Abstract
AVIATOR, a phase 2 clinical trial, evaluated ritonavir-boosted paritaprevir (a protease inhibitor), ombitasvir (an NS5A inhibitor), and dasabuvir (a nonnucleoside polymerase inhibitor) (the three-drug [3D] regimen) with or without ribavirin (RBV) for 8, 12, or 24 weeks in 406 HCV genotype 1 (GT1)-infected patients. The rate of sustained virologic response 24 weeks after treatment ranged from 88% to 100% across the arms of the 3D regimen with or without RBV; 20 GT1a-infected patients and 1 GT1b-infected patient experienced virologic failure (5.2%). Baseline resistance-conferring variants in NS3 were rare. M28V in GT1a and Y93H in GT1b were the most prevalent preexisting variants in NS5A, and C316N in GT1b and S556G in both GT1a and GT1b were the most prevalent variants in NS5B. Interestingly, all the GT1a sequences encoding M28V in NS5A were from the United States, while GT1b sequences encoding C316N and S556G in NS5B were predominant in the European Union. Variants preexisting at baseline had no significant impact on treatment outcome. The most prevalent treatment-emergent resistance-associated variants (RAVs) in GT1a were R155K and D168V in NS3, M28T and Q30R in NS5A, and S556G in NS5B. The single GT1b-infected patient experiencing virologic failure had no RAVs in any target. A paritaprevir-ritonavir dose of 150/100 mg was more efficacious in suppressing R155K in NS3 than a 100/100-mg dose. In patients who failed after receiving 12 or more weeks of treatment, RAVs were selected in all 3 targets, while most patients who relapsed after 8 weeks of treatment did so without any detectable RAVs. Results from this study guided the selection of the optimal treatment regimen, treatment duration, and paritaprevir dose for further development of the 3D regimen. (This study has been registered at ClinicalTrials.gov under registration number NCT01464827.)
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, single-stranded, positive-sense RNA virus in the Flaviviridae family that infects approximately 130 million to 150 million people worldwide (1, 2). Seven distinct HCV genotypes and 67 subtypes have been characterized (3). The level of nucleotide sequence diversity between genotypes is 30 to 35% and that between subtypes is 20 to 25% (4), leading to HCV genotype- and subtype-dependent variability in the treatment response to individual direct-acting antiviral agents (DAAs) (5–7). The RNA-dependent RNA polymerase of HCV is intrinsically error prone, and its lack of a proofreading function results in the introduction of approximately 1 nucleotide change per genome per replication cycle, leading to the presence of preexisting drug-resistant variants and their expansion under selective pressure (8). Understanding treatment-emergent resistance-associated variants (RAVs) as well as the impact of preexisting variants on treatment outcome in patients failing treatment with DAA therapy is important for the assessment of treatment and retreatment options.
In the AVIATOR phase 2b clinical study (study M11-652; ClinicalTrials.gov number NCT01464827), several combinations of three HCV DAAs with distinct mechanisms of action were evaluated (9). Paritaprevir (formerly ABT-450, identified by AbbVie and Enanta) is an inhibitor of the HCV NS3/4A protease and is coadministered with the pharmacokinetic enhancer ritonavir (paritaprevir/r). Amino acid variants conferring resistance to paritaprevir were detected in NS3 at position 155, 156, or 168 in vitro or following monotherapy in HCV genotype 1 (GT1)-infected subjects (10). Ombitasvir (formerly ABT-267) is an HCV NS5A inhibitor. NS5A variants conferring resistance to ombitasvir were selected in vitro or following monotherapy in GT1-infected subjects at amino acid position 28, 30, 31, 58, or 93 (11). Dasabuvir (formerly ABT-333) is a palm I site nonnucleoside HCV RNA-dependent RNA polymerase inhibitor. Variants conferring resistance to dasabuvir were selected in NS5B at amino acid position 316, 414, 448, 556, or 559 in vitro or following monotherapy in GT1-infected subjects (12, 13).
AVIATOR was an open-label study with 14 treatment arms that enrolled 571 GT1-infected patients without cirrhosis who were treatment naive or prior null responders to pegylated interferon and ribavirin (RBV). Patients were randomly assigned to one of several two-drug (2D) or three-drug (3D) regimens of paritaprevir/r combined with ombitasvir or dasabuvir, or both, for 8, 12, or 24 weeks (9). All treatment arms except one also included RBV. The rate of sustained virologic response 24 weeks after treatment (SVR24) ranged from 83% to 100% across the treatment arms, with optimal SVR24 rates being in the arms that contained a 3D rather than a 2D regimen (9). Among the treatment-naive patients who received a 3D regimen with RBV (with paritaprevir administered at 150 mg with 100 mg of ritonavir), SVR24 rates ranged from 88% to 97% among those who received 8 and 12 weeks of therapy; extending treatment to 24 weeks offered no further benefit (9). Across the study, the most frequent adverse events were fatigue, headache, nausea, and insomnia, and 1% of the patients discontinued treatment due to adverse events (9).
The objective of this analysis was to provide a comprehensive evaluation of viral resistance in patients in the AVIATOR study who received the 3D regimen with or without RBV (Table 1). This subpopulation included 406 treatment-naive or prior null-responder patients receiving 8, 12, or 24 weeks of therapy in 10 treatment arms (9). The overall SVR24 rate in the treatment-naive patients was 87.5% (189/216) for the GT1a-infected patients and 98% (100/102) for the GT1b-infected patients. The SVR24 rate in the prior null responders was 93% (51/55) for the GT1a-infected patients and 97% (32/33) for the GT1b-infected patients. Twenty-one patients (20 infected with GT1a and 1 infected with GT1b) experienced virologic failure (VF), due to on-treatment breakthrough or posttreatment relapse. Thirteen patients did not achieve an SVR24 for nonvirologic reasons, e.g., early discontinuations or missing viral load data at 24 weeks posttreatment. The results of the analysis of viral resistance in patients in the AVIATOR study who received a 2D regimen with RBV are presented in the supplemental material.
TABLE 1.
