Abstract
para-Aminosalicylic acid (PAS) entered clinical use in 1946 as the second exclusive drug for the treatment of tuberculosis (TB). While PAS was initially a first-line TB drug, the introduction of more potent antitubercular agents relegated PAS to the second-line tier of agents used for the treatment of drug-resistant Mycobacterium tuberculosis infections. Despite the long history of PAS usage, an understanding of the molecular and biochemical mechanisms governing the susceptibility and resistance of M. tuberculosis to this drug has lagged behind that of most other TB drugs. Herein, we discuss previous studies that demonstrate PAS-mediated disruption of iron acquisition, as well as recent genetic, biochemical, and metabolomic studies that have revealed that PAS is a prodrug that ultimately corrupts one-carbon metabolism through inhibition of the formation of reduced folate species. We also discuss findings from laboratory and clinical isolates that link alterations in folate metabolism to PAS resistance. These advancements in our understanding of the basis of the susceptibility and resistance of M. tuberculosis to PAS will enable the development of novel strategies to revitalize this and other antimicrobial agents for use in the global effort to eradicate TB.
INTRODUCTION
Mycobacterium tuberculosis is responsible for approximately 8.6 million new cases of active tuberculosis (TB) infection and 1.3 million deaths annually despite the existence of TB therapy (1). While this therapy has a high success rate in curing drug-susceptible TB infections, it is challenging, in part because it requires a minimum of 6 months of treatment with drugs that are associated with adverse reactions (2, 3). These factors contribute to treatment errors and noncompliance, which have been implicated in the emergence of drug-resistant strains of M. tuberculosis (4, 5). Further, subsequent relapse of the disease can occur and is associated with a high incidence of drug resistance (6). Together, these complications have enabled the emergent spread of multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of M. tuberculosis that require greater than 2 years of therapy with second-line drugs and threaten the efficacy of existing TB therapy (1, 7). Elucidating the mechanisms that govern the susceptibility and resistance of M. tuberculosis to existing antitubercular agents will facilitate the discovery of new therapeutic approaches to shorten treatment times and counter drug-resistant TB.
para-Aminosalicylic acid (PAS) entered clinical use as a bacteriostatic antitubercular agent in 1946 (8). Shortly before the introduction of PAS, the discovery of streptomycin as a therapeutic tool had dramatically improved TB survival rates (9). At that time, it was apparent that the rapid emergence of streptomycin-resistant M. tuberculosis strains posed a threat to this monotherapy strategy for TB infection (9). As PAS was effective against streptomycin-resistant strains of M. tuberculosis (10), it was soon recognized that combination therapy could reduce the emergence of drug resistance (11–13). In the early 1950s, isoniazid was found to be highly effective in treating M. tuberculosis infections and was often included in streptomycin-PAS treatment regimens (14). This three-drug combination was found to dramatically increase cure rates and further decrease the emergence of drug resistance (15, 16).
PAS treatment was commonly associated with gastrointestinal disturbance and was eventually replaced with a better-tolerated companion agent, ethambutol (17). Yet, with the development of improved formulations of PAS and the global spread of MDR and XDR strains of M. tuberculosis, this drug has re-entered antitubercular drug regimens as an important second-line agent (18). In response to the revitalization of PAS use in TB therapy, there have been recent critical advances in our understanding of the molecular details of susceptibility and resistance of M. tuberculosis to this drug.
In this minireview, we summarize the current understanding of the impact of PAS on M. tuberculosis metabolism. We focus much of this discussion on folate metabolism, as PAS activity is intimately associated with this essential metabolic pathway. We discuss the proposed modes of action of PAS, its bactericidal effects, and recently characterized resistance mechanisms. Finally, we summarize areas of investigation to further our understanding of PAS interaction with M. tuberculosis.
FOLATE METABOLISM AS A HIGH-VALUE DRUG TARGET.
