Abstract
Multidrug-resistant Pseudomonas aeruginosa is a major cause of severe hospital-acquired infections. Currently, polymyxin B (PMB) is a last-resort antibiotic for the treatment of infections caused by Gram-negative bacteria, despite its undesirable side effects. The delivery of drug combinations has been shown to reduce the required therapeutic doses of antibacterial agents and thereby their toxicity if a synergistic effect is present. In this study, we investigated the synergy between two cyclic antimicrobial peptides, PMB and gramicidin S (GS), against different P. aeruginosa isolates, using a quantitative checkerboard assay with resazurin as a growth indicator. Among the 28 strains that we studied, 20 strains showed a distinct synergistic effect, represented by a fractional inhibitory concentration index (FICI) of ≤0.5. Remarkably, several clinical P. aeruginosa isolates that grew as small-colony variants revealed a nonsynergistic effect, as indicated by FICIs between >0.5 and ≤0.70. In addition to inhibiting the growth of planktonic bacteria, the peptide combinations significantly decreased static biofilm growth compared with treatment with the individual peptides. There was also a faster and more prolonged effect when the combination of PMB and GS was used compared with single-peptide treatments on the metabolic activity of pregrown biofilms. The results of the present study define a synergistic interaction between two cyclic membrane-active peptides toward 17 multidrug-resistant P. aeruginosa and biofilms of P. aeruginosa strain PAO1. Thus, the application of PMB and GS in combination is a promising option for a topical medication and in the prevention of acute and chronic infections caused by multidrug-resistant or biofilm-forming P. aeruginosa.
INTRODUCTION
Pathogenic Gram-negative Pseudomonas aeruginosa possesses both intrinsic and adaptive resistance toward many currently available antibiotics and causes infections that are effectively untreatable (1–3). P. aeruginosa “superbugs” are resistant to fluoroquinolones, expanded-spectrum cephalosporins, carbapenems, aminoglycosides, and in a few cases, even polymyxins, a last-resort class of antibiotics used to treat P. aeruginosa infections (4–7). One important mechanism for the intrinsic antibiotic resistance of Gram-negative pathogens, especially multidrug-resistant (MDR) P. aeruginosa clinical isolates, is their ability to efflux antibacterial agents via tripartite efflux pumps located in the inner and outer membranes, a mechanism that limits the access of the drug to intracellular targets (8). However, the overexpression of the P. aeruginosa efflux pump protein MexAB was unable to confer resistance to the host defense peptides (HDPs) cathelicidin LL-37 and defensins (9). One way in which pathogens acquire adaptive resistance to positively charged antimicrobial peptides is to modify lipid A by substitution with aminoarabinose (6). The emergence of drug-resistant P. aeruginosa strains, which exhibit increased MICs even for polymyxin B (PMB) (10), requires the development of effective dual- and triple-drug combinations for medical treatment of these infections (7, 11). Despite the high effectiveness of polymyxins, the neuro- and nephrotoxicity associated with this treatment, especially during intravenous administration, increase the risk of using this drug (12, 13). However, daily subcutaneous administration of polymyxins resulted in decreased nephrotoxicity (14, 15), and inhaled polymyxin E, better known as colistin, showed no adverse effects (16). Because the bactericidal effect of PMB is concentration dependent (17), the synergistic effect of combination therapies can simultaneously enhance the effectiveness and reduce the required therapeutic doses of antibacterial agents. In a previous study, we demonstrated a synergistic effect of PMB and gramicidin S (GS) with silver nanoparticles against Gram-negative bacteria (18).
Another critical factor in chronic infections caused by P. aeruginosa is its ability to form highly drug-tolerant biofilms, leading to up to 1,000-fold-higher MICs for conventional antibiotics (3, 19, 20). Biofilm formation has been documented for P. aeruginosa strains isolated from patients with cystic fibrosis (21). Therefore, we aimed to study the synergistic effect of two membrane-active peptides on resistant P. aeruginosa strains grown both as planktonic cells and in biofilms. PMB and GS are cyclic nonribosomal peptides produced by the spore-forming soil bacteria Paenibacillus polymyxa (22, 23) and Aneurinibacillus migulanus (24, 25), respectively (Fig. 1). These peptides possess partial selectivity to Gram-negative (PMB) and Gram-positive (GS) bacteria because they target the outer or/and the inner bacterial membranes, respectively. PMB interacts specifically with the lipopolysaccharide (LPS) layer in the outer membrane of Gram-negative bacteria, in particular with phosphate groups on lipid A and the LPS core (26). Owing to these interactions, the divalent cations Ca2+ and Mg2+, which stabilize the integrity of the outer membrane, are replaced, which leads to destruction of the cell wall. The main target of GS is the inner prokaryotic membrane; therefore, this peptide is highly active against the Gram-positive bacteria Staphylococcus spp., Streptococcus spp., Enterococcus spp. (27), Micrococcus luteus, Mycobacterium vaccae (28), and Mycobacterium phlei (our unpublished data). However, our transmission electron microscopy images revealed that membrane protrusions were formed in Escherichia coli upon exposure to supra-MIC of GS (40 μg/ml) (29). In contrast to GS, PMB killed P. aeruginosa before the depolarization-inducing concentration was reached (30). However, the detailed mechanisms used by both peptides to cause bacterial death have not been entirely defined.
FIG 1.
Primary peptide structures of lipopeptide polymyxin B (A) and cyclic decapeptide gramicidin S (B).
MATERIALS AND METHODS
Bacterial strains.
E. coli strains DSM 1103 (ATCC 25922) and DSM 1116 (ATCC 9637), P. aeruginosa strain DSM 1117 (ATCC 27853), Aeromonas bestiarum strain DSM 13956T (ATCC 51108T), Staphylococcus aureus strain DSM 1104 (ATCC 25923), and Staphylococcus epidermidis strain DSM 1798 (ATCC 12228) were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ). P. aeruginosa PAO1 wild-type strain (mexAB+) was a kind gift from Terry Beveridge (Department of Molecular and Cellular Biology, University of Guelph, Ontario, Canada). The following clinical isolates of P. aeruginosa were obtained from the group of Susanne Häussler: 5497, 5517, 5520, 5521, 5522, 5523, 5524, 5525, 5529, and 5530. The susceptibility of five of these isolates (5497, 5520, 5522, 5524, and 5529) to ceftazidime, ciprofloxacin, meropenem, and tobramycin was previously studied using the broth microdilution assay (31). The MICs for ciprofloxacin, imipenem, and tobramycin for the additional five isolates were determined in this study using the modified broth microdilution assay (32). A disk diffusion susceptibility assay on Mueller-Hinton (MH) agar (10) with antibiotic disks from Mast Diagnostica (Reinfeld, Germany) was performed with all 10 isolates and with the DSM 1117 and PAO1 strains using the following antibiotics: 300 U of PMB, 10 μg of gentamicin, 5 μg of ciprofloxacin, 10 μg of imipenem, 30 μg of ceftazidime, 30 μg of aztreonam, 30 μg of amikacin, and 100/10 μg of piperacillin-tazobactam. The other P. aeruginosa isolates (49, 55, 56, 59, 910, 911, 912, 913, 914, 965, 966, 967, 968, 969, and 987) were enriched from clinical wastewater compartments (33). Their susceptibility to antibiotics was studied using the same disk diffusion assay, with the following antibiotics: 10 μg of gentamicin, 5 μg of ciprofloxacin, 10 μg of imipenem, 10 μg of ceftazidime, 20 μg of amikacin, 30 μg of azlocillin, and 30/10 μg of piperacillin-tazobactam. The criteria for determining resistant, intermediate, and sensitive strains were as defined using CLSI recommendations (10). Table 1 indicates the resistance of the P. aeruginosa strains to different antibiotics. MDR strains were characterized as those possessing resistance to at least one agent in three or more antimicrobial categories (34).
