Abstract
Invasive mycotic infections have become more common during recent decades, posing an increasing threat to public health. However, despite the growing needs, treatments for invasive fungal infections remain unsatisfactory and are limited to a small number of antifungals. The aim of this study was to identify novel fungal cell wall inhibitors from a library of small chemical compounds using a conditional protein kinase C (PKC)-expressing strain of Aspergillus nidulans sensitive to cell wall-active agents. Eight “hit” compounds affecting cell wall integrity were identified from a screen of 35,000 small chemical compounds. Five shared a common basic molecular structure of 4-chloro-6-arylamino-7-nitro-benzofurazane (CANBEF). The most potent compound, CANBEF-24, was characterized further and was shown to inhibit the growth of pathogenic Aspergillus, Candida, Fusarium, and Rhizopus isolates at micromolar concentrations but not to affect the growth of mammalian cell lines. CANBEF-24 demonstrated strong synergy in combination with caspofungin, an antifungal that inhibits cell wall biosynthesis. Genetic and biochemical analyses with Aspergillus nidulans and Saccharomyces cerevisiae indicated that CANBEFs selectively inhibit fungal rRNA maturation and protein synthesis, suggesting that their effect on the cell wall is indirect. CANBEFs were nontoxic in insect (Galleria mellonella, Drosophila melanogaster) and mouse models of fungal infection. Preliminary evidence showing no therapeutic benefit in these models suggests that further cycles of optimization are needed for the development of this novel class of compounds for systemic use.
INTRODUCTION
The number of life-threatening invasive fungal infections has risen dramatically over the past 30 years (1, 2). The vast majority of these infections are caused by species of the genera Candida, Aspergillus, Cryptococcus, and Coccidioides (3). Invasive aspergillosis has now overtaken candidiasis as the most frequent invasive fungal infection found after death in Europe and the United States (4, 5). Today, as many as 4% of all patients dying in modern tertiary care hospitals have invasive aspergillosis caused by fungal pathogen species that belong to the genus Aspergillus, while as many as 2% of these patients suffer from invasive candidiasis caused by Candida species (4, 6). However, despite the growing needs, treatments for invasive fungal infections remain unsatisfactory.
There are three main classes of antifungal drugs in common clinical use for the treatment of systemic mycoses: the polyene amphotericin B, which binds fungal membrane ergosterol, leading to cell lysis; azoles, which inhibit ergosterol biosynthesis (fluconazole, itraconazole, voriconazole [VRC], and posaconazole); and the newly introduced echinocandins, such as caspofungin (CAS), which inhibit fungal glucan biosynthesis. Most of these current systemic antifungal treatments interact unfavorably with other medications, have resistance problems, a narrow spectrum of activity, and limited formulations, and are fungistatic rather than fungicidal; some are often toxic (7). Therefore, there is an urgent need to develop additional, novel drugs that inhibit fungus-specific targets, such as the fungal cell wall.
To identify cell wall-destabilizing compounds, we took advantage of the Aspergillus nidulans alcA-PKC strain, which we have previously shown to display specific hypersensitivity to such compounds when grown under repressive conditions (with glucose) due to the involvement of protein kinase C (PKC) in regulating cell wall integrity (8, 9).
We screened a diverse chemical library of 35,000 drug-like molecules (ChemDiv Inc., San Diego, CA) in order to identify cell wall inhibitors. First, we identified compounds that inhibit the growth of a pathogenic isolate of Aspergillus fumigatus in a 96-well-based liquid assay. The resulting antifungal compounds were then tested for their effects on the growth of the alcA-PKC mutant, which exhibits enhanced sensitivity to cell wall damage under growth conditions that repress PKC expression. The mutant exhibited hypersensitivity to eight cell wall-active compounds under repressive conditions. Five of these compounds shared a common basic molecular structure of 4-chloro-6-arylamino-7-nitro-benzofurazane (CANBEF) and demonstrated promising in vitro antifungal activity against a panel of pathogenic fungi. We report on the detailed analysis of the antifungal CANBEFs, in particular CANBEF-24, the most potent and specific compound.
MATERIALS AND METHODS
Strains and preparation of inocula.
The strains used in this study are listed in Table 1. Conidia were harvested in 0.2% (vol/vol) Tween 80, resuspended in double-distilled water (DDW), and counted with a hemocytometer. Molds were grown either in a rich yeast extract–agar–glucose (YAG) medium, containing 0.5% (wt/vol) yeast extract, 1% (wt/vol) glucose, and 10 mM MgCl2, supplemented with a 0.1% (vol/vol) trace element solution and a 0.2% (vol/vol) vitamin mixture, or in a defined minimal medium (MM) containing 70 mM NaNO3, 1% (wt/vol) glucose, 12 mM potassium phosphate (pH 6.8), 4 mM MgSO4, 7 mM KCl, and trace elements. Yeasts were grown either in a rich yeast extract–peptone–dextrose (YPD) medium composed of 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) dextrose or in a synthetic complete (SC) medium containing 0.17% (wt/vol) yeast nitrogen base without amino acids (YNB), 0.5% (wt/vol) ammonium sulfate, 2% (wt/vol) dextrose, and a dropout mixture containing all possible supplements.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference | Source |
|---|---|---|---|
| Aspergillus nidulans | |||
| Strain GR-5 | wA3 pyrG89 pyroA4 | G. S. May | |
| Strain R153 | wA3 pyroA4 | G. S. May | |
| alcA-PKC strain | wA2 pyroA4 pyrG89::pyr4 alcA(p)::pkcAΔp | Our lab | |
| A. fumigatus | |||
| Af293 (FGSC A1100) | Wild type (patient isolate) | 29 | FGSCa |
| ATCC 13073 | Wild type (patient isolate) | FGSC | |
| Aspergillus niger | |||
| Strain #1 | Wild type (patient isolate) | 29 | I. Shalit |
| Strain #2 | Wild type (patient isolate) | 29 | I. Shalit |
| Strain #3 | Wild type (patient isolate) | 29 | I. Shalit |
| Aspergillus flavus | |||
| Strain #1 | Wild type (patient isolate) | 29 | I. Shalit |
| Strain #2 | Wild type (patient isolate) | 29 | I. Shalit |
| Fusarium solani | |||
| 603251 | Wild type (patient isolate) | 30 | I. Shalit |
| 600679 | Wild type (patient isolate) | 30 | I. Shalit |
| Fusarium oxysporum 600711 | Wild type (patient isolate) | 30 | I. Shalit |
| Rhizopus arrhizus 156 | Wild type (patient isolate) | I. Shalit | |
| Mucor racemosus | |||
| 167 | Wild type (patient isolate) | I. Shalit | |
| 3484 | Wild type (patient isolate) | I. Shalit | |
| 3465 | Wild type (patient isolate) | I. Shalit | |
| Candida albicans | |||
| ATCC 2901 | Wild type (patient isolate) | E. Segal | |
| ATCC 18804 (CBS562) | Wild type (patient isolate) | E. Segal | |
| ATCC 90028 | Wild type (patient isolate) | E. Segal | |
| 58455 | Wild type (patient isolate) | E. Segal | |
| Candida rugosa 3929 | Wild type (patient isolate) | E. Segal | |
| Candida parapsilosis ATCC 22019 | Wild type (patient isolate) | E. Segal | |
| Candida tropicalis ATCC 20336 | Wild type (patient isolate) | E. Segal | |
| Candida glabrata 59343 | Wild type (patient isolate) | E. Segal | |
| Candida krusei ATCC 6258 | Wild type (patient isolate) | E. Segal | |
| Saccharomyces cerevisiae BY4741 | S288C MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0 | 31 | M. Schuldiner |
FGSC, Fungal Genetics Stock Center.
