C using a Varian Inova 500 spectrometer operating at a proton resonance frequency of 499.98 MHz. The acetylated dibekacin structure (10 mg dissolved in 500 μl 99.996% D2O) was established by interpretation of 1D 1H and 13C{1H}, 2D homonuclear double-quantum-filtered correlation spectroscopy (DQF-COSY) and total correlation spectroscopy (TOCSY), heteronuclear 1H-13C heteronuclear single quantum coherence (HSQC) spectroscopy and heteronuclear multiple-bond correlation (HMBC), and 1H-15N HMBC spectra. All spectra were measured using standard pulse sequences (12–17) in the Bruker Topspin 3.2 p16 software. The 1D 1H, the 2D homonuclear, and the 1H dimension in 2D heteronuclear spectra were referenced to the residual H2O signal, δH = 4.7 ppm. The 1D 13C{1H} and the 13C and 15N dimensions in the 2D heteronuclear spectra were referenced indirectly (18).
Kinetic analysis of acetylation of dibekacin and kanamycin B.
The acetyltransferase activity was measured spectrophotometrically following production of CoASH, which is produced upon the transfer of the acetyl moiety to the aminoglycoside. The thiol group of CoASH reacts with DTNB [5,5′-dithiobis(2-nitrobenzoic acid); Sigma-Aldrich] to generate TNB (5-thio-2-nitrobenzoic acid) and a mixed disulfide. The generation of TNB can be monitored by absorbance at 412 nm (Δε412 = 14,150 M−1 cm−1). The acetylation solution contained 25 mM morpholineethanesulfonic acid (MES; pH 6.0), 80 μM acetyl-CoA, 500 μM DTNB, variable concentrations of dibekacin or kanamycin B (0 to 100 μM), and 300 nM enzyme in a total volume of 200 μl in a cuvette. The kinetic parameters for the consumption of acetyl-CoA were determined using a fixed concentration of dibekacin (200 μM) and varying the amounts of acetyl-CoA (0 to 120 μM). The real-time absorbance at 412 nm was monitored using a Cary 50 UV-visible (UV-Vis) spectrophotometer at room temperature for 5 min. The initial velocity was determined by linear regression and converted into micromolar per second using the molar extinction coefficient for TNB. The initial rates were plotted versus substrate concentration and were fitted by nonlinear regression to the Michaelis-Menten equation to determine Km and kcat.
Nucleotide sequence accession numbers.
The annotated genome sequence of C. striatum BM4687 has been deposited in the European Nucleotide Archive database under accession numbers CTEG01000001 to CTEG01000066.
RESULTS AND DISCUSSION
Identification of the putative aminoglycoside acetyltransferase.
Corynebacterium striatum clinical isolate BM4687 was resistant to gentamicin, tobramycin, sisomicin, dibekacin, and fortimicin but remained susceptible to kanamycin A, netilmicin, amikacin, apramycin, and neomycin B (Table 2), a resistance phenotype compatible with production of an aminoglycoside acetyltransferase (19). The strain was also resistant to tetracycline (MIC, 32 mg/liter), oxacillin (MIC, 12 mg/liter), and clindamycin (MIC, 256 mg/liter).
TABLE 2.
MICs of aminoglycosides against C. striatum and E. coli
| Strain | MIC (mg/liter) of antibiotica: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NEO B | KAN A | KAN B | TOB | DBK | AMK | GEN | SIS | NET | FOR | APR | |
| C. striatum ATCC 6940 | 0.06 | 0.250 | 0.250 | 0.125 | 0.250 | 0.250 | 0.125 | 0.125 | 0.06 | 0.250 | 0.125 |
| C. striatum BM4687 | 0.125 | 0.5 | 32 | 64 | >64 | 0.5 | 32 | 64 | 0.125 | 64 | 0.5 |
| E. coli/pUC18 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 4 |
| E. coli/pUC18Ωaac(3)-XI | 2 | 2 | 64 | >64 | >64 | 2 | 64 | >64 | 2 | >64 | 4 |
Abbreviations: AMK, amikacin; APR, apramycin; DBK, dibekacin; FOR, fortimicin; GEN, gentamicin; KAN A, kanamycin A; KAN B, kanamycin B; NEO B, neomycin B; NET, netilmicin; SIS, sisomicin; TOB, tobramycin.