Study design and treatment outcome of the AVIATOR 3D regimen with and without RBV
| Study population and arm | Inclusion of RBV with 3Da regimen | Duration (wk) of treatment | Paritaprevir dose (mg)b | % SVR24c |
No. of patients experiencing VFd |
||||
|---|---|---|---|---|---|---|---|---|---|
| Breakthrough |
Relapse |
||||||||
| GT1a | GT1b | GT1a | GT1b | GT1a | GT1b | ||||
| Treatment naive | |||||||||
| A | + | 8 | 150 | 83.9 (47/56) | 95.8 (23/24) | 9 | 1 | ||
| E | − | 12 | 150 | 82.7 (43/52) | 100 (25/25) | 1 | 4 | ||
| F | + | 12 | 100 | 96.3 (26/27) | 100 (12/12) | ||||
| G | + | 12 | 150 | 92.6 (25/27) | 100 (13/13) | 1 | |||
| H | + | 24 | 100 | 92.6 (25/27) | 92.3 (12/13) | 1 | |||
| I | + | 24 | 150 | 85.2 (23/27) | 100 (12/12) | ||||
| Treatment-experienced null responders | |||||||||
| K | + | 12 | 100 | 86.7 (13/15) | 100 (8/8) | 2 | |||
| L | + | 12 | 150 | 92.3 (12/13) | 100 (9/9) | 1 | |||
| M | + | 24 | 100 | 92.9 (13/14) | 88.9 (8/9) | 1 | |||
| N | + | 24 | 150 | 100 (13/13) | 100 (7/7) | ||||
The 3D regimen consisted of paritaprevir/r, ombitasvir, and dasabuvir.
The daily dose of paritaprevir (plus 100 mg of ritonavir) in each arm is indicated; ombitasvir was included at 25 mg once daily, dasabuvir was included at 400 mg twice daily, and RBV was included at 1,000 mg if the body was less than 75 kg or 1,200 mg if the body weight was 75 kg or more.
Data in parentheses represent the number of patients achieving SVR24/total number of patients in the treatment arm.
Patients not achieving SVR24 for nonvirologic reasons, e.g., early discontinuations and missing SVR24 data, were not considered VFs.
In the resistance analyses of the AVIATOR study, the prevalence of baseline variants at resistance-associated amino acid positions in NS3, NS5A, and NS5B and their impact on treatment outcome (SVR24) were evaluated. Treatment-emergent RAVs in the 21 patients who experienced VF and 2 of the patients who did not achieve SVR24 for nonvirologic reasons and for whom posttreatment samples were available were assessed.
MATERIALS AND METHODS
Clinical study design.
The AVIATOR study (Clinical.Trials.gov number NCT01464827) was a randomized, open-label, phase 2b study with 14 treatment arms that examined the safety and efficacy of combinations of paritaprevir/r, ombitasvir, dasabuvir, and RBV in patients with HCV GT1 infection. Details of the study and randomization procedure were previously described (9). Paritaprevir was administered once daily at a dose of 100 mg, 150 mg, or 200 mg with 100 mg of ritonavir. Ombitasvir was dosed at 25 mg once daily, and dasabuvir was dosed at 400 mg twice daily. The daily dose of RBV was 1,000 mg (divided into doses of 400 mg and 600 mg) if the body weight was less than 75 kg or 1,200 mg (600 mg twice daily) if the body weight was 75 kg or more. The treatment duration was 8, 12, or 24 weeks. Ten of the treatment arms included the 3D regimen with paritaprevir doses of 100 mg or 150 mg, as shown in Table 1. Four of the treatment arms included the 2D regimen with paritaprevir (100 mg, 150 mg, or 200 mg) dosed with either ombitasvir or dasabuvir plus RBV (see Table S1 in the supplemental material). The primary efficacy endpoint was SVR24 (which was considered an HCV RNA level below the lower limit of quantitation [25 IU/ml] at 24 weeks posttreatment).
All of the patients provided written informed consent. The study was performed in accordance with good clinical practice guidelines and the principles of the Declaration of Helsinki, and the study protocol was approved by the relevant institutional review boards and regulatory agencies.
Sample processing.
The subtype-specific primers for reverse transcription-PCR (RT-PCR), nested PCR, and sequencing were designed based on the alignments of GT1a or GT1b sequences in the European HCV database (14) in conserved regions specific to the gene of interest, with nucleotide degeneracies incorporated at positions where significant variability existed among the HCV sequences for the subtype. HCV RNA was purified from 550 μl of plasma samples using an Abbott m2000 instrument (Abbott Molecular, Des Plaines, IL) and eluted in a volume of 70 μl. The target genes, the NS3/4A, NS5A, and NS5B genes, were each amplified from 20 μl of the purified HCV RNA by RT-PCR using a SuperScript III one-step RT-PCR system with Platinum Taq high-fidelity DNA polymerase (Invitrogen, Carlsbad, CA), followed by nested PCR using primers appropriate for subtype 1a or 1b samples. Only samples with HCV RNA levels of ≥1,000 IU/ml were amplified in order to reduce the chance of oversampling bias. For patients who did not achieve SVR24, the sample with an HCV RNA level of ≥1,000 IU/ml obtained the closest in time after VF or treatment discontinuation was utilized. In this study, no patient samples were excluded due to a low viral titer. For samples with HCV RNA levels of ≤50,000 IU/ml, the RT-PCR was carried out in triplicate, and the products were pooled prior to their use as a template for nested PCR.
Population sequencing of NS3/4A, NS5A, and NS5B was conducted on the nested PCR products using gene- and subtype-specific primers. For clonal sequencing, the nested gel-purified NS3 or NS5A PCR products were cloned into the pJET1.2/blunt cloning vector using a CloneJET PCR cloning kit (Fermentas, Glen Burnie, MD), while NS5B was cloned into the GT1a or GT1b replicon shuttle vector cassette as described previously (10, 11, 13, 15, 16). Plasmid DNA was isolated from an average of 81 individual colonies per sample (range, 52 to 95), and the target gene was sequenced. At least two sequencing reads were performed in each direction across each target, providing a minimum of four sequencing reads.
Sequence analyses.