Screen for antifungal compounds.
The A. fumigatus wild-type (WT) strain Af293 was grown in 96-well plates at a concentration of 104 conidia/ml in YAG medium. Each well was supplemented with a 25 μM concentration of a compound from a chemical compound library (ChemDiv Inc., San Diego, CA) composed of 35,000 small drug-like molecules.
The compounds that completely inhibited fungal growth at 25 μM were selected for further characterization. To determine whether the selected compounds inhibit fungal growth by damaging the integrity of the cell wall, A. nidulans strain R153 and the isogenic conditional alcA-PKC mutant (8) were grown in 96-well plates at a concentration of 104 conidia/ml in MM or MMG (MMG contains 0.2% [wt/vol] glycerol instead of glucose). The wells were supplemented with 2-fold dilutions of each selected compound. After 24 h of incubation at 37°C, the MICs (the lowest drug concentrations to completely arrest germination and growth) and minimal effective concentrations (MECs) (the lowest drug concentrations to cause visibly aberrant growth or a significant reduction in growth) of wells with compounds were evaluated in comparison to those for nontreated wells. Caspofungin (Merck, NJ, USA) or voriconazole (Pfizer, NY, USA) was used as a control. For each compound, the “cell wall index” (CWI) was determined by dividing the MIC or MEC of the compound for each strain in derepressing medium (MIC-MMG or MEC-MMG) by its MIC or MEC for that strain in repressing medium (MIC-MM or MEC-MM) and then dividing the ratio for the alcA-PKC strain (a mutant strain hypersensitive to cell wall damage) by the ratio for R153 (isogenic control strain). Thus, the MIC cell wall index was calculated as [(MIC-MMG/MIC-MM)AlcA-PKC]/[(MIC-MMG/MIC-MM)R153], and the MEC cell wall index was calculated as [(MEC-MMG/MEC-MM)AlcA-PKC]/[(MEC-MMG/MEC-MM)R153]. A compound was considered cell wall specific when its MIC or MEC cell wall index was ≥4, which means that the alcA-PKC mutant exhibited a ≥4-fold decrease in the MIC or MEC when grown in a repressive medium relative to the MIC or MEC of R153. Compounds that fulfilled this condition were determined as “hit compounds.”
Panfungal and bacterial screen.
The fungal strains listed in Table 1 were tested for susceptibility according to a slight modification of CLSI standard M27-A3 or the CLSI M38-A2 protocol, respectively (10, 11). Briefly, strains were grown in 96-well plates at a concentration of 500 yeast cells/ml or 104 conidia/ml for filamentous fungi in MM supplemented with the hit compounds or antifungals in 96-well plates. MICs were evaluated after 24 h of incubation at 37°C. Bacteria were tested in LB broth composed of 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, and 1% (wt/vol) NaCl. The inoculation suspension was prepared by a 1:10 dilution of a growing bacterial culture at an optical density at 600 nm (OD600) of 0.1 (which equals approximately 108 cells/ml).
Structure-activity relationship (SAR) analysis was performed on all 45 commercially available CANBEF compounds from ChemDiv Inc., San Diego, CA. A. fumigatus conidia at 2 × 103/well were incubated in RPMI–morpholinepropanesulfonic acid (MOPS) medium at 37°C, and MIC and MEC values were measured after 24 h.
Cell culture.
Hit compounds were assessed for toxicity to mammalian cells by using the human cancer cell line A549 (ATCC CCL-185), derived from a human lung carcinoma, and the mouse embryo fibroblast cell line NIH 3T3 (ATCC CRL-1658).
The cell lines were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) in 10-cm tissue culture plates (Corning; Sigma-Aldrich, St. Louis, MO, USA). Cells were incubated at 37°C under a humidified atmosphere with 5.5% CO2. For the determination of MICs and MECs, cells were plated in 96-well plates at a concentration of 5 × 104/well. After 24 h of incubation, the medium was washed and replaced with DMEM without serum. The hit compounds were added and were incubated for 24 h as described above. Cell viability was then measured by the XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide, disodium salt] assay (Biological Industries, Beit HaEmek, Israel). XTT-based MICs were defined as the lowest drug concentrations to completely inhibit color formation.
Microscopy and staining.
The effects of the hit compounds on fungal ultrastructure were assessed by light and fluorescence microscopy after cell wall and vital staining. A. fumigatus conidia at a concentration of 104/ml were incubated for 24 h at 37°C on glass coverslips in 24-well plates (Nunclon; Nalge Nunc, Roskilde, Denmark) containing 1 ml MM/well in the presence of CANBEF-24 (0.8 μM). After 24 h, microscopy was performed with an Olympus BX40 microscope (equipped for fluorescence with a UV filter and a fluorescein isothiocyanate [FITC] filter) at a ×400 magnification. Images were recorded on an Olympus DP70 camera.
For calcofluor white staining, the fungus was stained for 45 min at room temperature and in darkness with calcofluor white (0.1 mg/ml in DDW). After staining, hyphae were washed twice with phosphate-buffered saline (PBS) and were analyzed. For DiBAC [bis-(1,3-dibutylbarbituric acid)] staining of dead conidia and hyphae, cells were stained with 2 μg/ml DiBAC in 100 mM MOPS buffer (pH 7.0) for 1 h at room temperature, washed twice with PBS, and analyzed.
For transmission electron microscopy (TEM), treated conidia or germlings were harvested by centrifugation and were fixed in 2.5% (wt/vol) glutaraldehyde (Merck Inc., Whitehouse Station, NJ) in PBS. They were then washed, postfixed in 1% OsO4 in PBS, and washed again. After dehydration in graded ethanol solutions, the cells were embedded in glycidyl ether 100 (Serva Electrophoresis GmbH, Heidelberg, Germany). Ultrathin sections were stained with uranyl acetate and lead citrate and were examined with a JEOL 1200 EX TEM.
Synergy checkerboard assay.
Checkerboard tests were carried out in standard 96-well plates (Costar; Corning, Corning, NY), according to the CLSI M38-A microdilution methodology (10) with some modifications as described below. After harvesting and counting with a hemocytometer, conidia were diluted to a final concentration of 2.5 × 104/ml in RPMI-MOPS medium. The plates were scanned with an inverted microscope (at a magnification of ×40) after 48 h of incubation at 37°C. MIC and MEC measurements were used to determine the fractional inhibitory concentration index (FICI) of the combinations of compounds. FICI values were interpreted as follows: ≤0.5, synergy; >0.5 to 4, no interaction; >4, antagonism.
Inhibition of [35S]methionine uptake in yeasts.