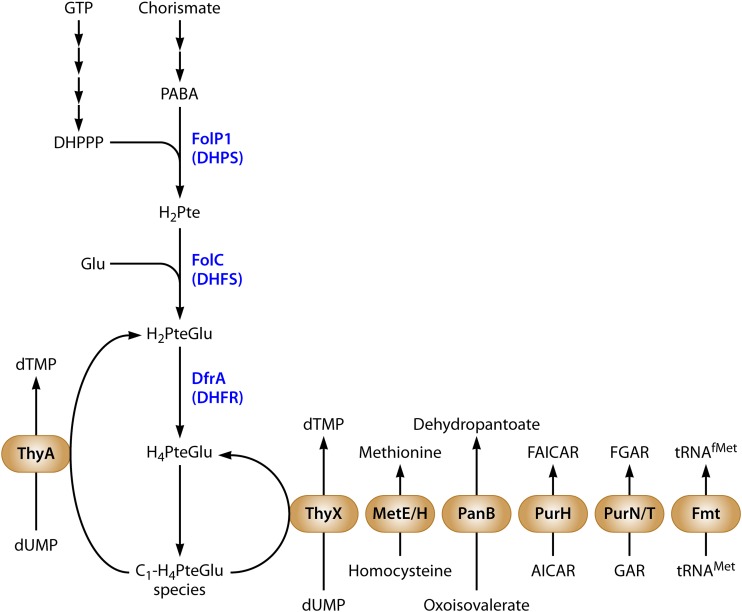
In prokaryotes and eukaryotes, reduced folate species serve as essential cofactors in the transfer of one-carbon groups in pathways for the synthesis of methionine, N-formylmethionyl-tRNA, glycine, serine, pantothenate, purines, and thymidine (Fig. 1) (19, 20). While mammals lack the de novo folate biosynthesis pathway and must obtain this nutrient from their diet, many microbes are unable to acquire folates from the external environment and rely on de novo folate synthesis to support one-carbon metabolism (21). The dichotomy in essentiality of this biosynthetic pathway in humans and microbial pathogens makes it an ideal target for the development of antimicrobial agents. There also exist structural differences in key enzymes of folate utilization that enable selective targeting of microbes (22). Indeed, antifolate drugs, such as sulfonamides and diaminopyrimidines (Fig. 2), have been widely used in the treatment of numerous bacterial and parasitic infections (20, 23–26).
FIG 1.
Schematic representation of M. tuberculosis folate metabolism. Enzymes of folate biosynthesis are blue, and enzymes of one-carbon metabolism are in beige ovals. Pathway intermediates are connected by black arrows. Abbreviations: GTP, guanosine-5′-triphosphate; DHPPP, 7,8-dihydropterin pyrophosphate; PABA, para-aminobenzoic acid; Glu, glutamate; H2Pte, dihydropteroate; H2PteGlu, dihydrofolate; H4PteGlu, tetrahydrofolate; C1-H4PteGlu, various single-carbon-modified species of H4PteGlu; DHPS, H2Pte synthase; DHFS, dihydrofolate synthase; DHFR, dihydrofolate reductase; dTMP, deoxythymidine monophosphate; FAICAR, 5-formamidoimidazole-4-carboxamide ribotide; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; FGAR, 5′-phosphoribosyl-N-formylglycinamide; GAR, 5′-phosphoribosylglycinamide; tRNAMet, methionyl-tRNA; tRNAfMet, N-formylmethionyl-tRNA.