Analyses for (i) the prevalence of variants at resistance-associated amino acid positions in NS3, NS5A, and NS5B at baseline, (ii) the impact of baseline HCV variants on treatment response (SVR24) using the chi-square test, and (iii) treatment-emergent RAVs were performed using SAS (version 9.3) software (SAS Institute, Inc., Cary, NC) under a UNIX operating system. On the basis of the results of in vitro studies with HCV subgenomic replicons and phase 2a clinical studies in HCV-infected patients, the following were identified to be signature resistance-associated amino acid positions in the baseline sequence analysis: 36, 56, 155, 156, and 168 in NS3 in GT1a and 155, 156, and 168 in NS3 in GT1b for paritaprevir; 28, 30, 31, 32, 58, and 93 in NS5A in GT1a and 28, 29, 30, 31, 32, 58, and 93 in NS5A in GT1b for ombitasvir; and 316, 414, 446, 448, 451, 553, 554, 555, 556, 558, 559, and 561 in NS5B in GT1a and 316, 368, 411, 414, 445, 448, 553, 556, 558, and 559 in NS5B in GT1b for dasabuvir (10–13). Although variants at amino acid residue 80 in NS3 are not associated with resistance to paritaprevir (10), position 80 was included in the GT1a analysis due to its impact on the NS3 protease inhibitor simeprevir (5, 7). Clonal sequencing validation experiments had previously established that variants identified in a single clone by clonal sequencing were not reproducibly detected in replicate experiments (P. Krishnan, unpublished data). Therefore, RAVs by clonal sequencing were defined as variants observed in 2 or more clones from a sample obtained at a baseline or postbaseline time point relative to the appropriate reference sequence GT1a strain H77 (GenBank accession number NC004102) or GT1b strain Con1 (GenBank accession number AJ238799).
Antiviral activity against a panel of resistant variants.
The methods for measuring the effects of individual amino acid variants on the activity of an inhibitor in HCV replicon cell culture assays were described previously (17). Variants in NS3, NS5A, or NS5B were each introduced into the GT1a strain H77 replicon using a Change-IT multiple-mutation site-directed mutagenesis kit (Affymetrix, Santa Clara, CA). After the presence of the variant(s) was confirmed by sequence analysis, the plasmid was linearized and a TranscriptAid T7 high-yield transcription kit (Fermentas, Glen Burnie, MD) was used to transcribe the HCV genomic RNA from the plasmid. In a transient-transfection assay, the replicon RNA containing the variant was transfected via electroporation into a Huh-7 cell line (16). The luciferase activity in the cells was measured using a Victor II luminometer (Perkin-Elmer, Waltham, MA). The 50% effective concentration was calculated using nonlinear regression curve fitting to the 4-parameter logistic equation and GraphPad Prism (version 4) software.
Compounds.
Paritaprevir [(2R,6S,12Z,13aS,14aR,16aS)-N-(cyclopropylsulfonyl)-6-{[(5-methylpyrazin-2-yl)carbonyl]amino}-5,16-dioxo-2-(phenanthridin-6-yloxy)-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamidehydrate], ombitasvir [dimethyl ([(2S,5S)-1-(4-tert-butylphenyl)pyrrolidine-2,5-diyl]bis{benzene-4,1-diylcarbamoyl(2S)pyrrolidine-2,1-diyl[(2S)-3-methyl-1-oxobutane-1,2-diyl]})biscarbamate hydrate], and dasabuvir (sodium N-{6-[3-tert-butyl-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-2-methoxyphenyl]naphthalen-2-yl} methanesulfonamide) were synthesized at AbbVie, and their structures have been previously disclosed (10, 13, 18).
Phylogenetic analysis of baseline sequences.
The population DNA sequences from baseline samples were included in the phylogenetic analyses to determine the genetic relatedness of the DAA target sequences according to geographic region: the United States or the European Union. One sample each from Canada and Australia was included in the analysis. Sequences encompassing nucleotides 1 to 1080 in NS3, 1 to 645 in NS5A, and 898 to 1773 in NS5B were aligned using the MAFFT method (19), and phylogenetic trees were constructed using the neighbor-joining tree-building method (20) with the HKY85 nucleotide substitution model (21). The reliability of the tree topology was examined using 1,000 bootstrapping replicates to generate a consensus tree with a 50% threshold cutoff for each phylogenetic analysis. Nucleotide sequence alignments and phylogenetic trees were generated using the Geneious (22) and MEGA (version 5) (23) software packages.
RESULTS
Baseline analyses.
The baseline amino acid variants at positions associated with resistance to NS3, NS5A, or NS5B inhibitors in GT1a and GT1b are shown in Tables 2 and 3, respectively.
TABLE 2.
Prevalence of baseline polymorphisms in GT1a-infected patients
| Target and variant | %a |
|---|---|
| NS3 (nb = 230) | |
| V36A | 0.9 |
| V36L | 1.3 |
| V36M | 0.9 |
| Q80K | 41 |
| Q80L | 2.2 |
| D168A | 0.4 |
| NS5 (n = 235) | |
| M28T | 0.4 |
| M28V | 6.0 |
| L31M | 0.4 |
| L31V | 0.4 |
| Q30H | 2.1 |
| Q30R | 1.3 |
| H58C | 0.4 |
| H58P | 1.7 |
| H58Q | 0.9 |
| H58R | 1.3 |
| H58Y | 0.4 |
| Y93C | 0.4 |
| Y93H | 1.3 |
| Y93N | 0.4 |
| NS5B (n = 258) | |
| C316Y | 0.8 |
| M414T | 0.4 |
| A553G | 0.4 |
| S556G | 3.1 |
| S556N | 0.4 |
| S556R | 0.4 |
Percentage of patients with the baseline polymorphism relative to the GT1a strain H77 reference sequence.
n, number of samples sequenced for that target.
TABLE 3.
Prevalence of baseline polymorphisms in GT1b-infected patients
| Target and variant | %a |
|---|---|
| NS3 (nb = 119), nonec | |
| NS5A (n = 130) | |
| R30Q | 8.5 |
| L31I | 2.3 |
| L31M | 6.2 |
| P58A | 0.8 |
| P58L | 0.8 |
| P58R | 0.8 |
| P58S | 3.8 |
| P58T | 2.4 |
| Y93H | 5.4 |
| NS5B (n = 125) | |
| C316H | 0.8 |
| C316K | 0.8 |
| C316N | 18.4 |
| C316W | 0.8 |
| S368A | 0.8 |
| M414L | 0.8 |
| C445F | 1.6 |
| S556G | 16.0 |
Percentage of patients with the baseline polymorphism relative to the GT1b strain Con1 reference sequence.
n, number of samples sequenced for that target.
None, baseline polymorphisms were not detected at resistance-associated amino acid positions.
Baseline polymorphisms in NS3 in GT1a at amino acid position V36, Q80, or D168 were detected in 45% (104/230) of the sequences. Variants at amino acid position 36 or 168 were rarely observed at the baseline. Polymorphisms at amino acid position 80, predominantly Q80K, were frequently observed but conferred ≤3-fold resistance to paritaprevir (10). Baseline polymorphisms at resistance-associated amino acid positions were not observed in the 119 NS3 sequences from GT1b-infected patients.