The genome of clinical isolate BM4687 consisted of a 2,870,042-bp chromosome with a mean G+C content of 59.4%, a value identical to that of C. striatum ATCC 6940 (GenBank accession no. ACGE00000000.1). Unexpectedly, using the MicroScope platform for annotation of the coding sequences (CDS) (6), no aminoglycoside acetyltransferases could be inferred from the sequence. Among the 12 CDS annotated as putative acetyltransferases or GCN5-related N-acetyltransferases, 11 were found to be also present in other Corynebacterium genomes deposited in GenBank, as found by a Delta-BLAST analysis. The last CDS, annotated as a GCN5-related N-acetyltransferase (tag U2A4042v1_500015), was not part of any Corynebacterium genome but had 55 to 60% amino acid identity with other acetyltransferases present in the high-GC-content Gram-positive Actinomycetales order, such as Nocardioides sp. strain J54 (accession no. WP_028656429.1), Actinomyces naeslundii (EJN83915), Gordonia polyisoprenivorans (AFA71328), and Salinispora tropica (ABP56629). This protein was considered a good candidate for the putative aminoglycoside resistance enzyme of C. striatum BM4687.
The corresponding gene was part of a 17,676-bp contig, bracketed by those for two transposases, which was found in part in the genomes of other Corynebacterium spp., such as Corynebacterium singulare IBS B52218 (99% identity for a query cover of 70%, GenBank accession number CP010827.1), Corynebacterium urealyticum DSM 7109 (97% identity for a query cover of 87%, GenBank accession number AM94244.1), Corynebacterium jeikeium K411 (94% identity for a query cover of 62%, GenBank accession number CR931997.1), and C. striatum M82B (99% identity for a query cover of 66%, GenBank accession number AF024666.2). This contig, among other CDS, included tet(AB) for an ABC transporter conferring resistance to tetracycline and to the structurally unrelated β-lactam oxacillin (20) and erm(X), which confers resistance to clindamycin by production of a 23S rRNA adenine N-6-methyltransferase (4). These genes have previously been described as parts of complex mobile DNA elements (4) that are capable of transposition into the chromosomes of various Corynebacterium spp., such as Corynebacterium glutamicum (21), C. urealyticum (22), and C. jeikeium (23).
Cloning and expression of the putative aminoglycoside acetyltransferase gene.
The sequence for the putative acetyltransferase was amplified by PCR using genomic DNA from C. striatum BM4687 as a template, cloned in pCRBlunt, and sequenced. The 473-bp EcoRI-BamHI fragment was ligated to plasmid pUC18 and conferred on E. coli TOP10 a phenotype indistinguishable from that of BM4687, that is, resistance to gentamicin, tobramycin, sisomicin, dibekacin, and fortimicin, the MICs of kanamycin A, netilmicin, amikacin, apramycin, and neomycin B remaining unchanged (Table 2).
The transfer of the acetyl moiety from [1-14C]acetyl-CoA to various aminoglycosides catalyzed by crude extracts of E. coli TOP10/pUC18Ωaac(3)-XI confirmed an aminoglycoside acetyltransferase activity. Tobramycin, dibekacin, kanamycin B, sisomicin, gentamicin C1A, and fortimicin were substrates for the enzyme, whereas kanamycin A, amikacin, netilmicin, neomycin B, and apramycin were not.
Kinetic properties of the enzyme and chemical characterization of the acetylation.
The protein was purified by a His tag affinity Ni-NTA column eluted with imidazole. Approximately 40 mg of pure protein was produced from 1 liter of culture. The enzyme activity was measured spectrophotometrically, monitoring the quantitative production of CoASH from acetyl-CoA for acetylation of dibekacin and kanamycin B. The steady-state kinetics parameters for acetylation are given in Table 3, and the values for the two substrates were fairly comparable. The enzyme achieves saturation at low-micromolar levels of the drugs (Km range of 9 to 13 μM). Whereas the first-order rate constants for turnover (kcat) are not large, the catalytic efficiencies for the two substrates, kcat/Km values of (1.8 ± 0.2) × 104 M−1 s−1 for dibekacin and (1.7 ± 0.2) × 104 M−1 s−1 for kanamycin B, were similar and respectably high for acetyltransferase enzymes.