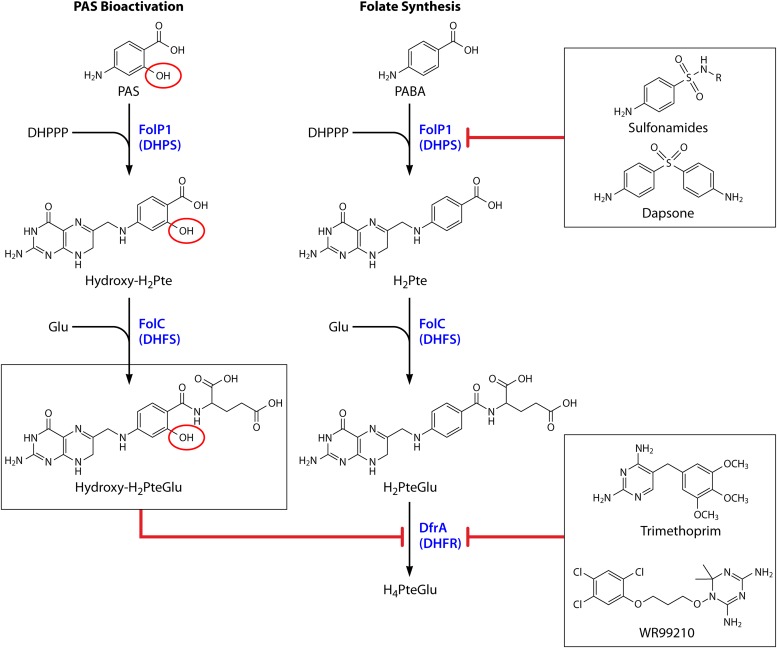
FIG 2.
Targets of antifolate drugs. As indicated by red blunted arrows, bioactivated PAS (hydroxy-H2PteGlu), trimethoprim, and WR99210 inhibit DfrA; 4-aminobenzene sulfonamides and dapsone inhibit FolP1. PAS is a prodrug that is activated through the folate biosynthetic pathway (shown on the left). PAS differs from PABA by the presence of a hydroxyl group in the ortho position (circled in red). The native folate synthesis pathway is represented on the right. During bioactivation, PAS serves as an alternate substrate to PABA and is sequentially converted to hydroxy-H2Pte and hydroxy-H2PteGlu by FolP1 and FolC, respectively. Relevant enzymes are blue. Abbreviations: DHPPP, 7,8-dihydropterin pyrophosphate; PABA, para-aminobenzoic acid; Glu, glutamate; H2Pte, dihydropteroate; H2PteGlu, dihydrofolate; H4PteGlu, tetrahydrofolate; DHPS, H2Pte synthase; DHFS, dihydrofolate synthase; DHFR, dihydrofolate reductase.
While antifolates are not part of the current first-line TB drug regimen, they have a long history of use in TB therapy and are of interest in ongoing TB drug discovery efforts. Shortly after their discovery, sulfonamides were used with limited success in the treatment of TB until more effective antitubercular drugs were introduced (27, 28). The need for novel therapeutic agents to meet the challenge of MDR and XDR TB has renewed interest in these and other antifolates. Several reports indicate that the vast majority of clinical isolates of M. tuberculosis, including MDR and XDR strains, are susceptible to a combination of sulfamethoxazole and the diaminopyrimidine trimethoprim (29–33). In addition, PAS has recently been shown to be an antifolate prodrug (34–36). Moreover, a series of folate structural analogs have recently been developed that show potent disruption of M. tuberculosis one-carbon metabolism through inhibition of folate reduction (37, 38). Thus, drugs that target folate metabolism show promise for the future treatment of drug-susceptible and drug-resistant TB infections.
The folate biosynthesis pathway of M. tuberculosis.
The folate biosynthetic pathway of M. tuberculosis begins with the synthesis of para-aminobenzoic acid (PABA) and 7,8-dihydropterin pyrophosphate (DHPPP) (Fig. 1). PABA is produced from chorismate by the concerted action of aminodeoxychorismate synthase (PabAB) and aminodeoxychorismate lyase (PabC) (39). While these enzymes have yet to be biochemically characterized in M. tuberculosis, genes corresponding to pabB and pabC were predicted to be essential for growth in high-throughput insertional transposon mutagenesis studies (40, 41). A gene corresponding to pabA, encoding chorismate-glutamine amidotransferase, is not predicted in the annotated complete genome sequence of strain H37Rv (42). Yet, it is possible that M. tuberculosis encodes an amphibolic glutamine amidotransferase (annotated as trpG) that is involved in the synthesis of both folate and tryptophan, as has been described for Bacillus subtilis (43). Similar to pabB and pabC, trpG has been predicted to be required for the growth of M. tuberculosis in a comprehensive assessment of gene essentiality (41).