Baseline polymorphisms in NS5A in GT1a at amino acid position M28, Q30, L31, H58, or Y93 were detected in 15% (35/235) of the sequences. M28V, which confers 58-fold resistance to ombitasvir in the GT1a replicon (11), was the most prevalent variant in GT1a. Baseline polymorphisms at amino acid position L28, R30, L31, P58, or Y93 in NS5A were detected in 25% (32/130) of the sequences in GT1b. Y93H, which confers 77-fold resistance to ombitasvir in the GT1b replicon (11), was the predominant resistance-conferring GT1b variant.
Baseline polymorphisms in NS5B in GT1a at amino acid position C316, M414, A553, or S556 were detected in 5% (13/258) of the sequences. S556G, which confers 30-fold resistance to dasabuvir (13), was the most prevalent variant in GT1a. Baseline polymorphisms amino acid position C316, S368, M414, C445, or S556 were detected in 28% (35/125) of the sequences in GT1b. The combination of variants C316N plus S556G, which confers 38-fold resistance to dasabuvir (13), was observed in 10.4% (13/125) of the samples.
By population sequencing analysis, none of the patients had baseline RAVs in all 3 targets, and only 2 patients (1 GT1a-infected patient and 1 GT1b-infected patient) had RAVs in NS5A as well as NS5B.
Association between baseline polymorphisms and treatment outcome.
The most prevalent amino acid variants in GT1a-infected patients at baseline were Q80K in NS3, M28V in NS5A, and S556G in NS5B; of these, Q80K confers minimal resistance (10). Other variants conferring high-level resistance to components of the 3D regimen, observed in a minority of GT1a-infected patients, were D168A in NS3, Q30R, L31V, or Y93C/H/N in NS5A, and C316Y in NS5B (10, 11, 13). Although the number of patients with baseline variants other than Q80K was small, there was no difference in SVR24 rates among GT1a-infected patients with any of these variants at the baseline from those among patients with the reference amino acid residue at the corresponding position (Table 4).
TABLE 4.
Observed SVR24 rate in the presence of baseline variants
| GT1a target | Variant | % SVR24a |
P valueb | |
|---|---|---|---|---|
| With variant | Without variant | |||
| NS3 | Q80K | 88 (78/89) | 94 (122/130) | 0.14 |
| D168A | 0 (0/1) | 92 (200/218) | 0.087 | |
| NS5A | M28T/V | 86 (12/14) | 92 (192/209) | 0.339 |
| Q30R | 100 (3/3) | 91 (201/220) | 1.0 | |
| L31V | 100 (1/1) | 91 (203/222) | 1.0 | |
| Y93C/N/H | 80 (4/5) | 92 (200/218) | 0.362 | |
| NS5B | S556G | 100 (7/7) | 92 (220/239) | 1.0 |
| C316Y | 50 (1/2) | 93 (226/244) | 0.149 | |
Data in parentheses represent the number of patients achieving SVR24/total number of patients who had a sequence available. Patients not achieving SVR24 for nonvirologic reasons, e.g., early discontinuations and missing SVR24 data, were excluded from this analysis.
P values were determined by the chi-square test.
Treatment-emergent variants in patients not achieving SVR24.
Of the 406 treatment-naive or prior null-responder patients receiving 8, 12, or 24 weeks of therapy with the 3D regimen, 21 patients experienced VF. Of these 21 patients, 5 experienced on-treatment breakthroughs and 16 were posttreatment relapsers (Table 1). Ten of the 21 patients, including the single GT1b-infected patient who experienced a posttreatment relapse, were in the 8-week arm of the study. The baseline variants and treatment-emergent RAVs identified by clonal sequencing in at least 2 clones (detection limit, 5 to 10%) in the patients experiencing VF are listed in Table 5.
TABLE 5.
RAVs relative to reference sequence in NS3, NS5A, and NS5B at time of VFa
| Study population and arm | GT | VF type | Variant(s) |
|||||
|---|---|---|---|---|---|---|---|---|
| NS3 |
NS5A |
NS5B |
||||||
| Baseline | Time of VF | Baseline | Time of VF | Baseline | Time of VF | |||
| Treatment naive | ||||||||
| A | 1a | Relapse at PTW 2 | Noneb | None | M28V | M28V | None | None |
| A | 1a | Relapse at PTW 4 | D168A/D | D168A | none | Q30R | S556G/S | S556G |
| A | 1a | Relapse at PTW 4 | None | None | None | None | None | None |
| A | 1a | Relapse at PTW 4 | None | None | None | None | None | None |
| A | 1a | Relapse at PTW 8 | None | None | L31L/M | None | S556G/S | None |
| A | 1a | Relapse at PTW 2 | None | None | None | M28T + H58P | S556G/S | None |
| A | 1a | Relapse at PTW 4 | None | None | None | None | None | None |
| A | 1a | Relapse at PTW 4 | None | None | None | None | None | None |
| A | 1a | Relapse at PTW 24 | None | D168V | None | Q30R | None | S556G |
| A | 1b | Relapse at PTW 4 | None | None | None | None | None | None |
| E | 1a | Relapse at PTW 8 | None | D168V | Y93N/S/Y | Y93N | None | None |
| E | 1a | Breakthrough at wk 12 | None | Y56H + D168Y, D168Y | None | M28V + Q30K | None | S556G |
| E | 1a | Relapse at PTW 8 | None | D168V | Y93N/Y | Y93N | None | M414T, S556G |
| E | 1a | Relapse at PTW 12 | None | D168V | None | Q30R | C316Y, S556G/S | C316Y + S556G |
| E | 1a | Relapse at PTW 2 | None | D168V | None | Q30R | None | S556G |
| G | 1a | Relapse at PTW 2 | None | R155K, D168V | M28M/V | M28T | None | G554S, S556G |
| H | 1a | Relapse at PTW 48 | None | None | None | None | None | None |
| Treatment-experienced null responders | ||||||||
| K | 1a | Breakthrough at wk 8 | None | V36M + R155K, R155K | None | M28T, Q30R | None | D559G |
| K | 1a | Breakthrough at wk 6 | None | D168V | Q30Q/R | Q30R | None | A553T |
| L | 1a | Breakthrough at wk 12 | None | D168Y | None | Q30R | None | S556G |
| M | 1a | Breakthrough at wk 16 | None | V36A/M + R155K | None | Q30R | M414I/M | G558R |
| Treatment naive | ||||||||
| I | 1a | D/C | None | None | M28V | M28V | None | S556G |
| I | 1a | D/C | None | None | None | None | None | None |
GT, genotype; D/C, premature study drug discontinuation; PTW, posttreatment week; VF, virologic failure; +, linked variants; slashes, mixture of variants.