Saccharomyces cerevisiae strain BY4741 was grown to an OD600 of 0.5 in SC medium (without methionine), and the culture was divided into 5-ml aliquots. [35S]methionine (>1,000 Ci/mmol; Institute of Isotopes, Hungary) at 10 μCi/ml was added to each aliquot in the presence of cycloheximide (10 μM) or CANBEF-13 (2 μM) for 15, 30, or 45 min. Cells were spun down for 5 min at 2,000 × g, and the reaction was terminated by the addition of 100 μl SDS sample buffer to the cell pellet, followed by vortexing and heating for 5 min at 95°C. Proteins were separated on an 8% SDS-PAGE gel and were analyzed by autoradiography.
Screening of an A. nidulans overexpression genomic library for resistance-conferring plasmids.
A library of A. nidulans genomic DNAs cloned into the multicopy nonintegrating pRG3 vector containing the AMA1 sequence of A. nidulans (12, 13) was screened for resistant strains. We have used this method successfully to identify the cellular targets of two antifungal drugs (14, 15). Transformation was performed by protoplasting as described previously (16). The A. nidulans GR5 transformants were plated onto 10 selective agar plates (without uracil and uridine) at a concentration of ∼5,000 colonies per plate. After 48 h of incubation at 37°C, spores were harvested and pooled. Pooled conidia were plated onto MM agar plates supplemented with 8 μM CANBEF-24 and were incubated for 48 h at 37°C. Resistant colonies were then reisolated on MM agar with 8 μM CANBEF-24 in order to avoid false-positive results. Resistance to CANBEF-24 was assessed by a standard broth microdilution assay. The resistance-conferring plasmid was isolated, and the resistance-conferring gene in the genomic-DNA insert was identified, as described previously (13).
Screening of S. cerevisiae overexpression libraries for resistant strains.
A conditional gene overexpression library of S. cerevisiae consisting of 5,800 strains was screened for resistance to CANBEF-13. Each strain contains a unique mRNA expressed under the control of the GAL1 promoter, which is induced by galactose, repressed by glucose, and neither induced nor repressed by raffinose (17).
Overexpression strains were first pooled and then plated at 105 CFU/plate on selective medium (SM) without uracil and with galactose as the sole carbon source (S. Gal −URA) for induction of overexpression. CANBEF-13 was added at 0.75 μM. The plates were incubated at 28°C, and resistant colonies were isolated after 24, 48, and 72 h on S. Gal −URA plates. Resistance was verified by broth microdilution as follows. Twenty-three isolates that displayed greater resistance to CANBEF-13 than the parental strain in inducing medium (S. Gal +Raf −URA) but not in noninducing medium (S. Raf −URA) were defined as resistant to CANBEF-13, and the overexpressed gene was amplified by PCR using primers on the constant region flanking the inserted open reading frame (ORF). Then PCR products were sequenced and were identified by BLAST on the S. cerevisiae Genome Project website (http://www.yeastgenome.org/).
In vivo toxicity. (i) Galleria mellonella model.
Groups of 10 caterpillars of the greater wax moth Galleria mellonella in the final instar larval stage, weighing 250 to 330 mg, were employed in all assays. Larvae were injected with 10 μl saline containing 1.8 to 14.4 mg/kg CANBEF-24. Larval survival was assessed daily for as long as 7 days posttreatment.
(ii) Drosophila melanogaster model.
Toll trans-heterozygotes (i.e., Tl−/− flies) were generated by crossing flies carrying a thermosensitive allele of Toll (Tlr632) with flies carrying a null allele of Toll (Tll-RXA). Female WT and Tl−/− flies aged 2 to 4 days were used; there were 25 flies per experimental group. Toll flies were fed with fly food containing 1.8 to 14.4 mg/ml of CANBEF-24 for 7 days. The flies were kept at 29°C and were transferred to fresh vials every 2 days. Flies that died 3 h after infection were excluded from the survival analysis. Survival was assessed until day 7 after infection. All experiments were performed on three different days at the same time of day in order to eliminate circadian-rhythm-associated variability.
(iii) Murine model.
A preliminary toxicity analysis of CANBEF-24 was carried out with female ICR mice aged 6 to 8 weeks. Groups of two mice were injected intraperitoneally with 14.4 mg/kg, 7.2 mg/kg, 3.6 mg/kg, or 1.8 mg/kg CANBEF-24 for four consecutive days. The animals were weighed once a day. Three days after the fourth injection, the mice were sacrificed, and their livers, kidneys, and spleens were weighed and observed.
RESULTS
Screen for cell wall-destabilizing antifungal compounds.
To identify compounds with antifungal activity, we screened 35,000 drug-like molecules from a diverse chemical compound library (ChemDiv Inc., San Diego, CA). Each compound was initially tested at a single concentration of 25 μM for general antifungal activity against wild-type A. fumigatus spores germinating in 96-well microtiter plates on rich YAG medium for 24 h of growth at 37°C. We identified 16 hit compounds, which completely inhibited fungal germination and growth at a concentration of 25 μM (see Table S1 in the supplemental material). To identify potential cell wall-specific compounds, the hits were further characterized with the alcA-PKC mutant under inducing (MMG) and repressing (MM) conditions. Eight of the 16 compounds (CW-1 to CW-8) yielded cell wall indices of >4 for MIC and MEC values (see Table S2 in the supplemental material), suggesting that they affect the cell wall. A noteworthy finding is that five of the cell wall-specific compounds, CW-1 to CW-5, share a basic molecular structure of 4-chloro-6-arylamino-7-nitro-benzofurazane (Fig. 1; see also Table S1 in the supplemental material) and were therefore called by the acronym CANBEF. These compounds were chosen for further analysis.
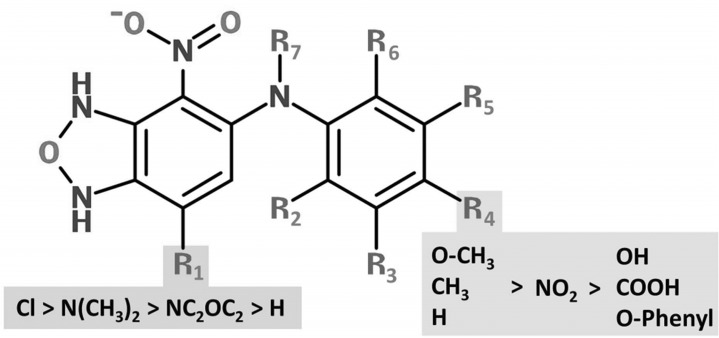
FIG 1.
Structure-activity relationships of the CANBEFs. The basic molecular structure of the CANBEFs is 4-chloro-6-arylamino-7-nitro-benzofurazane. Antifungal activity is highly sensitive to substitutions at position R4. Substitutions of the chlorine at position 4 (R1) reduce antifungal activity.
SAR analysis of the CANBEFs.