TABLE 3.
Steady-state kinetic parameters for the AAC(3)-XI-catalyzed reaction
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Acetyl-CoAa | 0.24 ± 0.01 | 2.7 ± 0.2 | (8.9 ± 0.8) × 104 |
| Dibekacin | 0.17 ± 0.01 | 9.2 ± 0.7 | (1.8 ± 0.2) × 104 |
| Kanamycin B | 0.23 ± 0.01 | 13.8 ± 1.2 | (1.7 ± 0.2) × 104 |
Kinetic parameters for the turnover of acetyl-CoA were determined using a fixed concentration of dibekacin, as described in Materials and Methods.
To elucidate the nature of the acetyltransferase reaction, we set out to characterize the structure of the product of turnover of dibekacin. A large-scale reaction was performed at room temperature over 10 h. The resulting product was isolated, purified by chromatography, and subjected to high-field NMR analysis for structure determination. The structure of acetylated dibekacin (Fig. 1) was determined by measurements and analyses of 1D 1H and 13C{1H}, 2D homonuclear DQF-COSY and TOCSY, heteronuclear 1H-13C HSQC and HMBC, and 1H-15N HMBC spectra. Proton-proton connectivities were obtained by analyzing the DQF-COSY and TOCSY spectra. 13C resonances corresponding to carbons with directly attached protons were assigned using the HSQC spectrum. The 1H-13C HMBC spectrum was then used to assign the carbonyl carbon resonance and to validate the carbon-carbon connectivities established by the other spectra, including connections of the pyran sugars to the cyclohexane unit. The position of the acetamido group was also verified by analysis of the 1H-15N HMBC spectrum. In the 13C-HMBC spectrum, the anomeric CH-1′ and CH-1″ proton signals, δ5.35 ppm and δ5.06 ppm, exhibited cross peaks with the carbon resonance signals at δ82.54 ppm and δ90.26 ppm, respectively, indicating that the 6′-aminomethyl pyran and the 3″-amino pyran units were connected to the cyclohexane unit through the C-4 and C-6 carbons, respectively. Simultaneously, the acetyl carbonyl resonance at δ170.13 (Table 4) exhibited cross peaks with the CH-3 and the methyl of the acetamido group proton signals, which revealed that the acetamido group was attached to the CH-3 carbon. This position of the acetamido group was further supported by examination of the 15N-HMBC spectrum in which the acetamido nitrogen resonance signal at δ132.62 displayed cross peaks with the CH-2, CH-3, CH-4, and acetamido methyl signals. The complete 1H, 13C, and 15N resonance assignments for acetylated dibekacin are given in Table 3. Thus, resistance to gentamicin and tobramycin in C. striatum BM4687 is due to the catalytic reaction of an aminoglycoside 3-N-acetyltransferase [AAC(3)].
FIG 1.