TABLE 1.
Resistance of studied P. aeruginosa strains to antibacterial agents
| Strain | Resistance profilea |
|---|---|
| DSM 1117 | Susceptibleb |
| PAO1 | Susceptible, MDEc |
| 49 | GENr CIPr IPMr CAZr AMKr AZLr TZPr |
| 55 | GENr CIPr IPMr CAZr AMKr AZLr TZPr |
| 56 | GENr CIPr IPMr CAZr AMKr AZLr TZPr |
| 57 | GENr CIPr IPMr CAZr AZLr TZPr |
| 59 | GENr CIPr IPMr CAZr AMKr AZLr TZPr |
| 910 | GENr CIPr IPMr CAZr AZLr |
| 911 | GENr CIPr IPMr CAZr AZLr |
| 912 | GENr CIPr IPMr CAZr |
| 913 | GENr CIPr IPMr CAZr AZLr TZPr |
| 914 | GENr CIPr IPMr |
| 965 | CIPr IPMr AZLr TZPr |
| 966 | CIPr IPMr AZLr TZPr |
| 967 | CIPr IPMr AZLr TZPr |
| 968 | CIPr IPMr AZLr TZPr |
| 969 | CIPr IPMr AZLr TZPr |
| 987 | CIPr IPMr AZLr TZPr |
| 5497 | Susceptible |
| 5517 | MEMr CIPr TOBr |
| 5520 | Susceptible |
| 5521 | MEMr CIPr TOBr |
| 5522 | PMBrc GENr TOBr |
| 5523 | Susceptible |
| 5524 | Susceptible |
| 5525 | MEMr CIPr TOBr |
| 5529 | TOBr |
| 5530 | CIPr TOBr |
GEN, gentamicin; CIP, ciprofloxacin; IPM, imipenem; CAZ, ceftazidime; AMK, amikacin; AZL, azlocillin; TZP, piperacillin-tazobactam; ATM, aztreonam; MEM, meropenem; TOB, tobramycin; PMB, polymyxin B.
Susceptibility to conventional antibiotics was tested for all strains in the disk diffusion assay, but additionally, the MICs were determined for the clinical isolates 5497, 5517, 5520, 5521, 5522, 5523, 5524, 5525, 5529, and 5530 in this study or in Müsken et al. (31).
Multidrug efflux (MDE) pump proteins were found in the PAO1 strain (69).
Resistance to PMB was shown only in the synergy assay (MIC, 8 μg/ml), not in the disk diffusion assay.
Standard procedure for preparing the inocula.
It is well-known that bacterial susceptibility to antibiotics depends on their growth phase and growth rate. To obtain a standard inoculation material from the bacterial strains, which were maintained at −80°C using the Cryobank system (Mast Diagnostica, Reinfeld, Germany), bacterial cells on single beads were recovered in 10 ml of MH broth (Becton, Dickinson and Co., Sparks, MD, USA) in an overnight incubation at 37°C and 200 rpm. Single colonies were obtained by streaking these cultures on LB-Lennox agar plates, which were then stored at 4°C. Subsequently, the colonies were used to inoculate 10 ml of MH broth to an optical density at 550 nm (OD550) of 0.02, and the cultures were grown overnight. The test cultures were prepared by inoculating 10 ml of MH medium to an OD550 of 0.2 with the overnight cultures and allowing them to grow until the bacteria reached the mid-exponential phase. For the synergy tests, the test cultures were diluted immediately prior to the experiment in MH broth to obtain final inoculation doses of 5 × 105 CFU/ml.
Checkerboard assay.
PMB was purchased from Sigma-Aldrich (St. Louis, MO, USA). GS was produced in-house by fermentation of A. migulanus strain DSM 5759 cultures, extracting GS from bacterial cells, purifying it by high-performance liquid chromatography (HPLC), and finally lyophilizing the drug (25). To generate stock solutions, PMB was dissolved in water, and GS, due to its low water solubility at concentrations of >240 μg/ml (35), was dissolved in 50% (vol/vol) ethanol. For broth microdilution, we used the not-cation-adjusted MH broth (BD Diagnostic Systems, Sparks, MD, USA) as recommended for the testing of cationic antimicrobial peptides (AMPs) (36). To set up the assay, 40 μl of double-concentrated MH broth and 40 μl of the peptide stock solutions were mixed in the upper wells of 96-well microtiter plates (Nunclon; Nunc GmbH & Co., Wiesbaden, Germany). For further peptide dilution, 40 μl of ordinary MH broth was added to the rest of the wells. GS was 2-fold serially diluted down the rows of the plate, and PMB was diluted in separate plates and then added to the test plate across the columns. The resulting checkerboard contained each combination of the two peptides in 7 doubly increasing concentrations, with wells containing the highest concentration of each antibiotic at opposite corners. As seen in Fig. 2, both the row and column with no GS or PMB contained only one drug and were prepared to evaluate the MIC of each antimicrobial alone in the same plate. Column pc was used as a positive growth control without drugs. All wells containing 80 μl of MH medium with single peptides or a combination of peptides were inoculated with 20 μl of the bacterial suspension. The wells of column nc, containing 100 μl of MH medium, served as a sterility control. The plates were incubated at 37°C under static conditions. To determine the respiratory activity of the bacteria, after 22 h of incubation, 20 μl of sterile 0.9 mM resazurin (a redox indicator) was added to the wells, and the plate was incubated for an additional 2 h. The reduction of the 0.15 mM resazurin to resorufin was measured with a microplate reader (FlashScan 550; Analytik Jena GmbH, Jena, Germany), which measures the whole absorption spectrum from 400 to 900 nm. The absorption values of resazurin (blue oxidized form) at 600 nm and absorption of resorufin (pink reduced form) at 570 nm were utilized to calculate differences in the absorption for each well, using the WinFlash program. In this analysis, negative absorption difference values indicated the absence of bacterial growth (Fig. 2). The MIC is defined as the lowest concentration of antibiotic that completely inhibits bacterial growth. The synergistic interactions were expressed as the fractional inhibitory concentration index (FICI), which is calculated as the sum of MICs of the combination (MICc) divided by the MICs of the peptides alone (MICa): FICI = (PMB MICc/PMB MICa) + (GS MICc/GS MICa). The mean FICIs and standard deviations were calculated from the results from at least five independent experiments performed with each bacterial strain. A synergistic effect was defined at an FICI of ≤0.5 and a nonsynergistic effect at an FICI between >0.5 and ≤4 (37).