We carried out a structure-activity relationship (SAR) analysis of all 45 commercially available CANBEF compounds. Both MICs and MECs were calculated for the A. fumigatus wild-type strain Af293 grown for 24 h on RPMI-MOPS medium (see Table S3 in the supplemental material). Twenty-six compounds were active against A. fumigatus. The SAR analysis of A. fumigatus clearly confirmed that a chloride residue (Cl) at the R1 position of the furazane ring is crucial for antifungal activity against A. fumigatus (Fig. 1). Of the 23 compounds lacking a chloride residue at R1, 20 were inactive (MIC, >25 μM), while only 3 (CANBEF-5, -7, and -27) were weakly active (Fig. 1; see also Table S3 in the supplemental material). Compounds remained satisfactorily active when an O-acetyl (CANBEF-14), O-methyl (CANBEF-23), or methyl (CANBEF-24) group, or hydrogen (CANBEF-22), was positioned at R4 of the benzene ring. Activity was partially lost when this residue was changed to a nitro group (CANBEF-16) or, even worse, to a hydroxyl (CANBEF-1), carboxyl (CANBEF-6), or O-phenyl (CANBEF-19) group. Methylation (CANBEF-12) or bromination (CANBEF-13) of R3 or methylation of R2 (CANBEF-9) did not improve activity over that of CANBEF-24. In summary, the most potent compounds contained a chlorine substitution at R1 and an O-acetyl, O-methyl, or methyl group at R4, while all other positions (R2, R3, R5 to R7) remained unsubstituted.
CANBEF compounds are specifically active against most pathogenic fungi.
Subsequently, three CANBEFs (CANBEF-13, -14, and -24), representative of the most specific cell wall compounds that were most active against A. fumigatus, were tested on a wide range of pathogenic fungal strains, mammalian cell lines, and bacteria in culture (Table 2). CANBEF-13 was active against Candida spp. (MIC, >6.25 μM and <12.5 μM) and, in particular, S. cerevisiae (MIC, 0.2 μM) but was ineffective against pathogenic molds. When performing the mechanism-of-action (MOA) studies in yeast, we used CANBEF-13 due to its strong activity against S. cerevisiae (Table 2). CANBEF-14 and CANBEF-24 had identical activity profiles; they were effective against yeasts and most molds (MIC, >3.13 μM and <12.5 μM). However, because the latter compound displayed a higher MIC CWI value (see Table S2 in the supplemental material), we concentrated our analysis on CANBEF-24. The fungicidal activity of CANBEF-24 was tested against A. fumigatus Af293 and S. cerevisiae BY4741, for which it exhibited minimal fungicidal concentrations (MFCs) of 8 μM and 0.5 μM, respectively, approximately 3- to 4-fold higher than the MICs for these organisms.
TABLE 2.
Susceptibilities of fungal pathogens, bacteria, and cultured mammalian cells to CANBEF-13, -14, and -24
| Strain | CANBEF-13 |
CANBEF-14 |
CANBEF-24 |
|||
|---|---|---|---|---|---|---|
| MIC (μM) | MEC (μM) | MIC (μM) | MEC (μM) | MIC (μM) | MEC (μM) | |
| Fungal pathogenic isolates | ||||||
| Candida albicans | ||||||
| ATCC 2901 | 12.50 | 12.50 | 6.25 | 12.50 | 6.25 | |
| ATCC 90028 | 12.50 | 12.50 | 6.25 | 12.50 | 6.25 | |
| 18804 | 12.50 | 12.50 | 6.25 | 12.50 | 6.25 | |
| Candida rugosa 3929 | 6.25 | 3.13 | 3.13 | 1.56 | 3.13 | 1.56 |
| Candida parapsilosis ATCC 22019 | 12.50 | 12.50 | 6.25 | 12.50 | 6.25 | |
| Candida tropicalis ATCC 20336 | 12.50 | 6.25 | 3.13 | 6.25 | 3.13 | |
| Candida glabrata 59343 | 12.50 | 6.25 | 3.13 | 6.25 | 3.13 | |
| Candida krusei ATCC 6258 | 12.50 | 3.13 | 1.56 | 3.13 | 1.56 | |
| Mucor racemosus | ||||||
| 3465 | ≤12.50 | ≤12.50 | 6.25 | 6.25 | 6.25 | 6.25 |
| 3484 | ≤50.00 | ≤50.00 | ≤50.00 | 25.00 | ≤50.00 | 25.00 |
| 167 | ≤50.00 | 25.00 | 6.25 | 3.13 | 6.25 | 3.13 |
| Rhizopus arrhizus 156 | ≤50.00 | ≤50.00 | ≤50.00 | 25.00 | ≤50.00 | 25.00 |
| Fusarium oxysporum 600711 | ≤50.00 | 25.00 | 25.00 | 3.13 | 25.00 | 3.13 |
| Fusarium solani | ||||||
| 603251 | ≤25.00 | 12.50 | 25.00 | 3.13 | 25.00 | 3.13 |
| 600679 | ≤25.00 | 12.50 | 3.13 | 1.56 | 3.13 | 1.56 |
| Aspergillus fumigatus | ||||||
| Af293 | 3.13 | 0.78 | 3.13 | 0.78 | 3.13 | 0.78 |
| ATCC 13073 | ≤12.50 | ≤12.50 | 6.25 | 3.13 | 6.25 | 3.13 |
| Aspergillus flavus | ||||||
| Strain #1 | ≤12.50 | 6.25 | 6.25 | 1.56 | 6.25 | 1.56 |
| Strain #2 | ≤12.50 | 6.25 | 6.25 | 1.56 | 6.25 | 1.56 |
| Aspergillus niger | ||||||
| Strain #1 | ≤12.50 | ≤12.50 | 6.25 | 1.56 | 6.25 | 1.56 |
| Strain #2 | ≤12.50 | ≤12.50 | 6.25 | 1.56 | 6.25 | 1.56 |
| Strain #3 | ≤12.50 | ≤12.50 | 3.13 | 1.56 | 3.13 | 1.56 |
| Saccharomyces cerevisiae | ||||||
| A2 | 0.20 | 1.56 | 1.56 | |||
| BY4741 | 0.20 | 1.56 | 1.56 | |||
| Bacterial species | ||||||
| Escherichia coli | ≤50.00 | ≤50.00 | ≤50.00 | |||
| Staphylococcus epidermidis | ≤50.00 | ≤50.00 | ≤50.00 | |||
| Staphylococcus aureus | 25.00 | ≤50.00 | 25.00 | |||
| Bacillus cereus | 6.25 | 25.00 | 12.50 | |||
| Mammalian cell lines | ||||||
| NIH 3T3 | ≤50.00 | ≤50.00 | ≤50.00 | |||
| A549 | ≤50.00 | ≤50.00 | ≤50.00 | |||
Importantly, CANBEF-13 and CANBEF-24 did not inhibit the proliferation of mammalian cells in culture and only weakly inhibited two of the four bacterial species tested, suggesting that these compounds are fungus specific (Table 2). CANBEF-13, -14, and -24 at concentrations as high as 50 μM did not induce sheep red blood cell hemolysis even after 24 h of incubation, indicating that they do not function as membrane-disrupting agents (data not shown). All CANBEFs were highly active in various media, including defined fungal MM or SC medium, rich YPD or YAG medium, and the defined cell culture media RPMI 1640 and DMEM. However, CANBEFs lost their antifungal activity (MIC, >32 μM) upon addition of 10% serum or albumin to the cell culture medium, indicating that they are tightly bound by the albumin.
CANBEF-24 causes morphological changes characteristic of damage to the cell wall of A. fumigatus.