None, variants at resistance-associated amino acid positions were not detected by clonal sequencing.
Of the 20 GT1a-infected VF patients, 12 had RAVs in NS3 at the time of VF. Seven patients from the 8-week treatment arm had no RAVs in NS3 at the time of failure, nor did one patient who was treated for 24 weeks and subsequently relapsed at posttreatment week 48. Among the other patients, the treatment-emergent RAV D168V was detected in 7 patients, D168Y alone or in combination with Y56H was detected in 2 patients, and R155K alone or in combination with V36A/M was detected in 3 patients. D168A was identified at baseline and at the time of failure in 1 patient. Of note, 2 of the 3 patients with treatment-emergent R155K received a regimen including paritaprevir at the lower dose of 100 mg.
Fourteen of the 20 GT1a-infected patients had RAVs in NS5A at the time of VF. The 6 patients with no RAVs in NS5A at the time of failure included 5 from the 8-week treatment arm, as well as the patient described above who relapsed at posttreatment week 48. Among the other patients, treatment-emergent NS5A variants M28V plus Q30K were detected in 1 patient, M28T was detected in 3 patients, and Q30R was detected in 7 patients. Four of the 6 patients with preexisting NS5A variants at baseline also had the same variant at the time of failure.
Eleven of the 20 GT1a-infected patients had RAVs in NS5B at the time of VF. Nine of these patients had no RAVs in NS5B at the time of failure, including 7 patients treated for 8 weeks, 1 patient treated for 12 weeks without RBV, and the patient described above who relapsed at posttreatment week 48. Treatment-emergent NS5B RAVs M414T, G554S, A553T, G558R, and D559G were each observed in 1 patient, and S556G was detected in 6 patients. Two of the 5 patients with preexisting NS5B variants at baseline had the same variant at the time of failure.
One GT1b-infected patient in the 8-week treatment arm experienced VF; this patient did not have any RAVs at baseline or at the time of VF in any of the three targets.
Among the 20 GT1a-infected patients with VF, treatment-emergent RAVs were detected in all 3 targets in 7 patients, in NS3 and NS5A in 1 patient, in NS3 and NS5B in 2 patients, and in only NS5A in 2 patients. One GT1a-infected patient had a preexisting NS5A variant but no treatment-emergent variants, and 7 GT1a-infected patients (including 6 in the 8-week treatment arm) had no RAVs in any target. The resistance profile was similar among patients experiencing on-treatment breakthrough and those who relapsed posttreatment.
Among the 13 patients not achieving SVR24 for nonvirologic reasons, 6 had missing data at the SVR24 time point. Postbaseline samples with HCV RNA levels of ≥1,000 IU/ml were available for 2 of the 13 patients (Table 5). Neither patient had treatment-emergent RAVs in NS3 or NS5A, and 1 had S556G in NS5B.
Activity of paritaprevir, ombitasvir, and dasabuvir against variants in GT1a H77 replicons.
The activity of paritaprevir, ombitasvir, and dasabuvir against variants observed in GT1a-infected patients experiencing VF was evaluated in the GT1a H77 replicon (Table 6). NS3 variants R155K and D168A conferred lower levels of resistance to paritaprevir than the D168V and D168Y variants. The combination of these variants with either V36M or Y56H conferred an additional 2-fold resistance to paritaprevir. The lower level of resistance conferred by R155K supports the observation that this variant appeared to be suppressed with higher doses of paritaprevir. NS5A variants L31M and H58P did not confer resistance to ombitasvir, and M28V conferred moderate levels of resistance, while M28T, Q30R, and Y93N each conferred at least 800-fold resistance to ombitasvir. Consistent with the phenotype, L31M was not enriched in the patient treated for 8 weeks who had a 31L/M mixture at baseline, and M28V was rarely detected as a treatment-emergent variant in patients experiencing VF. NS5B variants M414I/T and S556G conferred lower levels of resistance to dasabuvir than the C316Y, G554S, and A553T variants. Consistent with the phenotype, M414I was not enriched at the time of failure in the patient randomized to 24 weeks of treatment who had a mixture of 414I/M at baseline.
TABLE 6.
Activity of paritaprevir, ombitasvir, and dasabuvir against variants in GT1a H77 replicon
| NS3 |
NS5A |
NS5B |
|||
|---|---|---|---|---|---|
| Variant | Fold resistance to paritaprevir | Variant | Fold resistance to ombitasvir | Variant | Fold resistance to dasabuvir |
| V36A | 3 | M28T | 8,965 | C316Y | 1,472 |
| V36M | 2 | M28V | 58 | M414I | 8 |
| Y56H | 3 | Q30R | 800 | M414T | 32 |
| R155K | 37 | L31M | 2 | A553T | 152 |
| D168A | 50 | H58P | 0.5 | G554S | 198 |
| D168V | 96 | Y93N | 66,740 | S556G | 30 |
| D168Y | 219 | G558R | NDa | ||
| V36M + R155K | 79 | D559G | ND | ||
| Y56H + D168Y | 451 | ||||
ND, fold resistance could not be determined due to the low replication capacity of the variant.
Geographic and phylogenetic analysis of baseline sequences.