FIG 2.
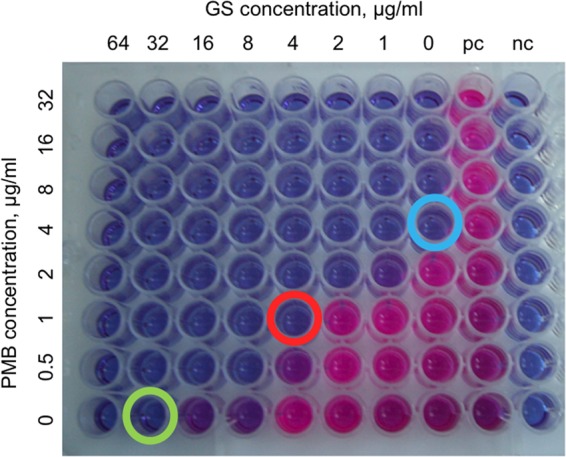
Evaluation of a checkerboard assay in the 96-well microtiter plate. The two rightmost columns contain the positive growth control (pc) and negative growth control (nc) to confirm bacterial growth without peptides and medium sterility, respectively. The green and blue circles show the peptide MICa. The red circle indicates the antibiotic combination that resulted in a synergistic effect on P. aeruginosa strain 55, for which the FICI calculation should be [1/4 + 4/32] = 0.375.
Prevention of P. aeruginosa PAO1 static biofilm formation.
The formation of static biofilms was studied in 96-well microtiter plates (Nunclon; Nunc GmbH & Co., Wiesbaden, Germany). In each well, 80 μl of MH broth (BD Diagnostic Systems, Sparks, MD, USA) containing single peptides or a combination of peptides was inoculated with 20 μl of a bacterial suspension at an OD550 of 0.1 (108 CFU/ml) from the overnight cultures. The plates were incubated at 37°C under static conditions for 24 h to allow biofilm formation. The medium was aspirated to remove planktonic cells, the wells were washed with water, and the formation of sessile biofilms was evaluated by crystal violet (CV) staining, as described previously (38). Briefly, 125 μl of CV solution (0.1% [wt/vol]) was transferred to each well. The plates were incubated for 10 min at room temperature and washed twice with water to remove excess dye. After drying the samples for 10 min, 200 μl of ethanol (95% [vol/vol]) was added to the wells to dissolve the dye. The plates were incubated for 20 min at room temperature (RT), and the intensity of CV at 595 nm was measured on the plate reader. The mean values obtained for the negative controls were subtracted from the results of the test wells. Student's t test P values were calculated from the average results from two independent experiments, each performed in triplicate.
Resazurin assay with P. aeruginosa PAO1 pregrown biofilm cells.
In order to investigate the effect of PMB and GS on sessile P. aeruginosa cells, we performed a modified resazurin assay with pregrown biofilms in 96-well plates. P. aeruginosa PAO1 overnight cultures were diluted to 106 CFU/ml (OD600, 0.001) in MH broth (Merck, Darmstadt, Germany) and used for the inoculation of polystyrene microtiter plates (100 μl/well). Biofilms were grown for 24 h at 37°C under static conditions. After two washes with 150 μl of MH medium, antibiotics (8 μg/ml PMB and 32 μg/ml GS), alone or in combination, and resazurin (0.1 mM final concentration) were added to the wells in a total volume of 150 μl, and the samples were incubated for an additional 7 h at 37°C. MH medium supplemented with 1% ethanol (vol/vol) served as a negative control, because the antibiotic stock solutions were dissolved in 50% (vol/vol) ethanol. At the indicated time points, 100 μl of the samples was transferred to a new 96-well microtiter plate, and the amount of reduced resazurin (resorufin) was determined by measuring the absorbance at 560 nm. Residual amounts of oxidized resazurin were quantified by measuring the absorbance at 620 nm, and corrected A560 values (AR560) were calculated using the following formulas from the manual for the alamarBlue assay (Life Technologies; version 1.1 PI-DAL1025-1100): AR560 = A560 − (A620 × RO) and RO = AO560/AO620, where A560 and A620 are the sample absorbances and AO560 and AO620 are the absorbances of MH broth containing 0.1 mM resazurin. Mean values and standard deviations were calculated from the results from three independent experiments, each performed in triplicate. Statistical analysis was done by the Mann-Whitney test.
RESULTS
Evaluation of PMB and GS synergy in checkerboard assays.
Our preliminary study on different Gram-negative and Gram-positive bacterial strains using the checkerboard assay identified combinations of PMB and GS that exerted synergistic or nonsynergistic effects on bacteria. The FICIs indicated that PMB-and-GS combinations had a distinct synergistic effect on P. aeruginosa strain DSM117 only (Table 2). A nonsynergistic effect (FICI, >0.5 and ≤ 1) was obtained for A. bestiarum DSM 13956T, E. coli DSM 1116, and S. aureus DSM 1104, and for the other E. coli DSM 1103 strain, which is often used as a control strain to measure the antimicrobial activity of antibiotics, or the S. epidermidis strain DSM 1798 (FICI, >1).
TABLE 2.
Preliminary results of the PMB-and-GS combination treatments on different Gram-negative and Gram-positive bacteria
| Bacterial strain | MIC (μg/ml) for peptide: |
FICI | |||
|---|---|---|---|---|---|
| Alone |
In combination |
||||
| GS | PMB | GS | PMB | ||
| A. bestiarum DSM 13956T | 4 | 2 | 1 | 1 | 0.625 |
| E. coli DSM 1103 | 16 | 0.5 | 4 | 1 | 2.250 |
| E. coli DSM 1116 | 32 | 1 | 8 | 0.5 | 0.750 |
| P. aeruginosa DSM 1117 | 32 | 4 | 4 | 1 | 0.375 |
| S. aureus DSM 1104 | 2 | 8 | 1 | 1 | 0.625 |
| S. epidermidis DSM 1798 | 1 | 8 | 1 | 2 | 1.250 |
Next, in follow-up experiments, we aimed to investigate the effect of combining PMB and GS on 28 different P. aeruginosa strains. The MIC of PMB for most strains is 4 μg/ml (intermediate resistant). However, a MIC of 8 μg/ml (resistant) was found for P. aeruginosa strain 5522, and P. aeruginosa strains 914, 5497, 5521, 5524, and 5530 were affected by a MIC of only 2 μg/ml (susceptible). The MIC for GS was significantly higher, generally 16 to 32 μg/ml, but for the strains isolated from the clinical wastewater compartments, it was 32 to 64 μg/ml.