The effects of CANBEF-24 on fungal structure were characterized further with A. fumigatus strain Af293. TEM, calcofluor white staining, and DiBAC staining were used to determine the effects of CANBEF-24 on cell wall polysaccharide deposition, ultrastructure, and viability, respectively. After 24 h of growth in the presence of subinhibitory concentrations (0.8 μM) of CANBEF-24, strain Af293 displayed defects in the ultrastructure of the cell wall characterized by distended hyphal growth, abnormal fragmentation of the outer cell wall, and increased chitin staining of the swollen cell bodies (Fig. 2A). DiBAC vital staining revealed numerous dead (fluorescing) hyphal segments (Fig. 2B).
FIG 2.
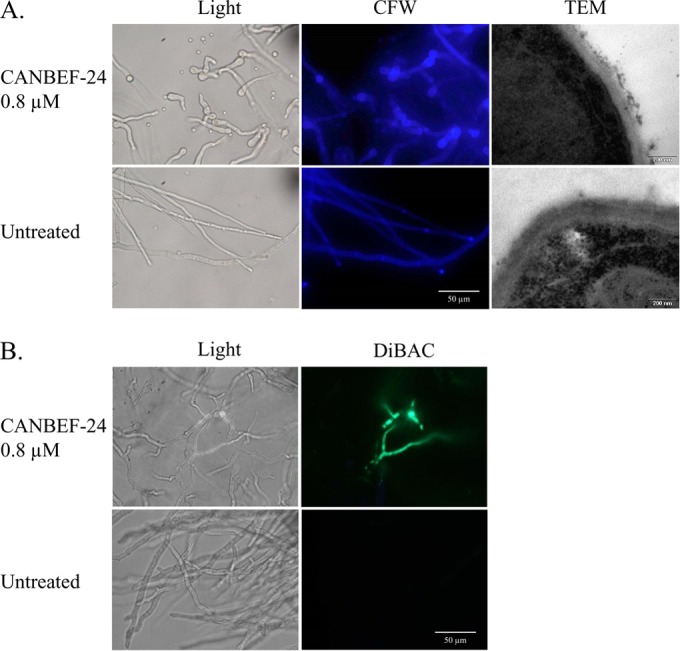
CANBEF-24 causes morphological changes characteristic of damage to the cell wall of A. fumigatus. A. fumigatus strain Af293 was incubated for 24 h in the presence of 0.8 μM CANBEF-24 and was analyzed microscopically, in comparison to untreated cells, by calcofluor white (CFW) staining and TEM (A) and DiBAC vital staining (B). Increased CFW staining of cell wall polysaccharides and abnormal cell wall morphology (TEM) reveal CANBEF-24-induced wall damage. DiBAC staining indicates partial cell death in discrete areas of the hypha.
CANBEF-24 and caspofungin interact synergistically.
We hypothesized that the mode of action of CANBEF-24 differs from that of existing antifungals and that consequently, they might display beneficial synergy when combined. Therefore, CANBEF-24 was tested for interactions with the antifungal drugs caspofungin (a glucan synthase inhibitor), voriconazole (an ergosterol biosynthesis inhibitor), and amphotericin B (a membrane-disrupting compound), and with the protein kinase C inhibitor staurosporine (which blocks the cell wall integrity pathway), by using a checkerboard modification of the guidelines presented in CLSI document M38-A (18). The MIC of each compound alone and in combination with the other compounds for A. fumigatus Af293 was determined. The results were used to determine the fractional inhibitory concentration index (FICI) of the combination of CANBEF-24 with amphotericin B, voriconazole, caspofungin, or staurosporine. FICI values were interpreted as follows: ≤0.5, synergy; ≥0.5 and ≤4, no interaction; >4, antagonism. While there was no interaction between CANBEF-24 and voriconazole, amphotericin B, or staurosporine, there was a synergistic interaction between CANBEF-24 and the cell wall inhibitor caspofungin (FICI, 0.31 [Table 3]). These results suggest that CANBEF-24 and caspofungin damage the fungal cell wall by inhibiting different targets, leading to synergy, or, alternatively, that wall damage allows either drug to enter the cell and act more efficiently.
TABLE 3.
In vitro activities of CANBEF-24 alone and in combination with caspofungin, voriconazole, amphotericin B, or staurosporine against A. fumigatus Af293
| Drug | Drug MIC (μg/ml) |
CANBEF-24 MIC (μM) |
FICI | ||
|---|---|---|---|---|---|
| Alone | Comb.a | Alone | Comb. | ||
| Caspofungin | 128 | 8.00 | 3.13 | 0.78 | 0.31 |
| Voriconazole | 1.00 | 2.00 | 3.13 | 1.56 | 2.50 |
| Amphotericin B | 0.50 | 0.06 | 6.25 | 3.13 | 0.63 |
| Staurosporine | 5.00 | 2.50 | 6.25 | 3.13 | 1.00 |
Comb., in combination.
Overexpression of AN2693 in A. nidulans confers resistance to CANBEF-24.
We screened a library of A. nidulans genomic DNAs cloned into the high-copy-number nonintegrating vector pRG3-AMA1-NotI. A. nidulans GR-5 transformants containing the AMA1-plasmid library were plated under selection with CANBEF-24 at 8 μM. First, we identified five resistant colonies (MIC, 32 μM) and isolated identical multicopy library vectors from two of them. Retransformation of this plasmid into a CANBEF-24-susceptible A. nidulans strain gave rise to resistance (MIC, 32 μM, in contrast to 4 μM for a control A. nidulans strain transformed with the empty library vector). Sequencing of the plasmids showed that all contained identical segments of A. nidulans chromosome IV, in which four hypothetical genes are encoded. Among these four genes is the HIT finger domain gene AN2693. Transposon mapping revealed that disruption of AN2693 in the plasmid resulted in loss of resistance. The AN2693 protein product is homologous to the essential S. cerevisiae protein Bcd1p (systematic gene name, YHR040W). This protein is required for the accumulation of box C/D snoRNA (19, 20), needed for the methylation and biogenesis of noncoding rRNA (21), suggesting that CANBEF-24 inhibits rRNA processing and maturation.
Overexpression of S. cerevisiae genes participating in ribosome assembly and regulation confers resistance to CANBEF-13.
To better understand the mechanism of action of the CANBEFs, we made use of the powerful genetic tools available in S. cerevisiae. A conditional gene overexpression library of S. cerevisiae consisting of ∼5,800 strains was screened for resistance to CANBEF-13. Each strain contains a unique ORF under the control of the galactose-inducible GAL1 promoter (17).
Overexpression strains were pooled and plated at 105 CFU/plate on SM with galactose as the sole carbon source for induction of overexpression. CANBEF-13 was added at 0.75 μM. Twenty-three isolates exhibiting greater resistance to CANBEF-13 than the parental strain in the inducing galactose-containing medium, but not in noninducing glucose medium, were defined as resistant to CANBEF-13 (MIC, 1 μM, in contrast to 0.2 μM). The overexpressed gene was amplified by PCR with primers for the constant region flanking the inserted ORF. The PCR products were sequenced and identified by BLAST. Among 23 sequenced isolates, 13 different genes conferring resistance when overexpressed were identified (Table 4). Interestingly, 6 of the 13 genes (RPL7B, NOP58, LIA1, YEF3, RIA1, and GON7) (P = 0.00017) are required for rRNA processing, ribosome assembly, and the control of protein synthesis, suggesting that CANBEF-13 may act by inhibiting these processes.