DHPPP is produced from GTP via a multistep process (Fig. 1). The first step is the conversion of GTP to dihydroneopterin phosphate and formate by GTP cyclohydrolase (FolE) (44). Dihydroneopterin phosphate is then dephosphorylated to dihydroneopterin by a nonspecific cytoplasmic phosphatase (45). Next, dihydroneopterin aldolase (FolB) converts dihydroneopterin to 6-hydroxymethy-7,8-dihydropterin and glycolaldehyde (46). The structure of M. tuberculosis FolB has been solved and was shown to form a novel tetramer that undergoes substrate-induced octamerization, which is essential for its catalytic activity (47). Finally, 6-hydroxymethy-7,8-dihydropterin is converted to DHPPP by the diphosphotransferase FolK (48, 49). While folE and folB have been predicted to be essential in M. tuberculosis (40, 41), disruption of folK is associated with a strong in vitro growth defect (41). The genes of the DHPPP pathway are organized in an apparent operon that includes one other gene involved in folate synthesis (folP1), as well as ftsH (involved in cell division) and a gene of unknown function (Rv3605c) (50, 51).
Production of dihydropteroate (H2Pte) from PABA and DHPPP is catalyzed by H2Pte synthase (DHPS; Fig. 1) (48, 49). While the M. tuberculosis genome contains two putative genes for DHPS, folP1 and folP2, biochemical analysis has revealed that only FolP1 is catalytically active in the production of H2Pte (52, 53). The physiological role of FolP2 has yet to be defined (52).
The final enzyme of the de novo folate synthesis pathway is dihydrofolate synthase (DHFS), which catalyzes the ATP-dependent addition of l-glutamate to H2Pte to generate dihydrofolate (H2PteGlu) (Fig. 1) (49). In M. tuberculosis, DHFS activity is encoded by folC (54) and has been predicted to be essential for in vitro growth (40, 41). In many species, DHFS is a bifunctional enzyme that also catalyzes the gamma linkage of additional l-glutamate residues to the fully reduced folate species tetrahydrofolate (H4PteGlu), producing polyglutamylated folates (H4PteGlun, where n refers to the number of glutamate residues) (55). In mammalian cells, it has recently been shown that polyglutamylation is important for the retention of folate species in subcellular compartments (56). Polyglutamylation of folates in bacterial systems is widespread, but its physiologic role has not been exhaustively studied (55). One important role for bacterial polyglutamylated folates lies in methionine synthesis. In contrast to most bacterial folate-dependent enzymes that can utilize H4PteGlu species, it has been demonstrated that the cobalamin-independent methionine synthase MetE preferentially utilizes 5-methyl-H4PteGlu3 for catalysis (57).
H2PteGlu is reduced to H4PteGlu by dihydrofolate reductase (DHFR) (58). H4PteGlu serves as a cofactor for serine hydroxylase in the synthesis of glycine and is the essential precursor for various one-carbon-carrying folate species (C1-H4PteGlu) used in one-carbon metabolism (Fig. 1). Since DHFR is essential in many organisms, therapeutic agents targeting DHFR have been developed for cancer, malaria, and toxoplasmosis, in addition to bacterial infections. In M. tuberculosis, DHFR is encoded by dfrA (22) and is essential for growth (41). Significant structural differences between the active sites of M. tuberculosis DHFR and human DHFR make it an attractive target for antitubercular drug development (22, 59).
Folate metabolism as a target for antitubercular agents.