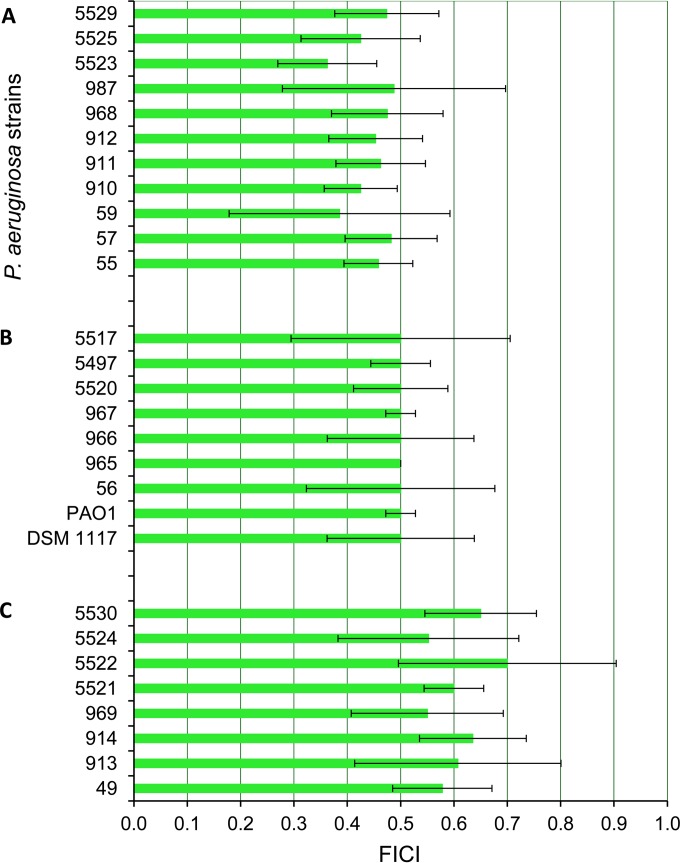
When the bacteria were treated with combinations of the peptides, we observed a 3- to 4-fold decrease in MICs for PMB and GS. The absolute values decreased from 2 to 8 μg/ml to 0.25 to 1 μg/ml for PMB and from 16 to 64 μg/ml to 2 to 8 μg/ml for GS. This decrease in effective inhibitory concentrations was calculated as an FICI and indicated a distinct synergy for 71.4% of the all studied strains and 68% of the MDR P. aeruginosa strains, with mean FICIs of ≤0.5 (Fig. 3). Remarkably, FICIs of >0.5 but ≤0.70 were observed for the 5521, 5522, 5524, and 5530 clinical isolates of P. aeruginosa, which revealed an adaptive colony morphological phenotype known as a small-colony variant (Fig. 3). This morphological phenotype plays an important role in chronic infections, the appearance of which is correlated with antimicrobial chemotherapy (39). The standard deviations of the FICIs obtained for several strains (P. aeruginosa 56, 59, 913, 987, 5517, 5522, and 5524) were as high as 0.2, which is presumably due to the phenotypic instability of P. aeruginosa and the high physiological divergence of some strains in general. For example, we observed that P. aeruginosa 5497 is dissociated into two circular colony phenotypes with diameters of 1 to 2 mm or <0.1 mm. P. aeruginosa DSM 1117 dissociated into two colony phenotypes: large colonies that were 4 to 5 mm in diameter and possessed undulating margins and circular colonies of 1 to 2 mm in diameter. The other strains grew on LB agar plates in circular colonies of 2 to 3 mm in diameter.
FIG 3.
The mean FICIs for 28 different strains and clinical isolates of P. aeruginosa (Pa), with standard deviations calculated from the results from at least five independent experiments. The first 11 strains showed an FICI of <0.5 (A), and the next 9 strains revealed an FICI of 0.5 that indicated a distinct synergistic effect of the PMB-GS combination toward these 20 strains. (C) FICIs in this group of P. aeruginosa clinical isolates are >0.5 and showed a nonsynergistic effect. The standard deviations of the FICIs for the strains P. aeruginosa 56, 59, 913, 987, 5517, 5522, and 5524 as high as 0.2 indicated the high physiological variability within these strains.
The checkerboard assays for the 26 MDR P. aeruginosa clinical isolates, biofilm-forming P. aeruginosa PAO1 strain, and control strain DSM 1117 were performed in at least five independent experiments, or up to 11 times in the case of two different phenotypes of P. aeruginosa DSM 1117. However, the results for both of the P. aeruginosa DSM 1117 phenotypes showed the same divergence and are presented in Fig. 3 as one result.
Synergistic effect on the formation of static biofilms.
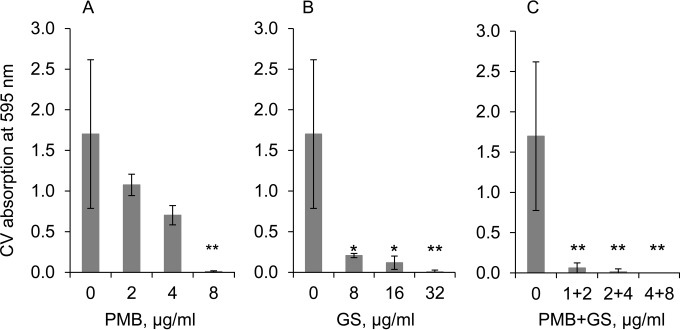
Given that the combination of PMB and GS showed a synergistic effect on the growth of planktonic P. aeruginosa PAO1 cells, we continued our study to evaluate whether the PMB-and-GS combination treatments could prevent biofilm formation. The concentration of PMB alone required to inhibit biofilm formation by P. aeruginosa PAO1 was 8 μg/ml (Fig. 4A). GS was less effective and required a concentration of 32 μg/ml to prevent P. aeruginosa PAO1 biofilm formation (Fig. 4B). Treatment with the two peptides together decreased the effective peptide concentrations required, from 8 μg/ml for PMB alone to 2 μg/ml in combination and from 32 μg/ml for GS alone to 4 μg/ml in combination (Fig. 4C). The FICI calculated from this decrease is 0.375, which indicates a synergistic effect of this treatment.
FIG 4.