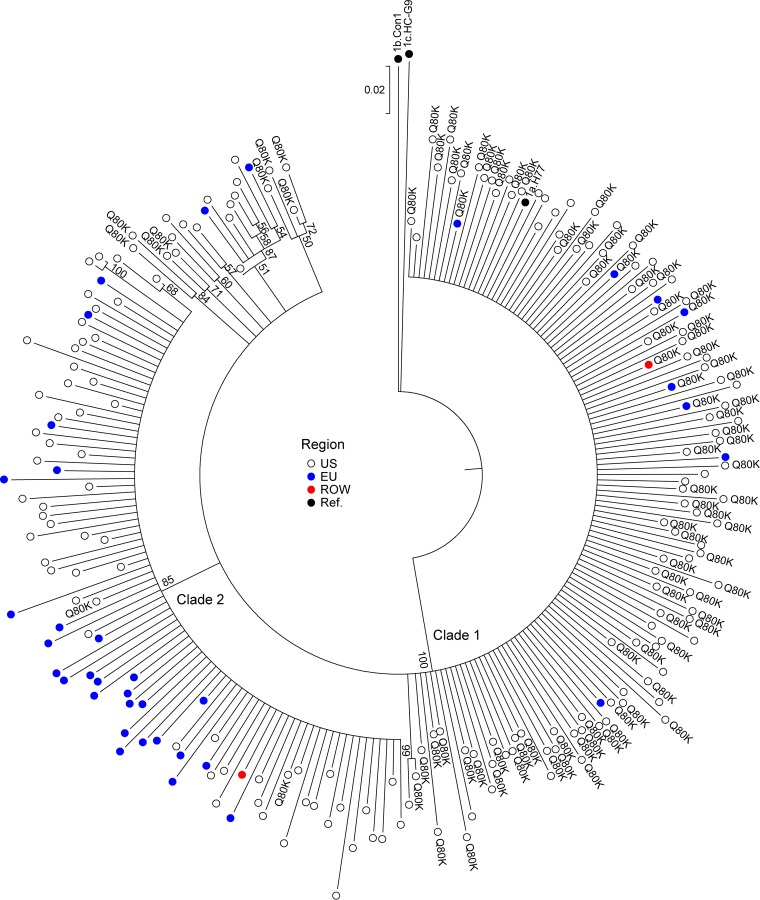
Phylogenetic analyses were conducted using baseline sequences from GT1a- and GT1b-infected patients to determine the genetic relatedness of the DAA target sequences according to geographic region. The analysis of NS3 in GT1a included 229 baseline sequences and 3 reference sequences. The geographic distribution of GT1a NS3 sequences included in the analysis was 83% (190/229) from the United States, 16% (37/229) from the European Union, and 1 sequence each from Canada and Australia. Four major phylogenetic groups are depicted in Fig. 1. The GT1b strain Con1 and GT1c strain HC-G9 reference sequences each sorted as separate groups, and GT1a baseline sequences from the AVIATOR study sorted into 2 distinct clades, with the GT1a H77 reference sequence sorting to clade 1. Both GT1a clades included sequences from the United States and the European Union. Clade 1 included 61% (140/229) of the GT1a baseline sequences, including 7 subgroups comprising 23 sequences. Clade 2 included 39% (89/229) of the GT1a baseline sequences, including 2 subgroups comprising 4 sequences. Baseline sequences from the United States clustered predominantly into clade 1 (68%, 129/190 sequences), while baseline sequences from the European Union clustered predominantly into clade 2 (73%, 27/37 sequences).
FIG 1.
Geographic distribution of HCV NS3 nucleotide sequences. The NS3 neighbor-joining phylogenetic tree is displayed in a circular format. The reliability of the tree topology was examined using 1,000 bootstrapping replicates, and bootstrap values of ≥50 are listed at appropriate nodes in the tree. The genetic distance scale bar indicates the number of nucleotide substitutions per site between sequences. Rest of world, 1 sequence each from Canada and Australia.
Phylogenetic analyses of NS5B GT1a baseline sequences also revealed a similar conservation of the 2 GT1a clades (data not shown). Phylogenetic analyses of NS5A in GT1a and NS3, NS5B, and NS5A baseline sequences in GT1b each revealed one major cluster of sequences, and specific clustering according to geographic region was not seen (data not shown).
Some of the polymorphisms in NS3, NS5A, and NS5B showed specific geographic distribution (Table 7).The majority of the GT1a sequences with Q80K in NS3 sorted into clade 1 (98%, 93/95), with the overall prevalence of Q80K being 66.4% in this clade. The prevalence of Q80K was 46.8% (89/190) in the United States, whereas it was 13.5% (5/37) in the European Union. All of the GT1a-infected patients with M28V in NS5A were from the United States. C316N and S556G in NS5B in GT1b were predominant among sequences from the European Union. Other variants were observed in sequences from the United States as well as the European Union, as shown in Table 7.
TABLE 7.
Distribution of prevalent baseline variants by geographic region
| Target, GT | Variant | Phylogenetic cluster | %a |
|
|---|---|---|---|---|
| United States | European Union | |||
| NS3, GT1a | Q80K | Clade 1 (n = 140) | 67.4 (87/129) | 50 (5/10) |
| Clade 2 (n = 89) | 3.3 (2/61) | 0 (0/27) | ||
| Total | 46.8 (89/190) | 13.5 (5/37) | ||
| NS5A, GT1a | M28V | 7 (14/191) | 0 (0/42) | |
| NS5B, GT1a | S556G | Clade 1 | 2.9 (4/139) | 6.2 (1/16) |
| Clade 2 | 3.1 (2/65) | 2.8 (1/36) | ||
| Total | 2.9 (6/204) | 3.9 (2/52) | ||
| NS5A, GT1b | Y93H | 2.9 (2/68) | 8.2 (5/61) | |
| NS5B, GT1b | C316N | 4.6 (3/65) | 32 (19/59) | |
| S556G | 6.2 (4/65) | 25 (15/59) | ||
The percentage of patients within each geographic region with the variant. Values in parentheses represent the number of patients with the variant/number of available sequences in the geographic region for each target.
DISCUSSION
The phase 2b AVIATOR trial assessed 2D or 3D combination regimens in 571 treatment-naive and treatment-experienced patients for 8, 12, and 24 weeks. On the basis of the differential SVR24 rates observed in this trial (9), a 3D regimen with and without RBV was selected for further clinical development. Among the 406 subjects across the various arms in the AVIATOR study administered the 3D regimen with or without RBV, there were low rates of discontinuation due to adverse events, and SVR24 rates ranged from 88% to 100% (9). There were 21 virologic failures, of which 10 were in the 8-week arm of the study.
In the AVIATOR trial, the prevalence of baseline RAVs varied by drug target, HCV subtype, and geographic region. The presence of RAVs in NS3 at baseline was rare in either subtype. M28V in GT1a and Y93H in GT1b were the most prevalent baseline RAVs in NS5A, while S556G in NS5B was the most prevalent baseline RAV in both GT1 subtypes. There was not a significant difference in SVR24 rates in patients with any preexisting RAV at baseline compared to patients without the RAV, suggesting that either the drug levels or the nonoverlapping resistance profiles of the drugs in the 3D regimen reduce the impact of baseline resistance on treatment response.