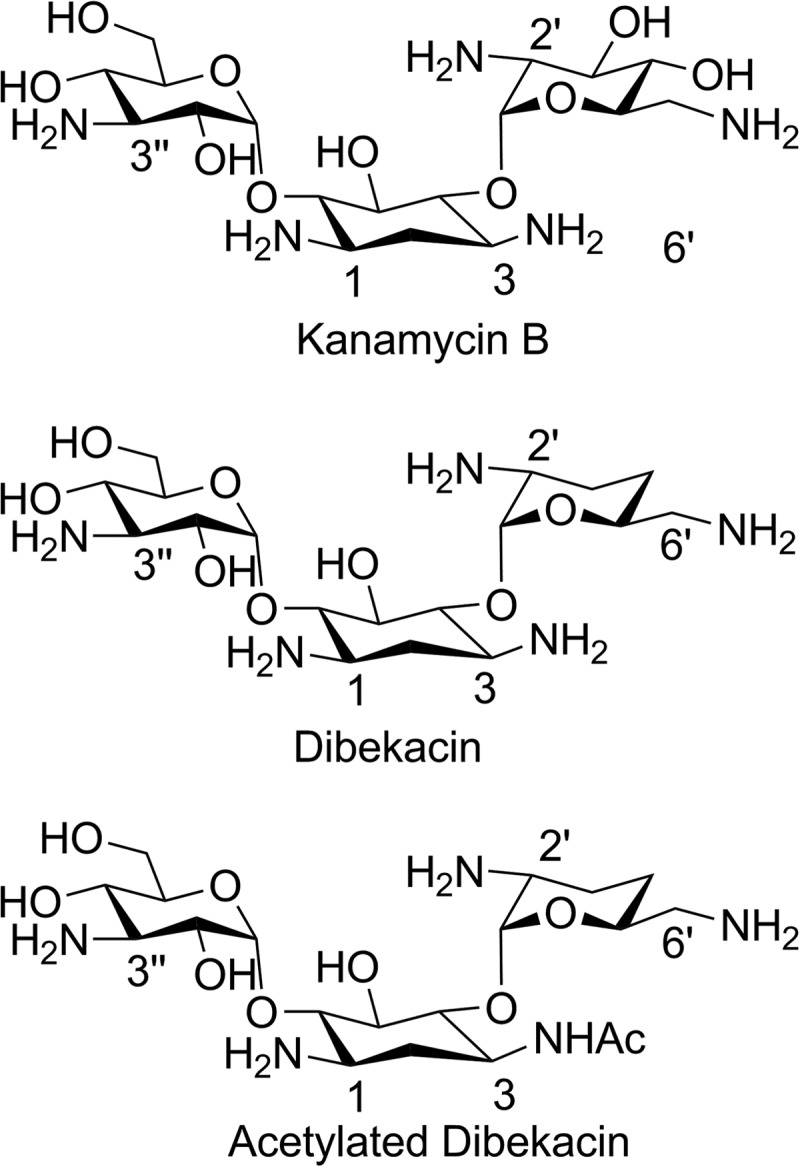
Structures of kanamycin B, dibekacin, and acetylated dibekacin.
TABLE 4.
1H and 13C NMR data for acetylated-dibekacin in D2O at 25°C
| Substructural unit | δC (ppm) | δH (ppm) | Proton multiplicity,a J(Hi, Hj) (Hz) | HMBCb |
|---|---|---|---|---|
| CH-1′ | 101.28 | 5.350 | d, 3.3 | 3′, 5′, 4 |
| CH-2′ | 52.32 | 2.866 | dt, 12.2, 3.6 | 1′, 3′ |
| CH2-3′ | 27.74 | 1.579 | ddd, 3.7, 12.9, 13.1 | 1′, 2′, 4′, 5′ |
| 1.738 | m | |||
| CH2-4′ | 29.57 | 1.441 | ddd, 2.8, 12.4, 13.2 | 3′, 6′ |
| 1.714 | m | |||
| CH-5′ | 71.55 | 3.752 | m, ΣJ = 26.5 | 1′, 4′, 6′ |
| CH2-6′ | 47.00 | 2.747 | dd, 6.1, 13.4 | 4′, 5′ |
| 2.783 | dd, 4.6, 13.4 | |||
| CH-1 | 53.04 | 2.961 | ddd, 4.2, 9.9, 12.3 | 2, 3, 5, 6 |
| CH2-2 | 36.80 | 1.397 | ddd, 12.7, 13.3, 13.3 | 1, 3, 4, 6 |
| 1.958 | dt, 4.2, 4.2, 13.1 | |||
| CH-3 | 50.93 | 3.938 | m | 1 |
| CH-4 | 82.54 | 3.561 | t, 9.2 | 1′, 2, 3, 5, 6 |
| CH-5 | 78.03 | 3.699 | t, 9.2 | 1, 3, 4, 6 |
| CH-6 | 90.26 | 3.268 | t, 9.6 | 1, 2, 5, 1″ |
| CH-1″ | 102.74 | 5.062 | d, 4.0 | 6, 3″, 5″ |
| CH-2″ | 74.47 | 3.523 | dd, 3.9, 10.4 | 3″ |
| CH-3″ | 56.88 | 3.031 | t, 10.0 | 1″, 2″, 4″, 5″ |
| CH-4″ | 71.93 | 3.347 | t, 10.0 | 2″, 3″, 5″, 6″ |
| CH-5″ | 74.81 | 3.929 | m | 1″, 3″, 4″, 6″ |
| CH-6″ | 62.98 | 3.787 | m | 4″ |
| Ac-methyl | 24.96 | 2.007 | s | |
| Ac-carbonyl | 170.13 | 3, Ac-methyl | ||
| NHAc | 132.62c | 2, 3, 4, Ac |
Due to numerous overlaps of several proton signals, the values of coupling constants or ΣJ could not be always extracted. d, doublet; dd, doublet of doublet; ddd, doublet of doublet of doublet; dt, doublet of triplet; m, multiplet; s, singlet; t, triplet.