The inhibitory effect of PMB or GS alone and in combination on the formation of P. aeruginosa PAO1 biofilms. Shown are the effect of PMB alone (A), the effect of GS alone (B), and the synergistic effect of PMB and GS in combination (C). Biofilms were quantified by crystal violet staining, and the absorbance at 595 nm was measured. The mean values obtained for negative controls were subtracted from the results for the test wells. Student's t test P values (*, P < 0.05; **, P < 0.01) were calculated for the average values from two independent experiments, each performed in triplicate.
Synergistic effect of the GS-and-PMB combination on the growth of pregrown PAO1 biofilms.
In addition to the staining of biofilm biomass with crystal violet, we monitored the respiratory activity of pregrown P. aeruginosa PAO1 biofilms treated with PMB (8 μg/ml) and GS (32 μg/ml) alone or in combination over time, using resazurin as a redox indicator. The peptide concentrations utilized were the MICs for biofilm growth previously determined for each peptide in the crystal violet biofilm assay (Fig. 4). Given that peptide stock solutions were prepared in 50% ethanol (vol/vol), the test wells and control wells contained residual ethanol concentrations of 1% (vol/vol), which did not affect bacterial growth, as verified by resazurin reduction (data not shown).
In experiments with pregrown biofilms, the respiratory activity of cells treated either with GS or PMB alone after 7 h of incubation was comparable to that of the control cultures without peptides. However, significantly lower AR560 values (P < 0.001, Mann-Whitney test) were exhibited by biofilms treated with a combination of PMB and GS (Fig. 5).
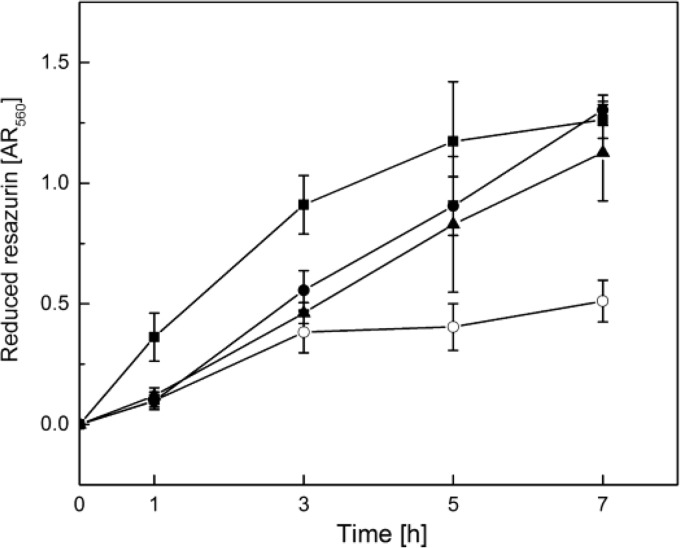
FIG 5.
PMB-and-GS combination treatment of pregrown P. aeruginosa PAO1 biofilms in comparison to treatment with these peptides alone. P. aeruginosa PAO1 biofilms were grown for 24 h in 96-well plates following the addition of PMB (8 μg/ml; closed squares), GS (32 μg/ml; closed triangles), or a combination of both peptides (open circles) and 0.1 mM resazurin, a redox indicator. Control wells (closed circles) contained 1% ethanol (vol/vol), because peptide stock solutions were prepared in 50% ethanol, resulting in final ethanol concentrations of 1% (vol/vol). At the indicated time points, resazurin reduction was quantified by measuring the absorbance at 560 nm. The figure shows means and standard deviations for the results of three independent experiments, each performed in triplicate.
When the biofilms were treated with GS alone, a nearly linear increase of resazurin reduction up to an AR560 of 1.1 was observed during the incubation time. This was comparable to that of the control cultures and indicated that GS treatment alone did not impair P. aeruginosa biofilm growth. In contrast, when GS and PMB treatments were combined, the AR560 values increased only during the first 3 h of incubation before reaching a plateau between 0.4 and 0.5. Treatment with PMB alone led to a continuous increase in redox activity during the first 5 h of incubation, resulting in AR560 values around 1.3, which were even higher than those of the control cultures. However, between 5 and 7 h of incubation, the amount of reduced resazurin increased only slightly, indicating that PMB inhibited bacterial growth at these time points. In summary, the combined treatment of pregrown P. aeruginosa PAO1 biofilms with PMB and GS at concentrations of 8 μg/ml and 32 μg/ml, respectively, led to a considerably faster inhibition of bacterial respiratory activity than the application of the peptides alone, whereby GS alone did not exert any lethal effect, and it appears that PMB actually triggered bacterial growth at early time points.
DISCUSSION
There are important clinical questions about how to treat P. aeruginosa infections, such as the possible benefits of combinatorial antimicrobial treatments versus monotherapies, which have to be answered (1). The majority of synergy studies have delineated the effect of combining conventional antibiotics or combining conventional antibiotics with antimicrobial peptides. In the case of peptide combinations, synergy occurs because there is better intracellular uptake of conventional antibiotics that subsequently have bactericidal effects (40, 41). For example, combining PMB with meropenem, amikacin, or rifampin was partially effective against extremely drug-resistant P. aeruginosa isolates (7). PMB and doxycycline combinations resulted in a ≥4-fold decrease in the MIC of PMB for resistant carbapenemase-producing Klebsiella pneumoniae (42). It was shown that PMB exerted a synergistic anti-P. aeruginosa effect in combination with chlorhexidine, another membrane-active compound (43). Even the antifungicidal effect of fluconazole against Cryptococcus and Candida species was higher in combination with PMB (44). Here, we studied the combination of two antimicrobial peptides, which target different membrane structures in the cell walls of Gram-negative bacteria. Additionally, both PMB and GS may prevent bacterial growth by disturbing the integrity and function of bacterial membrane proteins. Both peptides may directly inhibit membrane-associated respiratory proteins, such as the alternative bacterial NADH dehydrogenase isolated from Mycobacterium smegmatis, P. aeruginosa, and Gluconobacter oxydans. Whereas GS inhibits cytochrome bd quinol oxidase from E. coli, PMB was active against malate:quinone oxidoreductase from M. smegmatis and P. aeruginosa (45). Additionally, GS treatment caused the delocalization of some peripheral membrane proteins, such as the lipid II biosynthesis protein MyrG, the cell division regulator protein MinD, and the respiration protein cytochrome c (46). Thus, it is likely that if GS could more easily access the plasma membranes of Gram-negative cells, its effect on bacterial vitality would be significant.