Among the various enzymes of folate metabolism, only two, DHPS and DHFR, are currently used as targets for antimicrobial agents (Fig. 2). Many sulfonamides and the related compound dapsone (Fig. 2), are structurally similar to PABA and have been found to be competitive inhibitors of DHPS in various pathogens (23, 24, 60–65). Sulfonamides inhibit M. tuberculosis FolP1 enzymatic activity in cell-free assays and show variable antitubercular activity in culture (53, 60, 66). Dapsone, a first-line leprosy drug (67), also inhibits M. tuberculosis FolP1 enzymatic activity in cell-free assays (53, 60). Like some sulfonamides, dapsone lacks significant activity against whole cells because of the expression of an uncharacterized inactivation pathway (34).
Trimethoprim is a bacteriostatic antimicrobial agent that potently inhibits DHFR in various bacterial species. For example, the Ki of trimethoprim for Escherichia coli DHFR is in the low nanomolar range (68). Trimethoprim is frequently used in combination with sulfonamides because of the synergistic impact on the disruption of folate metabolism. Unlike that which is observed in many other bacteria, trimethoprim only weakly inhibits M. tuberculosis DHFR enzymatic activity in cell-free assays (50% inhibitory concentration of 16.5 μM) (69). Accordingly, M. tuberculosis is not regarded as being highly susceptible to trimethoprim alone (MIC, >128 μg/ml) (31, 70, 71). A recent structural study suggested that the Tyr100 residue in M. tuberculosis DHFR may be responsible for the weak binding of trimethoprim to M. tuberculosis DHFR and showed that a variant, Y100F, had increased affinity for trimethoprim (59). Despite this limited antitubercular activity of trimethoprim, several studies have demonstrated that combinations of trimethoprim and sulfamethoxazole are effective against drug-susceptible and MDR strains of M. tuberculosis (29–33). An evaluation of synergy between these drugs showed that subinhibitory concentrations of sulfamethoxazole conferred a significant reduction of the MIC of trimethoprim (fractional inhibitory concentration index of 0.5) (31). Yet, a parallel study reported that while sulfamethoxazole alone showed measurable activity against various M. tuberculosis isolates, no apparent improvement of trimethoprim activity was observed (70). Interestingly, it has been demonstrated that specific disruptions in the folate interconversion pathway can modulate the susceptibility of M. smegmatis to various trimethoprim-sulfonamide combinations (72), suggesting opportunities for potentiation of antifolate action in mycobacteria (73).
Several studies have focused on the identification of new M. tuberculosis DHFR inhibitors that have more potent antitubercular activity than trimethoprim (37, 71, 74, 75). Suling et al. screened a series of lipophilic deazapteridine derivatives with structural similarity to trimethoprim and identified several M. tuberculosis DHFR inhibitors with improved activity relative to that of trimethoprim in cell-free and whole-cell assays (71). In addition, the antimalarial lead compound WR99210 (76) was found to be effective against several species of mycobacteria (77, 78), including M. tuberculosis (37). The crystal structure of M. tuberculosis DHFR and the brominated analog of WR99210 revealed that Br-WR99210 binds within the active site of M. tuberculosis DHFR (22). Using a novel screening approach with a yeast strain expressing M. tuberculosis dfrA, Gerum et al. identified several promising WR99210 analogs that also showed improved antitubercular activity (37). The ability of these compounds to target tubercle bacilli in animal models of infection awaits further study.
MODE OF ACTION OF PAS
Since the discovery of PAS as an antitubercular agent, there have been multiple hypotheses regarding its antitubercular mode of action. In 1940 and 1941, Bernheim observed that salicylate stimulated increased oxygen consumption in M. tuberculosis (79), while some structural analogs of salicylate had a negative impact on oxygen uptake (80). Following up on these studies, Lehmann screened a panel of salicylate analogs and identified PAS as a potent antitubercular agent (8). These initial observations implied a role for PAS in the disruption of a salicylate-linked metabolic pathway; however, recent studies have revealed that the principal antitubercular action of PAS occurs through poisoning of folate metabolism.
PAS is a prodrug targeting folate metabolism.