HMBC correlations, optimized for 6 Hz, are to carbons/nitrogens from the indicated protons. Ac, acetyl.
15N chemical shift values.
Genomic environment and distribution of the aac(3)-XI gene in C. striatum.
Twenty C. striatum strains from our collection were screened for the presence of the new resistance determinant using primers AacF and AacR (Table 1). The aac(3)-XI gene was found in 10 clinical isolates from various origins: 2 strains isolated in 2000 at Limoges, France; 1 strain isolated in 2007 at Catania, Italy; and 4 strains (one per year) isolated in 1996, 2008, 2010, and 2013, and 3 strains isolated in 2014, at the Hôpital Saint-Joseph, Paris, France.
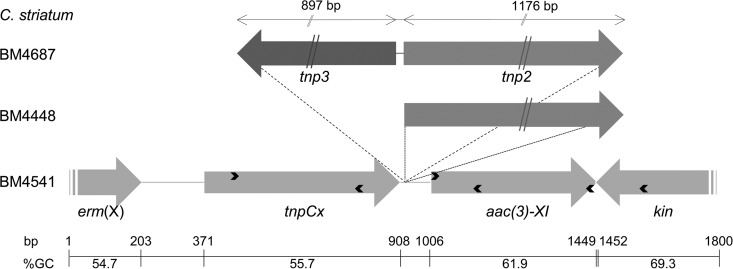
In strain BM4687, the aac(3)-XI gene was found to be located downstream from a gene for a transposase (TnpCx) and upstream from that for a two-component system sensor kinase (Fig. 2). Both genes were present in other sequences of Corynebacterium spp. deposited in GenBank. A weak putative ribosome-binding site (AAGG) was present eight nucleotides upstream from an ATG initiation codon leading to a 444-bp coding sequence. In silico analysis of the sequence upstream from aac(3)-XI did not reveal any putative promoter. Synthesis of cDNA from total RNA of C. striatum was carried out using either a random hexamer or an AacR-specific primer (Table 1; Fig. 2). PCR products were obtained with the two cDNAs and primers specific for tnp (TnpF) and aac (Aac intR) genes (Table 1; Fig. 2), indicating that the two genes were cotranscribed.
FIG 2.
Schematic representation of the genetic environment of aac(3)-XI. Arrows indicate open reading frames and directions of transcription. Only the 3′ ends of the erm(X) and of the kinase (kin) genes are represented. The primers used for amplification, reverse transcription, and sequencing are indicated by thin arrowheads within the genes. The sizes of the tnp2 and tnp3 transposase genes are indicated at the top. The G+C contents are shown under the scale.
To study the environment of aac(3), two primers were designed complementary to the transposase and to the kinase genes (Table 1; Fig. 2). A 1.2-kb PCR product corresponding to this region in C. striatum BM4687 was recovered in 8 out of the 10 clinical strains harboring an aac(3)-XI gene. PCR products of 2.5 and 3.7 kb were obtained from C. striatum BM4448 and BM4541, respectively. The fragments obtained using primer pairs (TnpF-AacR and AacF-KinR [Table 1]) indicated that insertions occurred upstream from the aac(3)-XI gene. In the two strains, the size of the product obtained with primers AacF-KinR was identical to that from BM4687; only those obtained with primers Tnp-AacR were different and corresponded to insertions of 1.3 kb in strain BM4448 and of 2.6 kb in BM4541. Sequencing of the 1.3-kb PCR product from BM4448 revealed the presence upstream from aac(3)-XI of gene tnp2 (Fig. 2) for a second transposase with a deduced sequence that was 100% identical to that of Corynebacterium resistens DSM45100 (GenPept accession number AEI08430.1). The 97-bp sequence upstream from the aac(3)-XI gene in BM4448 was 100% identical to that of BM4687.