We used a quantitative version of the checkerboard assay, in which bacterial growth was indicated by the sensitive redox indicator resazurin. This modification allowed us to better distinguish even slight growth differences, such as that from 5 × 105 to 5 × 106 CFU/ml, which cannot be detected by the naked eye. For each strain, the standard deviations of the FICIs were calculated for more than five independent experiments, which is in accordance with the requirements for synergy testing (47). Recently, distinct synergy was defined as an FICI of ≤0.5, and a nonsynergistic effect was defined as an FICI of between 0.5 and 4. However, previous papers demonstrated the mathematics of synergy and presented the pitfalls of using an FICI of <1 to define synergy (48, 49). Our results showed a range of FICIs between 0.36 and 0.70. At least 20 of the strains studied met the standard requirements of having an FICI of ≤0.5, which indicated distinct synergy. Among these 20 strains were 17 multidrug-resistant clinical isolates that showed a synergistic effect (Table 1). For isolate 5522, we even obtained resistance to PMB (MIC, 8 μg/ml) in the checkerboard dilution assay. In the disk diffusion assay, all strains were susceptible to PMB because of the diffusion peculiarities of positively charged molecules in MH agar. For the clinical isolates that grew as small-colony variants (5521, 5522, 5524, and 5530), the FICIs were mostly >0.5 and ≤0.70 (Fig. 3). Seven strains (56, 59, 913, 987, 5517, 5522, and 5524) showed the most divergent results (standard deviations from 0.16 to 0.2), which indicated their high intrinsic physiological variability. Since only two of them (5522 and 5524) are small-colony variants, the phenotypic variations could not be a single reason for this deviation.
The advantage of the checkerboard dilution assay is its relative ease of use. One disadvantage noted during testing was that some bacteriostatic concentrations had not been validated in clinical trials (21). However, it is well-known that the mode of action of both antimicrobial peptides PMB and GS is associated with a bactericidal effect (11, 29). In addition to depolarization, presumably via the formation of a short-term pore at sub-MICs (50), all ATP-dependent processes should be indirectly affected by GS, as it has a high affinity for ATP (51). Using scanning and transmission electron microscopy, we showed that even sub-MICs of GS were able to upset the osmoregulatory properties of E. coli cells that were exposed to the peptides for 1 h (29).
Standardization of growth conditions, especially for the clinical isolates, is a critical requirement for determining MICs and the corresponding synergy effects. Visual determination of growth can lead to the omission of slow bacterial growth, which was overcome in our studies by using the redox indicator resazurin. In addition, alterations in Mg2+ concentrations can also impact the MIC or synergy values. It was demonstrated previously that the MIC of PMB for PAO1 is strongly dependent on the presence of divalent cations in the defined minimal medium with glucose and that MICs ranged from 16 μg/ml at low (20 μM) Mg2+ concentrations to 1 μg/ml at high (2 mM) Mg2+ concentrations (6). The increased resistance at low Mg2+ concentrations was explained by a cell response resulting in aminoarabinosylation of the lipid A moiety and a loss of negative charges (5). A similar effect of Mg2+ and Ca2+ ions on the lytic activity of GS was observed for Micrococcus lysodeikticus protoplasts (52). In our experiments, MH broth was not supplemented with Mg2+ and Ca2+ salts, according to recommendations for AMPs (36), which might explain the intermediate resistance (MIC, 4 μg/ml) to PMB observed for the majority of the studied strains. Remarkably, the concentrations of divalent cations, such as Mg2+ and Ca2+, determined in four different brands of MH broth, were different (53). Therefore, the cationic specification mentioned by the suppliers might be very useful for the determination of the MICs of AMPs.
In this study, we focused on the synergistic activity of PMB-and-GS combination treatment of P. aeruginosa isolates growing both in planktonic and biofilm states. MDR P. aeruginosa causes relapsing and persistent lung and skin infections and medical implant-related infections, which usually contain biofilms. With regard to the molecular structure, the antibiofilm mechanism of cyclic antimicrobial peptides (AMPs) and α-helical HDPs seems to be different (54). Biofilm suppression by application of AMPs can be achieved in three ways, namely: (i) reduction in the planktonic population, (ii) prevention of the initial adhesion of cells to the surface, and (iii) removal of the established biofilm (55). Human HDP LL-37 and the bovine neutrophil peptide indolicidin were able to prevent P. aeruginosa PAO1 biofilm formation at subinhibitory concentrations by downregulating the genes essential for cell attachment and biofilm formation (56). Our data indicate that combination treatment with the cyclic bacterial AMPs PMB and GS was effective for the prevention of biofilm formation, presumably due to a reduction in the initial planktonic population, considering the bactericidal action of these peptides.
It is known that the extent of biofilm clearance also depends on the medium composition. The MIC of PAO1 biofilm was higher at low Mg2+ concentrations (20). Biofilm maturation is another point that can impact eradication results. “Young” biofilms were more sensitive to attack by neutrophils than were mature biofilms (57). Our testing of PMB and GS activity against pregrown biofilms showed that a strong inhibition of respiration occurred only when biofilms were treated with PMB and GS in combination, but not if the peptides were applied separately. These results suggest that PMB enables better translocation of GS through the outer membrane of P. aeruginosa, which is an additional barrier blocking GS access to the inner membrane (58).
The lipopeptide PMB remains a last-resort antibiotic in the treatment of Gram-negative infections, despite its associated nephro- and neurotoxicity. Medical use of GS is limited to topical application, due to its hemolytic activity. However, hemolytic activity depends on the buffer system, peptide solvent, hematocrit concentration in the reaction sample, incubation time, and health state of the donor. The hemolytic concentrations obtained for GS were 10 to 39 μg/ml, which correspond to 11 to 35 μM (59–62). Some chemically synthesized GS derivatives with high antibacterial and low hemolytic activity (62, 63) could be utilized in place of the natural GS for applications other than topical, but a subsequent study will be necessary.
Surprisingly, the cytotoxicity of the human HDP LL-37 was comparable to that of GS at similar concentrations of 13 to 25 μM (64). Similar hemolytic properties of LL-37 suggest that hemolytic activity does not play a significant role upon direct application of AMPs to the site of infection. Topical application is suitable for several different diseases, especially when parenteral application of conventional antibiotics may lead to the dysfunction of the natural intestinal microbiota and have a negative impact on the status of a patient. PMB and GS alone were successfully administered directly to the respiratory tracts of critically ill patients (16, 65, 66). Recently, it was shown that P. aeruginosa is a part of normal human skin microbiota (67). When the integument is compromised, skin bacteria are able to gain access to underlying tissues where the conditions are optimal for colonization and growth. P. aeruginosa and S. aureus were among the most common organisms isolated from both acute and chronic wounds of various etiologies, demonstrating their prevalence (68). In vitro evaluation of PMB-and-GS combination treatments against MDR P. aeruginosa strains showed mostly a 4-fold reduction in the MICs for both antibiotics, which indicates a synergistic effect. Considering all infection types, such as skin infections, including wounds, intratissue, lung, and medical device infections, which are especially common after surgical procedures, it may be preferential to prevent these by topical application of the bactericidal peptides PMB and GS to avoid subacute and chronic infections and to prevent more serious diseases. GS activity toward S. aureus might provide an additional positive impact, especially in the case of polymicrobial wound infections resulting from P. aeruginosa and S. aureus. PMB-and-GS combination treatment administered as an aerosol, irrigation, or dressing may be successfully used in pulmonology, dermatology, and surgery/traumatology to treat acute and chronic postsurgery infections.