Among the 21 patients who experienced VF, the majority of those who received 12 weeks or more of treatment showed the emergence of RAVs across all 3 targets. The most prevalent treatment-emergent RAVs in GT1a were R155K and D168V in NS3, M28T and Q30R in NS5A, and S556G in NS5B. Consistent with the lower levels of resistance conferred by M28V in NS5A to ombitasvir in the GT1a H77 replicon, this variant, which had a 6% baseline prevalence, did not impact treatment outcome and was rarely treatment emergent in virologic failures. The paucity of RAVs at the time of failure among samples from patients who relapsed after 8 weeks of treatment, as detected by clonal sequencing (detection limit, 5 to 10%), suggested that for those patients the duration of treatment was insufficient to fully suppress the wild-type virus population. Virologic failure was rarely seen in patients with GT1b, and the single GT1b-infected patient experiencing VF in the 8-week arm of the study had no RAVs at the time of failure.
Among the 165 patients receiving a 2D regimen with RBV, 17 experienced VF, all with GT1a infection (see Table S1 in the supplemental material). The pattern of treatment-emergent RAVs in these patients was similar to that observed with the 3D regimen (see Table S2 in the supplemental material).
The AVIATOR trial also helped to establish the optimal dosage of paritaprevir. Although comparable efficacies were seen with all doses of paritaprevir/r studied, the infrequent emergence of R155K in patients with VF at a dose of 150/100 mg suggests that this dose was more efficacious at suppressing this variant, which confers 37-fold resistance, than the 100/100-mg dose; hence, in the phase 3 development program, paritaprevir/r was dosed at 150/100 mg.
The persistence of treatment-emergent RAVs is currently being followed to posttreatment week 48 among patients treated with paritaprevir/r-, ombitasvir-, and dasabuvir-based regimens in a pooled analysis of phase 2 and 3 clinical studies and in a 3-year follow-up study that enrolled a subset of subjects from the phase 2 and 3 clinical studies. Resistance analyses with other combination DAA regimens generally suggest that NS5A variants are persistent, while NS3 variants generally decay through posttreatment week 48 (24).
Given the heterogeneity of the HCV genome, phylogenetic analyses were conducted on NS3, NS5A, and NS5B baseline sequences from patients infected with GT1a and GT1b to compare the genetic relatedness of the sequences according to geographic region. Analysis of NS3 and NS5B revealed conservation of 2 major clusters of HCV GT1a sequences. Clade 1 contained a larger proportion of sequences from the United States and clade 2 contained a larger proportion of sequences from the European Union for both targets. NS3 and NS5B sequences from the United States or the European Union did not form specific subgroup clusters in either clade, suggesting similar genetic relatedness between sequences from the United States and the European Union within each clade of the phylogenetic analysis. A similar divergence of GT1a isolates into 2 distinct clades was described by Pickett et al., using full-genome sequencing data (25). The clustering of GT1a isolates was not attributed to geography or time of isolation, but a number of clade-informative sites were identified within the NS3 protease, NS5A domains 2 and 3, and NS5B (25), which is consistent with our observations. The inclusion of only domain 1 of NS5A in our phylogenetic analysis may account for the observed lack of clustering of GT1a NS5A sequences into separate clades.
In this study, all GT1a sequences encoding M28V in NS5A were from the United States, and GT1b sequences encoding C316N and S556G in NS5B were predominant in the European Union. As Q80K in GT1a NS3 impacts treatment outcome with the NS3 protease inhibitor simeprevir (5, 7), the prevalence of this polymorphism was evaluated in this study. The majority of sequences encoding Q80K in NS3 (98%) were found within one GT1a cluster. A phylogenetic analysis of GT1a sequences by geographic region and time of sample collection by McCloskey et al. indicated that the majority of the Q80K-carrying sequences (96%) have descended from a single substitution event that occurred over 50 years ago in the United States (26). This evolutionary history may account for the geographic differences in Q80K prevalence. In this study, Q80K was observed at a prevalence of 46.8% among sequences from the United States and at a prevalence of 13.5% among the sequences from the European Union.
In conclusion, while RAVs in NS5A and NS5B were observed at baseline, they did not appear to affect treatment response, suggesting that this multitargeted HCV GT1 antiviral regimen affords a high barrier to resistance. Overall VF rates were low at 5.2%. RAVs were typically selected in all 3 targets in patients who failed after receiving 12 or more weeks of treatment, while most patients who relapsed after 8 weeks of treatment did so without any detectable RAVs. The results from the AVIATOR study were used to determine the optimal treatment regimen, duration, and paritaprevir dose for further development of paritaprevir/r, ombitasvir, and dasabuvir in treatment-naive and treatment-experienced patients with chronic HCV GT1 infection, including those with compensated cirrhosis (27–31).
Supplementary Material
ACKNOWLEDGMENTS
We thank the trial participants, investigators, and coordinators who made this study possible. We thank Barbara McGovern for critical review of the manuscript.
Design, study conduct, and financial support for this study were provided by AbbVie. AbbVie participated in the interpretation of the data, review, and approval of the publication. We are all employees of AbbVie and may own AbbVie stock.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00998-15.