In strain BM4541, sequencing of the 2.6-kb PCR product indicated that the tnp2 gene was present and located at the same position. However, an additional gene, tnp3 (Fig. 2), for a third transposase of 299 amino acids, 96% identical to that of Corynebacterium jeikeium (GenPept accession number WP_034993276.1), was identified. The latter transposase was located downstream from the transposase (TnpCx) and in the opposite orientation (Fig. 2).
The 64% average GC content of the 17,676-bp contig was significantly higher than that of the BM4687 genome (average, 59.4 mol%). Moreover, the base composition of the predicted coding regions ranged from 55% [erm(X) and tnpCx genes] to 69% [tet(AB) genes], suggesting horizontal gene transfer from bacteria of various species. The GC content of the aac(3) gene (61.9 mol%) was significantly different from those of the upstream- (tnpCx, 55.7 mol%) and downstream-flanking regions (kinase sensor gene, 69.3 mol%), respectively, and significantly lower than those of the acetyltransferases found in the aminoglycoside producers (ca. 75 mol%).
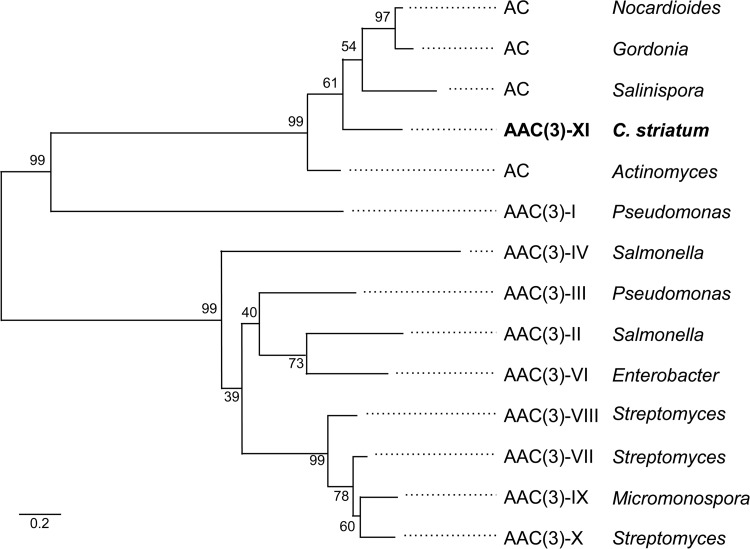
Aminoglycoside 3-N-acetyltransferases [AAC(3)] are widely distributed in Gram-negative bacteria and are categorized in five types (I, II, III, IV, and VI) (24). To date, only four have been identified in the Gram-positive aminoglycoside producers: AAC(3)-VII in Streptomyces rimosus (25) (GenPept accession number P30180), AAC(3)-VIII in Streptomyces fradiae (25) (GenPept accession number P29809), AAC(3)-IX in Micromonospora chalcea (26) (GenPept accession number P29810), and AAC(3)-X in Streptomyces griseus (GenPept accession number BAA78619). These enzymes exhibit 62 to 72% amino acid identity. Using the programs MUSCLE (7) and PhyML (8), comparative analysis of the various types of acetyltransferases found in Gram-negative and -positive bacteria allowed us to clearly identify the known types and, importantly, to designate the new type XI in C. striatum BM4687 (Fig. 3). The levels of identity of AAC(3)-XI with the AAC(3)-VII, -VIII, -IX, and -X types were between 9.6 and 15.3%. The AAC(3)-XI enzyme was more closely related to the AAC(3)-I type (16 to 22% identity) and to the predicted annotated acetyltransferases found in environmental bacteria such as Salinispora tropica (53.2% identity, 36.1% similarity), Nocardioides (55.1% identity, 67.7% similarity), and Gordonia polyisoprenivorans (55.4% identity, 68.2% similarity), a rare but emerging human pathogen (27).