TABLE 4.
S. cerevisiae BY4741 overexpression strains with increased resistance to CANBEF-13
| Process | Gene namea | ORF | Function |
|---|---|---|---|
| Ribosomal assembly and control | RPL7B | YPL198W | Large-subunit protein, rRNA processing, nucleolus |
| NOP58 | YOR310C | 18S rRNA synthesis, nucleolus | |
| RIA1 (6) | YNL163C | Ribosomal biogenesis, cytoplasm | |
| LIA1 (2) | YJR070C | eIF5A modification, translation initiation | |
| YEF3 (3) | YLR249W | Translational elongation factor, gamma subunit | |
| GON7 | YJL184W | t6A tRNA modification; may be involved in transcription and in osmotic stress response | |
| Proteasomal degradation | CUE2 | YKL090W | Unknown; binds ubiquitin |
| DMA2 | YNL116W | Ubiquitin ligase E3 | |
| Cell wall | PIR3 (2) | YKL163W | O-glycosylated, covalently bound |
| Others | INO80 | YGL150C | Nucleosome spacing factor |
| YJL147C | Unknown, mitochondrial | ||
| YBL086C | Unknown, cell periphery | ||
| SRB8 (2) | YCR081W | Subunit of the RNA polymerase II mediator complex |
For strains that were isolated more than once, the number of independent isolations is written in parentheses.
To test if CANBEF-13 inhibits protein synthesis in S. cerevisiae, cells were incubated in the presence of [35S]methionine and 2 μM CANBEF-13 or 10 μM cycloheximide (a known inhibitor of protein synthesis) for 15 to 45 min. The results showed that CANBEF-13 strongly and rapidly inhibits protein synthesis (Fig. 3, top).
FIG 3.
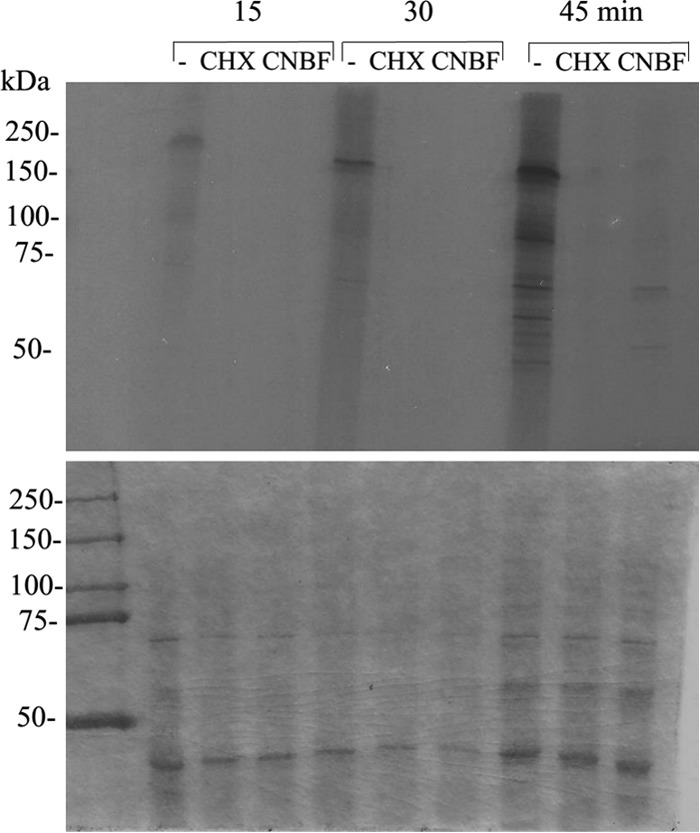
CANBEF-13 blocks protein translation in S. cerevisiae. S. cerevisiae strain BY4741 was incubated for 15, 30, or 45 min in the presence of [35S]methionine either alone (−) or together with cycloheximide (CHX; 10 μM) or CANBEF-13 (CNBF; 2 μM). Cells were subsequently lysed and were analyzed by SDS-PAGE. (Top) The autoradiogram shows inhibition of [35S]methionine incorporation into nascent proteins by both CHX (a known protein synthesis inhibitor) and CANBEF-13 relative to that for the untreated control (−). (Bottom) Coomassie protein stain.
CANBEF-24 is nontoxic to Galleria mellonella larvae, Drosophila melanogaster Tl flies, and mice.
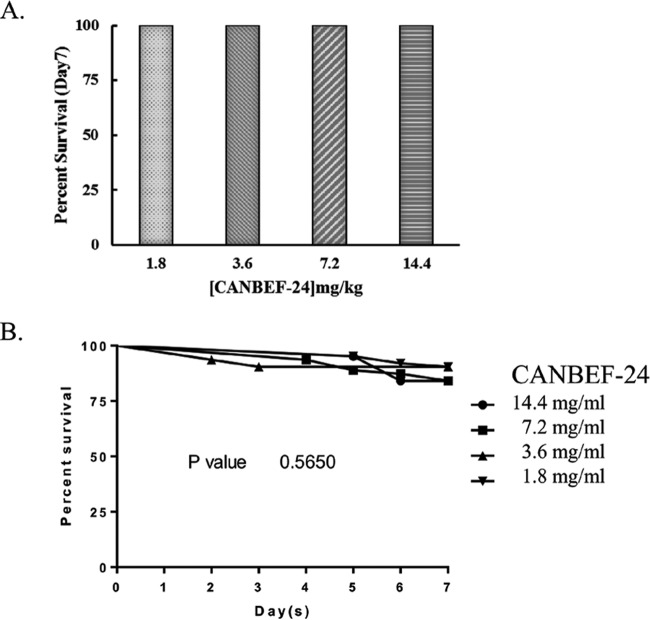
The toxicity of CANBEF-24 was tested in vivo in two insect models currently used to study fungal infection (Galleria mellonella larvae and Drosophila melanogaster Tl flies). CANBEF-24 was injected once into Galleria larvae (n, 10/group) at 1.8, 3.6, 7.2, or 14.4 mg/kg of body weight (equivalent to 6, 12, 24, and 48 μM, respectively). All survived unchanged for 7 days in the presence of CANBEF-24 (Fig. 4A). Toxicity in D. melanogaster Tl flies was tested by adding CANBEF-24 at increasing concentrations (1.8, 3.6, 7.2, and 14.4 mg/ml) in the fly food. CANBEF-24 was nontoxic to Tl flies at concentrations as high as 14.4 mg/ml for 7 days (Fig. 4B). Preliminary in vivo toxicity studies were also performed in mice. CANBEF-24 was injected intravenously on four consecutive days into immunocompetent ICR outbred female mice (n, 2/group) at 1.8, 3.6, 7.2, and 14.4 mg/kg (equivalent to 6, 12, 24, and 48 μM, respectively), and mice were sacrificed on day 7. No change in body weight occurred during the experiment. Internal organs (spleen, liver, kidneys) were unchanged in appearance and weight (data not shown).
FIG 4.