Initial hints toward an interaction of PAS with folate metabolism came from the observation that its antitubercular activity could be antagonized by supplementation with exogenous PABA (10) or methionine (81, 82). This link to folate metabolism was further solidified by the observation that loss-of-function mutations in thyA, encoding a folate-dependent thymidylate synthase, conferred resistance to PAS (83). On the basis of both the structural similarity of PAS to many antimicrobial sulfonamides and the ability of PABA to antagonize the antitubercular activity of these drugs, it was predicted that these compounds possess a conserved mode of action (10). However, despite these structural and functional similarities, there was no measurable cross-resistance between PAS and sulfonamides in M. tuberculosis (84). Further, it was shown that, in contrast to sulfonamides, PAS imposed only weak inhibition of purified recombinant M. tuberculosis FolP1 enzymatic activity (60). Thus, as described below, for PAS to disrupt M. tuberculosis folate metabolism, it must do so downstream of FolP1.
Recently, Chakraborty et al. used a metabolomic approach to investigate the impact of PAS treatment on M. tuberculosis folate metabolism (34). In this innovative study, it was revealed that PAS could be bioconverted to the folate intermediate analogs hydroxy-H2Pte and hydroxydihydrofolate (hydroxy-H2PteGlu) via the M. tuberculosis folate synthesis pathway (34). Further, by using cell-free assays with purified recombinant proteins, it was demonstrated that PAS and hydroxy-H2Pte were competent substrates for M. tuberculosis DHPS and DHFS, respectively (Fig. 2) (34). These data suggested for the first time that PAS was likely a prodrug that required activation via the folate synthesis pathway. This concerted conversion of PAS to hydroxy-H2Pte and hydroxy-H2PteGlu was confirmed by two subsequent studies (35, 36). Taken together, these findings suggested that PAS is bioactivated to a hydroxylated folate species analog that is disruptive for a downstream target(s) in folate metabolism.
To further probe into this metabolic disruption, Chakraborty et al. then profiled the change in abundance of folate-linked metabolites in PAS-treated M. tuberculosis. It was found that PAS treatment resulted in the rapid, dose-responsive accumulation of precursors of folate-dependent metabolites such as 5-amino-1-(5-phospho-d-ribosyl)imidazole-4-carboxamide, dUMP, homocysteine, and serine (34). This metabolic disruption could be antagonized by exogenously supplied PABA and mirrored metabolic responses to bona fide folate antagonists (34, 38). These data indicated that the ultimate bioactivation product of PAS was broadly disruptive of one-carbon metabolism, likely at an early step in folate activation (34).
Subsequently, Zheng et al. provided the instrumental observation that PAS-mediated growth inhibition of M. tuberculosis could be circumvented by overexpression of DHFR or by expression of a structurally distinct enzyme (RibD) with DHFR activity (35). Further, the authors demonstrated that small-molecule extracts from PAS-treated M. tuberculosis contained an inhibitory activity against purified recombinant M. tuberculosis DHFR (35). Synthesis of this inhibitory activity could be blocked by the treatment of bacilli with the DHPS inhibitor sulfathiazole (35). Collectively, these data strongly support a model in which PAS is bioactivated within the folate synthesis pathway to the H2PteGlu analog hydroxy-H2PteGlu, which then disrupts folate metabolism through potent inhibition of DHFR (Fig. 2).
Bactericidal effects of PAS.
