Abstract
To investigate the presence and structure of AbaR-type genomic islands (GIs) in non-Acinetobacter baumannii isolates, a total of 155 non-baumannii Acinetobacter isolates from a South Korean hospital were analyzed. GIs were found in three Acinetobacter nosocomialis and two Acinetobacter seifertii isolates. Their structures were similar to those in A. baumannii isolates from Asian countries, including South Korea. The existence of AbaR-type GIs in non-baumannii Acinetobacter isolates is believed to be due to interspecies transfer of GI.
TEXT
Recently, Acinetobacter spp. have emerged as important opportunistic nosocomial pathogens. In addition to Acinetobacter baumannii, which is the most important species in clinical settings, especially in intensive care units, other Acinetobacter spp., such as A. nosocomialis and A. pittii, are frequently isolated in hospitals (1, 2). A resistance island, termed AbaR1, was identified by whole-genome sequence comparisons with a multidrug-resistant (MDR) A. baumannii strain, AYE (3). AbaR1 is integrated into the ATPase gene (now called comM) and contains a large cluster of antimicrobial and heavy metal resistance genes. Some studies have revealed diverse related AbaR resistance islands in the same region of comM in A. baumannii isolates (4–8). Although most AbaR resistance islands have been reported in A. baumannii isolates, recently, AbaR4 was reported in A. nosocomialis isolates from South Korea and Thailand (9). However, the prevalence and characteristics of AbaR-type genomic islands (GIs) in non-baumannii Acinetobacter isolates are not well known.
In the present study, the prevalence of AbaR-type GIs was investigated among non-baumannii Acinetobacter isolates from a South Korean hospital. In addition, the structure of AbaR-type GIs found in non-baumannii Acinetobacter isolates was analyzed.
In a previous study (10), 155 non-baumannii Acinetobacter strains were isolated from patients with bloodstream infections admitted to a tertiary care hospital in South Korea between August 2003 and February 2010. Species identification of the isolates using rpoB and 16S rRNA gene sequences revealed 93 A. nosocomialis, 28 Acinetobacter seifertii (formerly Acinetobacter genomic species “close to 13TU”), 15 A. pittii, three Acinetobacter calcoaceticus, six Acinetobacter bereziniae, four Acinetobacter genomic species 16, three Acinetobacter ursingii, two Acinetobacter parvus, and one Acinetobacter junii strain. In vitro antimicrobial susceptibility testing was also performed using a broth microdilution method, according to CLSI guidelines, in a previous study (10, 11). All of these 155 isolates were used in the present study.
Transposon insertion into comM was investigated using previously published primers for all Acinetobacter isolates (12, 13). Amplification of intact comM (982 bp) indicated that no GI interrupted comM, while no amplification indicated the possibility of GI interrupting comM. The integration of GI was confirmed by using two primer sets amplifying AbaR-comM (RH927 and RH797) and comM-AbaR (RH916 and RH928) (12). The structure of AbaR-type GIs in non-baumannii Acinetobacter isolates was identified by sequential PCR amplification (amplicon sizes, 4 to 5 kb) and sequencing using additional 20 primers.
Among the 155 non-baumannii Acinetobacter isolates, GIs were identified in five isolates, of which three were A. nosocomialis and two were A. seifertii. On the other hand, the comM gene was yielded by primers used in this study in A. pittii, A. calcoaceticus, A. junii, A. parvus, A. ursingii, A. bereziniae, and Acinetobacter genomic species 16. Among the five GI-positive non-baumannii Acinetobacter isolates, three and four were resistant to imipenem and meropenem, respectively (Table 1). All GI-positive non-baumannii Acinetobacter isolates were resistant to ciprofloxacin, piperacillin-tazobactam, and ampicillin-sulbactam. A. nosocomialis strain H06-681 was resistant to all antimicrobial agents, excluding imipenem, polymyxins, and tigecycline. Only A. seifertii strain C066 was resistant to colistin.
TABLE 1.