Toxicity analyses of CANBEF-24. For analysis of toxicity, Galleria mellonella larvae were injected once with as much as 14.4 mg/kg CANBEF-24 (A), and Drosophila Tl−/− flies were fed with fly food containing as much as 14.4 mg/ml of CANBEF-24 for 7 days (B). Mortality was assessed daily for 7 days.
The ability of CANBEF-24 to reduce mortality in Galleria mellonella larvae and Drosophila melanogaster Tl flies infected with A. fumigatus or in mice infected with Candida albicans was also tested. CANBEF-24 at concentrations as high as 14.4 mg/kg did not reduce the mortality of infected larvae (P = 0.97) or flies (P = 0.88) (see Fig. S1A and B in the supplemental material). Similarly, CANBEF-24 did not reduce mortality in immunocompetent mice infected intravenously with C. albicans (P = 0.92) (see Fig. S1C in the supplemental material). The significance of these results is discussed below.
DISCUSSION
Current treatments for invasive fungal infection are still associated with considerable patient mortality (1, 2). Clinically used antifungals inhibit a limited number of cellular pathways, and their administration is associated with toxicity and emerging resistance (22).
Attempts to identify novel antifungal compounds have been described for several model fungi, such as S. cerevisiae (23) and C. albicans (24), for which advanced molecular genetic tools are available. Few screens have focused on identifying compounds compromising fungal cell wall integrity, despite the fact that the cell wall constitutes a unique and essential organ synthesized and maintained by hundreds of gene products (25).
In this report, we describe a whole-cell screening strategy to identify cell wall-perturbing compounds. In the first screening phase, a diverse chemical compound library was screened for antifungal activity against the pathogenic mold A. fumigatus. Sixteen hits were identified and were further analyzed. We used an inducible alcA-PKC mutant of A. nidulans hypersensitive to cell wall damage in order to identify which of the 16 compounds affects cell wall integrity, as described previously (8, 9). Eight compounds were identified, including five CANBEF derivatives and three other compounds that were later dropped due to toxicity for cultured mammalian cells (see Table S1 in the supplemental material). CANBEFs have not been described previously in the scientific literature as having antimicrobial or antifungal activity. The 4-chloro-7-nitrobenzofurazan moiety of the molecule is a highly sensitive chromogenic and fluorogenic reagent and an inhibitor of purified plant and fungal vacuolar ATPases (26).
The most potent CANBEFs displayed excellent in vitro characteristics: they damaged the cell wall (TEM, synergy with CAS), inhibited most pathogenic yeasts and molds at low concentrations (3.1 to 12.5 μM), were fungicidal at concentrations 3- to 4-fold higher than their MICs, and did not inhibit the growth of cultured mammalian cells at concentrations as high as 50 μM.
To determine the mode of action of the CANBEFs, we used two complementary overexpression screens in A. nidulans and S. cerevisiae. In A. nidulans, high-copy-number expression of AN2693, homologous to the essential S. cerevisiae protein Bcd1p, conferred 8-fold-increased resistance to CANBEF. This protein is required for the accumulation of box C/D snoRNA (19, 20), needed for the methylation and biogenesis of noncoding rRNA (21). In S. cerevisiae, overexpression of 13 genes conferred 5-fold-increased CANBEF resistance. Six of the 13 genes are required for rRNA processing (RPL7B, NOP58), ribosome assembly (RIA1), and the control of protein synthesis (LIA1, YEF3, and GON7). Interestingly, both AN2693/Bcd1p, identified in the A. nidulans screen, and NOP58, identified in the yeast screen, are box C/D snoRNA binding proteins participating in the methylation of 18S rRNA, strongly suggesting a key role for the CANBEFs in inhibiting the maturation of this essential ribosomal component. Inhibition of 18S rRNA maturation would be expected to block protein synthesis rapidly, and indeed, we show that in yeasts treated with CANBEF-13, [35S]methionine incorporation into newly synthesized proteins was rapidly inhibited. However, at this point, it is not possible to determine if these compounds inhibit protein synthesis directly or if they do so indirectly, by inhibiting ribosome maturation. Distinguishing between these possibilities will require more-specialized analyses, such as rRNA labeling and fractionation, and polysome profiling, to detect changes in the abundances of the ribosomal subunits.
Selective inhibition of protein synthesis in fungi is not an obvious target for the development of antifungals, considering the high degree of conservation of this system in all eukaryotes, including fungi and mammals. However, an entire class of antifungals, the sordarins, that selectively inhibits fungal translation has been identified and developed (27). The sordarins target fungal elongation factor 2 (EF-2), rapidly blocking translation. Nonetheless, because they display a poor pharmacokinetic profile, sordarin derivatives have not entered clinical use (28).
It is somewhat surprising that we initially identified the CANBEFs for their ability to damage the cell wall. How could inhibition of protein synthesis lead to cell wall damage? There is precedent for this result: gene deletions affecting protein synthesis in yeast (e.g., deletions of EGD1, JJJ1, RSA1, SDA1, SSZ1, YEF3, and ZUO1) result in a pleiotropic phenotype that includes compromised cell wall integrity as well as defects in numerous other cellular functions, such as cell cycle progression, cytokinesis, transport, and endocytosis (http://www.yeastgenome.org/). A decrease in protein synthesis rates or translational fidelity can rapidly affect the production of proteins necessary for numerous cellular functions, including construction of the cell wall.
Preliminary in vivo experiments carried out in insect and murine models revealed that CANBEF compounds are apparently nontoxic, even at concentrations equivalent to 10-fold the MIC in vitro. The fact that the CANBEF compounds we tested were not effective as treatments for disseminated mycoses in insect and mouse models of infection is not surprising, since primary screen “hit” compounds are rarely effective in vivo and usually require considerable additional derivatization and screening cycles for optimization in vivo. Such improvement cycles were necessary for the optimization of both the azole and the candin antifungals, which were too toxic for in vivo administration during their early development.
Although we showed that serum or albumin abolished CANBEF activity in vitro, this is not necessarily the reason they are inactive in vivo. Frequently, there is a very poor correlation between the in vitro binding affinity of a drug for serum and its in vivo activity. Many of the most highly prescribed drugs have a high binding affinity for serum that reduces their in vitro activity, yet they are nevertheless highly effective in vivo (29). This is because in vivo activity is only weakly affected by drug serum binding and is more strongly affected by several other processes, such as the rate of clearance, compartmentalization, target binding, and transport of the drug (29). Further cycles of optimization to increase the bioavailability of the CANBEFs are warranted. At this stage, however, CANBEF derivatives may prove useful in the treatment of fungal skin infections or as plant fungicides, where the issue of bioavailability does not arise. For example, the antifungal tavaborole, an inhibitor of aminoacyl-tRNA synthetase and protein synthesis, was recently authorized as an effective topical treatment for fungal nail infections (onychomycosis) (30).
In summary, we have identified and characterized a novel class of antifungal compounds, the CANBEFs, which selectively interfere with the process of protein synthesis in fungi. While CANBEFs display fungal specificity and a wide spectrum of antifungal activity in vitro and are not toxic in vivo, further development is needed to optimize them for in vivo efficacy.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Binational Science Foundation (BSF) grant 2011322 to N.O. and D.P.K.
We thank Maya Schuldiner for generously providing us with the yeast libraries and strains.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00850-15.