REFERENCES
- 1.Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect 17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J Gen Virol 85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson I, Dore G, Foster G, Fried M, Radu M, Rafalskiy V, Moroz I, Craxi A, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Kalmeijer R, Beumont-Mauviel M. 2013. Simeprevir (TMC435) with peginterferon/ribavirin for chronic HCV genotype-1 infection in treatment-naive patients: results from QUEST-1, a phase III trial. J Hepatol 58:S574. doi: 10.1016/S0140-6736(14)60494-3. [DOI] [Google Scholar]
- 6.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 7.Manns M, Marcellin P, Poordad F, Affonso de Araujo ES, Buti M, Horsmans Y, Janczewska E, Villamil F, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, Kalmeijer R, Beumont-Mauviel M. 2014. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 384:414–426. doi: 10.1016/S0140-6736(14)60538-9. [DOI] [PubMed] [Google Scholar]
- 8.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A 110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowdley KV, Lawitz E, Poordad F, Cohen DE, Nelson DR, Zeuzem S, Everson GT, Kwo P, Foster GR, Sulkowski MS, Xie W, Pilot-Matias T, Liossis G, Larsen L, Khatri A, Podsadecki T, Bernstein B. 2014. Phase 2b trial of interferon-free therapy for hepatitis C virus genotype 1. N Engl J Med 370:222–232. doi: 10.1056/NEJMoa1306227. [DOI] [PubMed] [Google Scholar]
- 10.Pilot-Matias T, Tripathi R, Cohen D, Gaultier I, Dekhtyar T, Lu L, Reisch T, Irvin M, Hopkins T, Pithawalla R, Middleton T, Ng T, McDaniel K, Or YS, Menon R, Kempf D, Molla A, Collins C. 2015. In vitro and in vivo antiviral activity and resistance profile of the hepatitis C virus NS3/4A protease inhibitor ABT-450. Antimicrob Agents Chemother 59:988–997. doi: 10.1128/AAC.04227-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan P, Beyer J, Mistry N, Koev G, Reisch T, DeGoey D, Kati W, Campbell A, Williams L, Xie W, Setze C, Molla A, Collins C, Pilot-Matias T. 2015. In vitro and in vivo antiviral activity and resistance profile of ombitasvir, an inhibitor of hepatitis C virus NS5A. Antimicrob Agents Chemother 59:979–987. doi: 10.1128/AAC.04226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Middleton T, He Y, Beyer J, Menon R, Klein C, Cohen D, Collins C. 2010. Resistance profile of ABT-333 and relationship to viral load decrease in patients treated in combination with peg-interferon and ribavirin for 28 days. J Hepatol 52:S296–S297. doi: 10.1016/S0168-8278(10)60764-7. [DOI] [Google Scholar]
- 13.Kati W, Koev G, Irvin M, Beyer J, Liu Y, Krishnan P, Reisch T, Mondal R, Wagner R, Molla A, Maring C, Collins C. 2015. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob Agents Chemother 59:1505–1511. doi: 10.1128/AAC.04619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deleage G. 2007. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton T, He Y, Pilot-Matias T, Tripathi R, Lim BH, Roth A, Chen CM, Koev G, Ng TI, Krishnan P, Pithawalla R, Mondal R, Dekhtyar T, Lu L, Mo H, Kati WM, Molla A. 2007. A replicon-based shuttle vector system for assessing the phenotype of HCV NS5B polymerase genes isolated from patient populations. J Virol Methods 145:137–145. doi: 10.1016/j.jviromet.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Tripathi RL, Krishnan P, He Y, Middleton T, Pilot-Matias T, Chen CM, Lau DT, Lemon SM, Mo H, Kati W, Molla A. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antiviral Res 73:40–49. doi: 10.1016/j.antiviral.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Mo H, Lu L, Pilot-Matias T, Pithawalla R, Mondal R, Masse S, Dekhtyar T, Ng T, Koev G, Stoll V, Stewart KD, Pratt J, Donner P, Rockway T, Maring C, Molla A. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob Agents Chemother 49:4305–4314. doi: 10.1128/AAC.49.10.4305-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeGoey DA, Randolph JT, Liu D, Pratt J, Hutchins C, Donner P, Krueger AC, Matulenko M, Patel S, Motter CE, Nelson L, Keddy R, Tufano M, Caspi DD, Krishnan P, Mistry N, Koev G, Reisch TJ, Mondal R, Pilot-Matias T, Gao Y, Beno DW, Maring CJ, Molla A, Dumas E, Campbell A, Williams L, Collins C, Wagner R, Kati WM. 2014. Discovery of ABT-267, a pan-genotypic inhibitor of HCV NS5A. J Med Chem 57:2047–2057. doi: 10.1021/jm401398x. [DOI] [PubMed] [Google Scholar]
- 19.Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 22.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPhee F, Hernandez D, Yu F, Ueland J, Monikowski A, Carifa A, Falk P, Wang C, Fridell R, Eley T, Zhou N, Gardiner D. 2013. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology 58:902–911. doi: 10.1002/hep.26388. [DOI] [PubMed] [Google Scholar]
- 25.Pickett BE, Striker R, Lefkowitz EJ. 2011. Evidence for separation of HCV subtype 1a into two distinct clades. J Viral Hepat 18:608–618. doi: 10.1111/j.1365-2893.2010.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCloskey RM, Liang RH, Joy JB, Krajden M, Montaner JS, Harrigan PR, Poon AF. 2014. Global origin and transmission of hepatitis C virus nonstructural protein 3 Q80K polymorphism. J Infect Dis 211:1288–1295. doi: 10.1093/infdis/jiu613. [DOI] [PubMed] [Google Scholar]
- 27.Andreone P, Colombo MG, Enejosa JV, Koksal I, Ferenci P, Maieron A, Mullhaupt B, Horsmans Y, Weiland O, Reesink HW, Rodrigues L Jr, Hu YB, Podsadecki T, Bernstein B. 2014. ABT-450, ritonavir, ombitasvir, and dasabuvir achieves 97% and 100% sustained virologic response with or without ribavirin in treatment-experienced patients with HCV genotype 1b infection. Gastroenterology 147:359–365. doi: 10.1053/j.gastro.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1594–1603. doi: 10.1056/NEJMoa1315722. [DOI] [PubMed] [Google Scholar]
- 29.Ferenci P, Bernstein D, Lalezari J, Cohen D, Luo Y, Cooper C, Tam E, Marinho RT, Tsai N, Nyberg A, Box TD, Younes Z, Enayati P, Green S, Baruch Y, Bhandari BR, Caruntu FA, Sepe T, Chulanov V, Janczewska E, Rizzardini G, Gervain J, Planas R, Moreno C, Hassanein T, Xie W, King M, Podsadecki T, Reddy KR. 2014. ABT-450/r-ombitasvir and dasabuvir with or without ribavirin for HCV. N Engl J Med 370:1983–1992. doi: 10.1056/NEJMoa1402338. [DOI] [PubMed] [Google Scholar]
- 30.Poordad F, Hezode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM, Forns X, Lovell SS, Da Silva-Tillmann B, Collins CA, Campbell AL, Podsadecki T, Bernstein B. 2014. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med 370:1973–1982. doi: 10.1056/NEJMoa1402869. [DOI] [PubMed] [Google Scholar]
- 31.Zeuzem S, Jacobson IM, Baykal T, Marinho RT, Poordad F, Bourliere M, Sulkowski MS, Wedemeyer H, Tam E, Desmond P, Jensen DM, Di Bisceglie AM, Varunok P, Hassanein T, Xiong J, Pilot-Matias T, DaSilva-Tillmann B, Larsen L, Podsadecki T, Bernstein B. 2014. Retreatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med 370:1604–1614. doi: 10.1056/NEJMoa1401561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.