FIG 3.
Phylogeny of acetyltransferases. To assess the robustness of the topology, 100 bootstrap experiments were performed on the sequences. GenBank accession numbers are as follows: acetyltransferases (AC) from Nocardioides sp. strain J54, W_028656429; Gordonia polyisoprenivorans, AFA71328; Salinispora tropica, ABP56629; and Actinomyces naeslundii, EJN83915; aminoglycoside 3-N-acetyltransferases from Gram-negative and -positive bacteria, AAC(3) type I, P23181; type II, P0A255; type III, P29808; type IV, P08988; type VI, AAA16194; type VII, P30180; type VIII, P29809; type IX, P29810; and type X, BAA78619. The scale bar represents a 20% amino acid sequence difference.
Corynebacterium striatum, previously considered a saprophyte of skin and nasal mucosa, is increasingly associated with serious infections (1); the identification of AAC(3)-XI is therefore of clinical importance. In addition, this enzyme by its particular substrate profile contributes to expanding our knowledge on aminoglycoside 3-N-acetyltransferases. The substrates of AAC(3)-XI, like those of other AAC(3) enzymes, preferentially belong to the 4,6-disubstituted 2-deoxystreptamines (gentamicin and kanamycin classes) rather than the 4,5-disubstituted 2-deoxystreptamines (neomycin class) with the exception of the AAC(3)-IIIa, -IIIb, and -IIIc enzymes. Within the 4,6-aminoglycoside group, an NH2 at position 2′ is required for acetylation (kanamycin B but not kanamycin A is inactivated) as well as one at position 1 (sisomicin is a substrate, but netilmicin, i.e., ethyl-sisomicin, is not). This determinant is likely, for now, confined in C. striatum; however, the fact that the aac(3)-XI gene was found located on putative mobile elements and variably present among members of the species suggests an acquisition from environmental Actinomycetales. However, because of the absence of sequences homologous to aac(3)-XI in the data banks, the identity of the progenitor for the gene remains presently unknown.
ACKNOWLEDGMENTS
This work was supported in part by an unrestricted grant from Reckitt-Benckiser and by the France Génomique National Infrastructure (funded as part of the “Investissements d'Avenir” program managed by the Agence Nationale pour la Recherche [contract ANR-10-IBNS-09]) for the sequence of C. striatum BM4687.
We thank the LABGeM team for annotation.
REFERENCES
- 1.Lee PP, Ferguson DA Jr, Sarubbi FA. 2005. Corynebacterium striatum: an underappreciated community and nosocomial pathogen. J Infect 50:338–343. doi: 10.1016/j.jinf.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Weiss K, Labbé AC, Laverdière M. 1996. Corynebacterium striatum meningitis: case report and review of an increasingly important Corynebacterium species. Clin Infect Dis 23:1246–1248. doi: 10.1093/clinids/23.6.1246. [DOI] [PubMed] [Google Scholar]
- 3.Renom F, Gomila M, Garau M, Gallegos MD, Guerrero D, Lalucat J, Soriano JB. 2014. Respiratory infection by Corynebacterium striatum: epidemiological and clinical determinants. New Microbes New Infect 2:106–114. doi: 10.1002/nmi2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tauch A, Krieft S, Kalinowski J, Pühler A. 2000. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol Gen Genet 263:1–11. doi: 10.1007/PL00008668. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. A07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli C, Thil Smith AA, Weiman M, Médigue C. 2013. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 9.Chastanet A, Fert J, Msadek T. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol Microbiol 47:1061–1073. doi: 10.1046/j.1365-2958.2003.03355.x. [DOI] [PubMed] [Google Scholar]
- 10.Haas MJ, Dowding JE. 1975. Aminoglycoside-modifying enzymes. Methods Enzymol 43:611–628. doi: 10.1016/0076-6879(75)43124-X. [DOI] [PubMed] [Google Scholar]