AbaR-type genomic island and antimicrobial resistance in non-baumannii Acinetobacter isolates harboring genomic islands
| Species | Isolate | Type | MIC (mg/liter) ofa: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | TET | CIP | RIF | AMK | CPM | CTR | CAZ | P/T | A/S | PB | COL | TIG | |||
| A. nosocomialis | H06-681 | Tn6022ΔtniD | 2 | 16 | >64 | >64 | 4 | >128 | >64 | >128 | >64 | >256/4 | 64/32 | 1 | 2 | 4 |
| H09-1045 | AbaR4 | >64 | >64 | 64 | 64 | 2 | 32 | 64< | 16 | 8 | >256/4 | 64/32 | 2 | 2 | 4 | |
| E09-34 | AbaR4 | >64 | >64 | >64 | 8 | 4 | 32 | 64< | 16 | 8 | >256/4 | >64/32 | 2 | 2 | 4 | |
| A. seiffertii | C044 | Tn6166 | 1 | 1 | >64 | >64 | 4 | >128 | 16 | >128 | >64 | 256/4 | 64/32 | 1 | 2 | 2 |
| C066 | Tn6022ΔtniD | 16 | 32 | 8 | >64 | 2 | 16 | >64 | >128 | >64 | >256/4 | 64/32 | 2 | 8 | 4 | |
IMP, imipenem; MEM, meropenem; TET, tetracycline; CIP, ciprofloxacin; RIF, rifampin; AMK, amikacin; CPM, cefepime; CTR, ceftriaxone; CAZ, ceftazidime; P/T, piperacillin-tazobactam; A/S, ampicillin-sulbactam; PB, polymyxin B; COL, colistin; TIG, tigecycline. MICs in bold indicate resistance. The breakpoints of resistance are from CLSI guidelines (11) for most antimicrobial resistance (resistance defined as ≥16 mg/liter for both imipenem and meropenem). The criteria recommended by the CLSI for staphylococci were applied for rifampin (resistance defined as ≥4 mg/liter), and the criteria of the U.S. Food and Drug Administration (FDA) for Enterobacteriaceae were used for tigecycline (resistance defined as ≥8 mg/liter) (18).
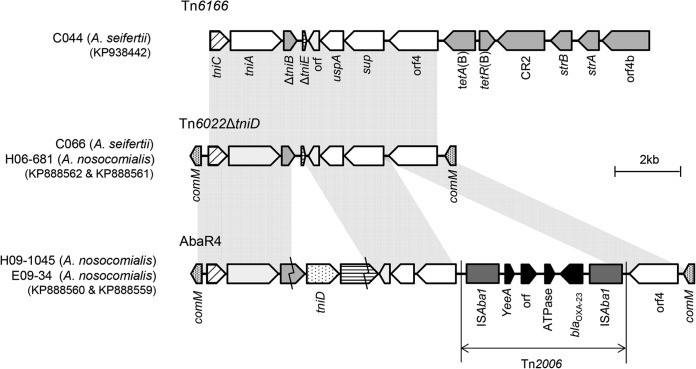
The structure of AbaR-type GIs in three A. nosocomialis and two A. seifertii isolates was determined (Fig. 1). A. nosocomialis H06-681 and A. seifertii C066 carried Tn6022 with a deletion of tniD (Tn6022ΔtniD), although they belong to different species (Fig. 1). Although a blaOXA-23-like gene was identified with ISAba1 in A. nosocomialis H06-681, it was not detected within the GI. Thus, the blaOXA-23-like gene was assumed to be located in another region of the chromosome of A. nosocomialis H06-081, along with ISAba1. On the other hand, no blaOXA-23-like gene was identified in A. seifertii C066. A. nosocomialis strains H09-1045 and E09-34, which showed very high MICs for carbapenems (Table 1), harbored AbaR4, which is composed of Tn6022 with Tn2006 (14). In A. seifertii strain C044, Tn6166 was also identified (Fig. 1). However, the Tn6166 identified in this isolate, which lacked tniD and Tn2006, included tetA(B), tetR(B), CR2, strB, strA, and orf4b instead and did not interrupt the comM gene.
FIG 1.
Structure of GIs in five non-baumannii Acinetobacter isolates from Korea. Tn6166 of C044 is different from Tn6022ΔtniD of C066 and H06-681 in that six genes, tetA(B), tetR(B), CR2, strB, strA, and orf4b, are present at the 5′end of orf4. In AbaR4 of H09-1045 and E09-34, tniD was intact and Tn2006, including blaOXA-23, was incorporated, comparing it with Tn6022ΔtniD.
Since the discovery of AbaR1 in A. baumannii strain AYE in 2006 (3), it has been known that the resistance island may play a significant role in the antimicrobial resistance of A. baumannii. Since then, several of its variants have been identified and used in epidemiological studies (4–8, 12, 15–17).
One of the most interesting findings in this study was that GIs were identified in five isolates of A. nosocomialis and A. seifertii, which belong to the A. calcoaceticus/A. baumannii (ACB) complex or A. baumannii complex. None of them are clonal, judging from 16S rRNA and rpoB gene sequences. Non-baumannii Acinetobacter species of A. baumannii complex, such as A. nosocomialis, A. pittii, and A. seifertii, are increasingly reported to cause human infections with the introduction of molecular identification tools (2). The AbaR-type GIs of five non-baumannii Acinetobacter isolates identified in this study were shared with those of A. baumannii isolates. Tn6022ΔtniD, which was detected in A. nosocomialis H06-681 and A. seifertii C066, contains a Tn6022 backbone and is the simplest GI (14, 16). The same GI structure found in different non-baumannii Acinetobacter species may be evidence that horizontal transfer of GIs occurred several times. AbaR4, which was identified in two isolates of A. nosocomialis, has been reported in A. baumannii sequence type 75 (ST75) isolates from South Korea, in A. nosocomialis strain Th01-06 from Thailand, and in many A. baumannii isolates from South Korea (9, 15). In addition, the Tn6166 structure of C044 is identical to that described by Nigro and Hall (6). These data imply that the GIs are possibly transferred among Acinetobacter species and suggest the increased frequency of AbaR-type GI in non-baumannii Acinetobacter isolates in the future.