REFERENCES
- 1.Ben-Ami R, Lewis RE, Kontoyiannis DP. 2009. Invasive mould infections in the setting of hematopoietic cell transplantation: current trends and new challenges. Curr Opin Infect Dis 22:376–384. doi: 10.1097/QCO.0b013e32832db9f3. [DOI] [PubMed] [Google Scholar]
- 2.Yapar N. 2014. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag 10:95–105. doi: 10.2147/TCRM.S40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Denning DW. 2000. Early diagnosis of invasive aspergillosis. Lancet 355:423–424. doi: 10.1016/S0140-6736(00)82003-6. [DOI] [PubMed] [Google Scholar]
- 5.McNeil MM, Nash SL, Hajjeh RA, Phelan MA, Conn LA, Plikaytis BD, Warnock DW. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. Clin Infect Dis 33:641–647. doi: 10.1086/322606. [DOI] [PubMed] [Google Scholar]
- 6.Lewis RE, Cahyame-Zuniga L, Leventakos K, Chamilos G, Ben-Ami R, Tamboli P, Tarrand J, Bodey GP, Luna M, Kontoyiannis DP. 2013. Epidemiology and sites of involvement of invasive fungal infections in patients with haematological malignancies: a 20-year autopsy study. Mycoses 56:638–645. doi: 10.1111/myc.12081. [DOI] [PubMed] [Google Scholar]
- 7.Paiva JA, Pereira JM. 2013. New antifungal antibiotics. Curr Opin Infect Dis 26:168–174. doi: 10.1097/QCO.0b013e32835ebcb7. [DOI] [PubMed] [Google Scholar]
- 8.Mircus G, Hagag S, Levdansky E, Sharon H, Shadkchan Y, Shalit I, Osherov N. 2009. Identification of novel cell wall destabilizing antifungal compounds using a conditional Aspergillus nidulans protein kinase C mutant. J Antimicrob Chemother 64:755–763. doi: 10.1093/jac/dkp270. [DOI] [PubMed] [Google Scholar]
- 9.Ronen R, Sharon H, Levdansky E, Romano J, Shadkchan Y, Osherov N. 2007. The Aspergillus nidulans pkcA gene is involved in polarized growth, morphogenesis and maintenance of cell wall integrity. Curr Genet 51:321–329. doi: 10.1007/s00294-007-0129-y. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2008. M38-A2. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.CLSI. 2008. M27-A3. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Osherov N, Mathew J, May GS. 2000. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet Biol 31:181–188. doi: 10.1006/fgbi.2000.1236. [DOI] [PubMed] [Google Scholar]
- 13.Osherov N, May G. 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, May GS, Lionakis MS, Lewis RE, Kontoyiannis DP. 2004. Extra copies of the Aspergillus fumigatus squalene epoxidase gene confer resistance to terbinafine: genetic approach to studying gene dose-dependent resistance to antifungals in A. fumigatus. Antimicrob Agents Chemother 48:2490–2496. doi: 10.1128/AAC.48.7.2490-2496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osherov N, Kontoyiannis DP, Romans A, May GS. 2001. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J Antimicrob Chemother 48:75–81. doi: 10.1093/jac/48.1.75. [DOI] [PubMed] [Google Scholar]
- 16.Osmani SA, May GS, Morris NR. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol 104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, Lee AY, Ryan O, Gottert H, Surendra A, van Dyk D, Giaever G, Boone C, Nislow C, Andrews BJ. 2012. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda) 2:1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinel-Ingroff A, Fothergill A, Ghannoum M, Manavathu E, Ostrosky-Zeichner L, Pfaller M, Rinaldi M, Schell W, Walsh T. 2005. Quality control and reference guidelines for CLSI broth microdilution susceptibility method (M 38-A document) for amphotericin B, itraconazole, posaconazole, and voriconazole. J Clin Microbiol 43:5243–5246. doi: 10.1128/JCM.43.10.5243-5246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, Snydsman BE, Bradley P, Muller EG, Fields S, Baker D, Yates JR III, Davis TN. 2003. Assigning function to yeast proteins by integration of technologies. Mol Cell 12:1353–1365. doi: 10.1016/S1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- 20.Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, Mitsakakis N, Ryan OW, Datta N, Jojic V, Pal C, Canadien V, Richards D, Beattie B, Wu LF, Altschuler SJ, Roweis S, Frey BJ, Emili A, Greenblatt JF, Hughes TR. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919–933. doi: 10.1016/S0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- 21.Weinstein LB, Steitz JA. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol 11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 22.Jucker E, Polack A. 2003. Antifungal agents: advances and problems. Progress in drug research. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 23.Roemer T, Davies J, Giaever G, Nislow C. 2011. Bugs, drugs and chemical genomics. Nat Chem Biol 8:46–56. doi: 10.1038/nchembio.744. [DOI] [PubMed] [Google Scholar]
- 24.Singh SB, Ondeyka J, Harris G, Herath K, Zink D, Vicente F, Bills G, Collado J, Platas G, Gonzalez del Val A, Martin J, Reyes F, Wang H, Kahn JN, Galuska S, Giacobbe R, Abruzzo G, Roemer T, Xu D. 2013. Isolation, structure, and biological activity of phaeofungin, a cyclic lipodepsipeptide from a Phaeosphaeria sp. using the genome-wide Candida albicans fitness test. J Nat Prod 76:334–345. doi: 10.1021/np300704s. [DOI] [PubMed] [Google Scholar]
- 25.Tada R, Latge JP, Aimanianda V. 2013. Undressing the fungal cell wall/cell membrane—the antifungal drug targets. Curr Pharm Des 19:3738–3747. doi: 10.2174/1381612811319200012. [DOI] [PubMed] [Google Scholar]
- 26.Bowman EJ, Mandala S, Taiz L, Bowman BJ. 1986. Structural studies of the vacuolar membrane ATPase from Neurospora crassa and comparison with the tonoplast membrane ATPase from Zea mays. Proc Natl Acad Sci U S A 83:48–52. doi: 10.1073/pnas.83.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calugi C, Trabocchi A, Guarna A. 2011. Novel small molecules for the treatment of infections caused by Candida albicans: a patent review (2002–2010). Expert Opin Ther Pat 21:381–397. doi: 10.1517/13543776.2011.551116. [DOI] [PubMed] [Google Scholar]
- 28.Hanadate T, Tomishima M, Shiraishi N, Tanabe D, Morikawa H, Barrett D, Matsumoto S, Ohtomo K, Maki K. 2009. FR290581, a novel sordarin derivative: synthesis and antifungal activity. Bioorg Med Chem Lett 19:1465–1468. doi: 10.1016/j.bmcl.2009.01.051. [DOI] [PubMed] [Google Scholar]
- 29.Smith DA, Di L, Kerns EH. 2010. The effect of plasma protein binding on in vivo efficacy: misconceptions in drug discovery. Nat Rev Drug Discov 9:929–939. doi: 10.1038/nrd3287. [DOI] [PubMed] [Google Scholar]
- 30.Hu QH, Liu RJ, Fang ZP, Zhang J, Ding YY, Tan M, Wang M, Pan W, Zhou HC, Wang ED. 2013. Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase. Sci Rep 3:2475. doi: 10.1038/srep02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.