- 11.Kim C, Hesek D, Zajíček J, Vakulenko SB, Mobashery S. 2006. Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3″)-Ii/AAC(6′)-Id from Serratia marcescens. Biochemistry 45:8368–8377. doi: 10.1021/bi060723g. [DOI] [PubMed] [Google Scholar]
- 12.Bax A, Morris GA. 1981. An improved method for heteronuclear chemical shift correlation by two-dimensional NMR. J Magn Reson 42:501–505. [Google Scholar]
- 13.Bax A, Summers MF. 1986. Proton and carbon-13 assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J Am Chem Soc 108:2093–2094. doi: 10.1021/ja00268a061. [DOI] [Google Scholar]
- 14.Bodenhausen G, Freeman R, Niedermeyer R, Turner DL. 1977. Double Fourier transformation in high-resolution NMR. J Magn Reson 26:133–164. [Google Scholar]
- 15.Griesinger C, Otting G, Wuthtrich K, Ernst RR. 1988. Clean TOCSY for 1H spin system-identification in macromolecules. J Am Chem Soc 110:7870–7872. doi: 10.1021/ja00231a044. [DOI] [Google Scholar]
- 16.Sattler M, Schwalbe H, Griesinger C. 1992. Stereospecific assignment of leucine methyl groups with carbon-13 natural abundance or with random 13C labeling. J Am Chem Soc 114:1126–1127. doi: 10.1021/ja00029a072. [DOI] [Google Scholar]
- 17.Wijmenga SS, Hallenga K, Hilbers CW. 1989. A three-dimensional heteronuclear multiple quantum coherence homonuclear Hartmann-Hahn experiment. J Magn Reson 84:634–642. [Google Scholar]
- 18.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140. [DOI] [PubMed] [Google Scholar]
- 19.Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tauch A, Krieft S, Pühler A, Kalinowski J. 1999. The tetAB genes of the Corynebacterium striatum R-plasmid pTP10 encode an ABC transporter and confer tetracycline, oxytetracycline and oxacillin resistance in Corynebacterium glutamicum. FEMS Microbiol Lett 173:203–209. doi: 10.1111/j.1574-6968.1999.tb13503.x. [DOI] [PubMed] [Google Scholar]
- 21.Tauch A, Kassing F, Kalinowski J, Pühler A. 1995. The erythromycin resistance gene of the Corynebacterium xerosis R-plasmid pTP10 also carrying chloramphenicol, kanamycin and tetracycline resistances is capable of transposition in Corynebacterium glutamicum. Plasmid 33:168–179. doi: 10.1006/plas.1995.1018. [DOI] [PubMed] [Google Scholar]
- 22.Tauch A, Trost E, Tilker A, Ludewig U, Schneiker S, Goesmann A, Arnold W, Bekel T, Brinkrolf K, Brune I, Götker S, Kalinowski J, Kamp PB, Pereira Lobo F, Viehoever P, Weisshaar B, Soriano F, Dröge M, Pühler A. 2008. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. J Biotechnol 136:11–21. doi: 10.1016/j.jbiotec.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, Albersmeier A, Bekel T, Bischoff N, Brune I, Chakraborty T, Kalinowski J, Meyer F, Rupp O, Schneiker S, Viehoever P, Pühler A. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol 187:4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Cabrera M, Pérez-González JA, Heinzel P, Piepersberg W, Jiménez A. 1989. Isolation and nucleotide sequencing of an aminocyclitol acetyltransferase gene from Streptomyces rimosus forma paromomycinus. J Bacteriol 171:321–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salauze D, Pérez-González JA, Piepersberg W, Davies J. 1991. Characterisation of aminoglycoside acetyltransferase-encoding genes of neomycin-producing Micromonospora chalcea and Streptomyces fradiae. Gene 101:143–148. doi: 10.1016/0378-1119(91)90237-6. [DOI] [PubMed] [Google Scholar]
- 27.Moser BD, Pellegrini GJ, Lasker BA, Brown JM. 2012. Pattern of antimicrobial susceptibility obtained from blood isolates of a rare but emerging human pathogen, Gordonia polyisoprenivorans. Antimicrob Agents Chemother 56:4991–4993. doi: 10.1128/AAC.01251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]