ACKNOWLEDGMENTS
We thank Susanne Häussler and Terry Beveridge for providing the P. aeruginosa clinical isolates. We thank the Bio Interfaces (BIF) program of the Karlsruhe Institute of Technology (KIT) in the Helmholtz Association for their support of this work.
REFERENCES
- 1.Giamarellou H, Poulakou G. 2009. Multidrug-resistant Gram-negative infections: what are the treatment options? Drugs 69:1879–1901. doi: 10.2165/11315690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Naas T, Fortineau N, Poirel L. 2007. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr Opin Microbiol 10:436–440. doi: 10.1016/j.mib.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, Rafailidis PI, Matthaiou DK. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updat 13:132–138. doi: 10.1016/j.drup.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocincova D, Lam JS, Martinez JL, Hancock RE. 2012. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim TP, Lee W, Tan TY, Sasikala S, Teo J, Hsu LY, Tan TT, Syahidah N, Kwa AL. 2011. Effective antibiotics in combination against extreme drug-resistant Pseudomonas aeruginosa with decreased susceptibility to polymyxin B. PLoS One 6:e28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. 2009. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A 106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rieg S, Huth A, Kalbacher H, Kern WV. 2009. Resistance against antimicrobial peptides is independent of Escherichia coli AcrAB, Pseudomonas aeruginosa MexAB and Staphylococcus aureus NorA efflux pumps. Int J Antimicrob Agents 33:174–176. doi: 10.1016/j.ijantimicag.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2013. Performance standards for antimicrobial susceptibility testing. CLSI M100-S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 12.Dai CS, Li JC, Lin W, Li GX, Sun MC, Wang FX, Li J. 2012. Electrophysiology and ultrastructural changes in mouse sciatic nerve associated with colistin sulfate exposure. Toxicol Mech Method 22:592–596. doi: 10.3109/15376516.2012.704956. [DOI] [PubMed] [Google Scholar]
- 13.Weinstein L, Doan TL, Smith MA. 2009. Neurotoxicity in patients treated with intravenous polymyxin B: two case reports. Am J Health Syst Pharm 66:345–347. doi: 10.2146/ajhp080065. [DOI] [PubMed] [Google Scholar]
- 14.Arnold TM, Forrest GN, Messmer KJ. 2007. Polymyxin antibiotics for Gram-negative infections. Am J Health Syst Pharm 64:819–826. doi: 10.2146/ajhp060473. [DOI] [PubMed] [Google Scholar]
- 15.Abdelraouf K, Braggs KH, Yin TJ, Truong LD, Hu M, Tam VH. 2012. Characterization of polymyxin B-induced nephrotoxicity: implications for dosing regimen design. Antimicrob Agents Chemother 56:4625–4629. doi: 10.1128/AAC.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalopoulos A, Fotakis D, Virtzili S, Vletsas C, Raftopoulou S, Mastora Z, Falagas ME. 2008. Aerosolized colistin as adjunctive treatment of ventilator-associated pneumonia due to multidrug-resistant Gram-negative bacteria: a prospective study. Respir Med 102:407–412. doi: 10.1016/j.rmed.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Tam VH, Schilling AN, Vo G, Kabbara S, Kwa AL, Wiederhold NP, Lewis RE. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3624–3630. doi: 10.1128/AAC.49.9.3624-3630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruden S, Hilpert K, Berditsch M, Wadhwani P, Ulrich AS. 2009. Synergistic interaction between silver nanoparticles and membrane-permeabilizing antimicrobial peptides. Antimicrob Agents Chemother 53:3538–3540. doi: 10.1128/AAC.01106-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 20.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saiman L. 2007. Clinical utility of synergy testing for multidrug-resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis: ‘the motion for.’. Paediatr Respir Rev 8:249–255. [DOI] [PubMed] [Google Scholar]
- 22.Stansly PG, Schlosser ME. 1947. Studies on polymyxin: isolation and identification of Bacillus polymyxa and differentiation of polymyxin from certain known antibiotics. J Bacteriol 54:549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalopoulos A, Falagas ME. 2008. Colistin and polymyxin B in critical care. Crit Care Clin 24:377–391. doi: 10.1016/j.ccc.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Gause GF, Brazhnikova MG. 1944. Gramicidin S origin and mode of action. Lancet 247:715–716. [Google Scholar]
- 25.Berditsch M, Afonin S, Ulrich AS. 2007. The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl Environ Microbiol 73:6620–6628. doi: 10.1128/AEM.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindler M, Osborn MJ. 1979. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 27.Salgado J, Grage SL, Kondejewski LH, Hodges RS, McElhaney RN, Ulrich AS. 2001. Membrane-bound structure and alignment of the antimicrobial beta-sheet peptide gramicidin S derived from angular and distance constraints by solid state 19F-NMR. J Biomol NMR 21:191–208. doi: 10.1023/A:1012946026231. [DOI] [PubMed] [Google Scholar]
- 28.Afonin S, Glaser RW, Berditchevskaia M, Wadhwani P, Guehrs KH, Mollmann U, Perner A, Ulrich AS. 2003. 4-Fluorophenylglycine as a label for 19F NMR structure analysis of membrane-associated peptides. ChemBioChem 4:1151–1163. doi: 10.1002/cbic.200300568. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Dhillon P, Yan H, Farmer S, Hancock RE. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3317–3321. doi: 10.1128/AAC.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müsken M, Di Fiore S, Römling U, Häussler S. 2010. A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc 5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 32.Babii O, Afonin S, Berditsch M, Reibetaer S, Mykhailiuk PK, Kubyshkin VS, Steinbrecher T, Ulrich AS, Komarov IV. 2014. Controlling biological activity with light: diarylethene-containing cyclic peptidomimetics. Angew Chem Int Ed Engl 53:3392–3395. doi: 10.1002/anie.201310019. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz T, Volkmann H, Kirchen S, Kohnen W, Schön-Hölz K, Jansen B, Obst U. 2006. Real-time PCR detection of Pseudomonas aeruginosa in clinical and municipal wastewater and genotyping of the ciprofloxacin-resistant isolates. FEMS Microbiol Ecol 57:158–167. doi: 10.1111/j.1574-6941.2006.00100.x. [DOI] [PubMed] [Google Scholar]
- 34.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 35.Fomicheva GK, Komarov EV. 1984. Acidity and solubility of gramicidin S in water. Antibiotiki 29:353–357. (In Russian.) [PubMed] [Google Scholar]