In this study, we identified the AbaR-type GIs in five non-baumannii Acinetobacter isolates, and their structures were determined. The structure of GIs in non-baumannii Acinetobacter isolates suggests the interspecies transfer of GIs.
Nucleotide sequence accession numbers.
Sequences determined in this study have been deposited in GenBank under accession no. KP938442, KP888559, KP888560, KP888561, and KP888562.
ACKNOWLEDGMENTS
The Acinetobacter isolates used in this study were provided by Sook-In Jung (Chonnam National University Medical School, Gwangju, South Korea).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (grant NRF-2013R1A2A2A0101413).
REFERENCES
- 1.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, Wenzel RP, Seifert H. 2012. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect 64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Park YK, Jung SI, Park KH, Kim DH, Choi JY, Kim SH, Ko KS. 2012. Changes in antimicrobial susceptibility and major clones of Acinetobacter calcoaceticus-baumannii complex isolates from a single hospital in Korea over 7 years. J Med Microbiol 61:71–79. doi: 10.1099/jmm.0.033852-0. [DOI] [PubMed] [Google Scholar]
- 3.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet 2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krizova L, Dijkshoorn L, Nemec A. 2011. Diversity and evolution of AbaR genomic resistance island in Acinetobacter baumannii strains of European. Antimicrob Agents Chemother 55:3201–3206. doi: 10.1128/AAC.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nigro SJ, Hall RM. 2012. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J Antimicrob Chemother 67:1342–1346. [DOI] [PubMed] [Google Scholar]
- 6.Nigro SJ, Hall RM. 2012. Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J Antimicrob Chemother 67:335–338. doi: 10.1093/jac/dkr447. [DOI] [PubMed] [Google Scholar]
- 7.Šeputienė V, Povilonis J, Suźiedėlienė E. 2012. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob Agents Chemother 56:1969–1973. doi: 10.1128/AAC.05678-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kochar M, Crosatti M, Harrison EM, Rieck B, Chan J, Constantinidou C, Pallen M, Ou HY, Rajakumar K. 2012. Deletion of TnAbaR23 results in both expected and unexpected antibiogram changes in a multidrug-resistant Acinetobacter baumannii strain. Antimicrob Agents Chemother 56:1845–1853. doi: 10.1128/AAC.05334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Choi JY, Jung SI, Thamlikitkul V, Song JH, Ko KS. 2012. AbaR4-type resistance island including the blaOXA-23 gene in Acinetobacter nosocomialis isolates. Antimicrob Agents Chemother 56:4548–4549. doi: 10.1128/AAC.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park YK, Jung SI, Park KH, Park KH, Kim SH, Ko KS. 2012. Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea. Int J Antimicrob Agents 39:81–85. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:1162–1170. [DOI] [PubMed] [Google Scholar]
- 13.Turton JF, Baddal B, Perry C. 2011. Use of accessory genome for characterization and typing of Acinetobacter baumannii. J Clin Microbiol 49:1260–1266. doi: 10.1128/JCM.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 form an Australian hospital. J Antimicrob Chemother 66:2484–2491. doi: 10.1093/jac/dkr356. [DOI] [PubMed] [Google Scholar]
- 15.Kim DH, Park YK, Ko KS. 2012. Variations of AbaR4-type resistance island in Acinetobacter baumannii isolates from South Korea. Antimicrob Agents Chemother 56:4544–4547. doi: 10.1128/AAC.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, Peck KR, Thamlikitkul V, So TM, Yasin RM, Hsueh PR, Carlos CC, Hsu LY, Buntaran L, Lalitha MK, Song JH, Ko KS. 2013. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 Asia and AbaR-type resistance. Antimicrob Agents Chemother 57:5239–5246. doi: 10.1128/AAC.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saule M, Samuelsen Ø, Dumpis U, Sundsfjord A, Karlsone A, Balode A, Miklasevics E, Karah N. 2013. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob Agents Chemother 57:1069–1072. doi: 10.1128/AAC.01783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyeth Pharmaceuticals, Inc. 2014. Tygacil–tigecycline injection, powder, lyophilized, for solution. Wyeth Pharmaceuticals, Inc., Philadelphia, PA: http://www.pfizerpro.com/hcp/tygacil. [Google Scholar]