- 36.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 37.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 38.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, Kaever V, Landmann R, Jenal U. 2010. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog 6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fassi Fehri L, Wróblewski H, Blanchard A. 2007. Activities of antimicrobial peptides and synergy with enrofloxacin against Mycoplasma pulmonis. Antimicrob Agents Chemother 51:468–474. doi: 10.1128/AAC.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le T, Bayer AS. 2003. Combination antibiotic therapy for infective endocarditis. Clin Infect Dis 36:615–621. doi: 10.1086/367661. [DOI] [PubMed] [Google Scholar]
- 42.Elemam A, Rahimian J, Doymaz M. 2010. In vitro evaluation of antibiotic synergy for polymyxin B-resistant carbapenemase-producing Klebsiella pneumoniae. J Clin Microbiol 48:3558–3562. doi: 10.1128/JCM.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Najjar AR, Quesnel LB. 1979. Synergism between chlorhexidine and polymyxins against Pseudomonas aeruginosa. J Appl Bacteriol 47:469–476. doi: 10.1111/j.1365-2672.1979.tb01208.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhai B, Zhou H, Yang L, Zhang J, Jung K, Giam CZ, Xiang X, Lin X. 2010. Polymyxin B, in combination with fluconazole, exerts a potent fungicidal effect. J Antimicrob Chemother 65:931–938. doi: 10.1093/jac/dkq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mogi T, Kita K. 2009. Gramicidin S and polymyxins: the revival of cationic cyclic peptide antibiotics. Cell Mol Life Sci 66:3821–3826. doi: 10.1007/s00018-009-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, Gust R, Albada HB, Penkova M, Kramer U, Erdmann R, Metzler-Nolte N, Straus SK, Bremer E, Becher D, Brotz-Oesterhelt H, Sahl HG, Bandow JE. 2014. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A 111:E1409–E1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rand KH, Houck HJ, Brown P, Bennett D. 1993. Reproducibility of the microdilution checkerboard method for antibiotic synergy. Antimicrob Agents Chemother 37:613–615. doi: 10.1128/AAC.37.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wareham DW, Bean DC. 2006. In vitro activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:825–826. doi: 10.1128/AAC.50.2.825-826.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berenbaum MC. 1978. A method for testing for synergy with any number of agents. J Infect Dis 137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Scott MG, Yan H, Mayer LD, Hancock RE. 2000. Interaction of polyphemusin I and structural analogs with bacterial membranes, lipopolysaccharide, and lipid monolayers. Biochemistry 39:14504–14514. doi: 10.1021/bi0011173. [DOI] [PubMed] [Google Scholar]
- 51.Krauss EM, Chan SI. 1983. Complexation and phase transfer of nucleotides by gramicidin S. Biochemistry 22:4280–4291. [DOI] [PubMed] [Google Scholar]
- 52.Bulgakova VG, Kostrova OM, Koroleva PN, Sazykina S, Polin AN. 1988. Effect of salts on the lytic activity of gramicidin S and its derivatives. Antibiot Khimioter 33:661–665. (In Russian.) [PubMed] [Google Scholar]
- 53.Girardello R, Bispo PJ, Yamanaka TM, Gales AC. 2012. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol 50:2414–2418. doi: 10.1128/JCM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berditsch M, Afonin S, Vladimirova T, Wadhwani P, Ulrich AS. 2012. Antimicrobial peptides can enhance the risk of persistent infections. Front Immunol 3:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorge P, Lourenço A, Pereira MO. 2012. New trends in peptide-based antibiofilm strategies: a review of recent achievements and bioinformatic approaches. Biofouling 28:1033–1061. doi: 10.1080/08927014.2012.728210. [DOI] [PubMed] [Google Scholar]
- 56.Overhage J, Campisano A, Bains M, Torfs EC, Rehm BH, Hancock RE. 2008. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect Immun 76:4176–4182. doi: 10.1128/IAI.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Günther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, Wagner C, Hänsch GM. 2009. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN). Mol Immunol 46:1805–1813. doi: 10.1016/j.molimm.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 58.Bulgakova VG, Korolev PN, Konoshenko GI, Novozhilova T, Polin AN. 1990. Stability of Escherichia coli to the membranotropic antibiotic gramicidin S. Mikrobiologiia 59:702–704. (In Russian.) [PubMed] [Google Scholar]
- 59.Kondejewski LH, Farmer SW, Wishart DS, Kay CM, Hancock RE, Hodges RS. 1996. Modulation of structure and antibacterial and hemolytic activity by ring size in cyclic gramicidin S analogs. J Biol Chem 271:25261–25268. doi: 10.1074/jbc.271.41.25261. [DOI] [PubMed] [Google Scholar]
- 60.Semrau S, Monster MW, van der Knaap M, Florea BI, Schmidt T, Overhand M. 2010. Membrane lysis by gramicidin S visualized in red blood cells and giant vesicles. Biochim Biophys Acta 1798:2033–2039. doi: 10.1016/j.bbamem.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Hackl EV, Berest VP, Gatash SV. 2012. Effect of cholesterol content on gramicidin S-induced hemolysis of erythrocytes. Int J Pept Res Ther 18:163–170. doi: 10.1007/s10989-012-9289-9. [DOI] [Google Scholar]
- 62.Tamaki M, Fujinuma K, Harada T, Takanashi K, Shindo M, Kimura M, Uchida Y. 2012. Fatty acyl-gramicidin S derivatives with both high antibiotic activity and low hemolytic activity. Bioorg Med Chem Lett 22:106–109. doi: 10.1016/j.bmcl.2011.11.062. [DOI] [PubMed] [Google Scholar]
- 63.Tamaki M, Sasaki I, Kokuno M, Shindo M, Kimura M, Uchida Y. 2010. Antimicrobially active cycloundecapeptides related to gramicidin S having a novel turn structure with cis d-Phe-Pro peptide bond. Org Biomol Chem 8:1791–1797. doi: 10.1039/b922159j. [DOI] [PubMed] [Google Scholar]
- 64.Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem 273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- 65.Falagas ME, Kasiakou SK. 2007. Local administration of polymyxins into the respiratory tract for the prevention and treatment of pulmonary infections in patients without cystic fibrosis. Infection 35:3–10. doi: 10.1007/s15010-007-6104-1. [DOI] [PubMed] [Google Scholar]
- 66.Sergiev PG. 1944. Clinical use of gramicidin S. Lancet 244:717–718. [Google Scholar]
- 67.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. 2013. The microbiome extends to subepidermal compartments of normal skin. Nat Commun 4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. 2013. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]