PAS is generally regarded as a bacteriostatic agent for M. tuberculosis (8), yet bactericidal effects on metabolically active populations of bacilli have been noted (85). While antifolate drugs disrupt multiple biosynthetic pathways, limitation for most of these metabolites results in stasis. However, in metabolically active populations, limitation for dTMP typically imposes a unique microbicidal effect known as thymineless death (86–91). In organisms that express a thymine salvage pathway, thymineless death can be circumvented via supplementation with exogenous thymine or thymidine. Although the molecular mechanism leading to this loss of cell viability is still under investigation, several cellular changes have been causally linked with loss of dTMP and include an increase in single- and double-stranded DNA breaks, impaired Okazaki fragment assembly, and loss of origin-of-replication integrity (92–94). Accumulation of dUMP in M. tuberculosis cells treated with PAS was observed (34), indicating that PAS treatment interferes with dTMP synthesis. Further, induction of several DNA repair genes was observed in M. tuberculosis during treatment with other DHFR-inhibiting antifolates (38). These data suggest that thymineless death may contribute to loss of cell viability of M. tuberculosis during PAS treatment.
Folate metabolism is also intimately tied to the activated methyl cycle, which is involved in the biosynthesis of S-adenosylmethionine (SAM). SAM-dependent methyltransferases are essential for many cellular functions in M. tuberculosis, including DNA methylation, biotin synthesis, modification of mycolic acids, and methylation of rRNA (95–97). Loss of the ability to regenerate SAM has been shown to be bactericidal in Borrelia burgdorferi (98). Antifolate treatment has been demonstrated to reduce the intracellular abundance of SAM in M. tuberculosis (38). It was also recently shown that SAM can antagonize the antitubercular activity of WR99210. This finding suggests that WR99210 promotes cell death in M. tuberculosis through the depletion of SAM pools (38). Since WR99210 and bioactivated PAS act on the same cellular target, it is likely that PAS-mediated cell death is also linked to depletion of SAM abundance.
Direct evidence of the contribution of dTMP and SAM limitation to the bactericidal activity of PAS has yet to be described. Thymineless death is difficult to assess directly because of the lack of a thymidine salvage pathway in M. tuberculosis (99). Without a salvage pathway, supplementation with exogenous salvage pathway intermediates such as thymine or thymidine will not provide a source of dTMP. Further, it is unlikely that dTMP can be taken up directly by M. tuberculosis because of the limited permeability of the cell wall (100). Effects stemming from disruption of the activated methyl cycle have yet to be fully characterized in M. tuberculosis. Thus, further studies are essential to elucidate the mechanism of cell death in M. tuberculosis caused by PAS.
Other biological impacts of PAS.
While folate metabolism is a principal target of PAS action in M. tuberculosis, there is compelling evidence that PAS can also interfere with mycobacterial iron acquisition (101–103). Salicylate is a structural analog of PAS and is an essential moiety of the mycobacterial siderophores mycobactin and carboxymycobactin (104–106). It has been proposed that PAS noncompetitively inhibits the incorporation of salicylate into both mycobactin and carboxymycobactin, thereby disrupting high-affinity iron scavenging of M. tuberculosis (101, 102). This model stems from the observation that PAS inhibited mycobactin production in Mycobacterium smegmatis and Mycobacterium bovis under iron-restricted growth conditions (102, 107). Interestingly, M. smegmatis mutant strains with the salicylate synthesis pathway genes trpE2, entC, and entD deleted were found to have enhanced susceptibility to PAS (101). In addition, it was found that treatment of M. smegmatis with PAS led to a reduction of the specific activity of some iron-containing enzymes, such as aconitase, glycerol dehydrogenase, and NADH oxidase (107). Further, Mycobacterium avium showed a modest enhancement of PAS susceptibility when cultivated under iron-limiting conditions (108). Interestingly, in contrast to the robust antagonistic activity of PABA on PAS-mediated growth inhibition, salicylate was found to be a relatively weak antagonist (109), and mycobactin did not show measurable antagonism of PAS action (107). Given the dispensability of mycobactins for the growth of M. tuberculosis under iron-replete culture conditions, yet the essentiality of mycobactins for growth in iron-restricted niches of the mammalian host (110), it will be important to evaluate the impact of PAS on M. tuberculosis fitness in the context of iron restriction.
MECHANISMS OF PAS RESISTANCE
Resistance to PAS was described shortly after its introduction into clinical use and was most prevalent when it was used in monotherapy (111). Recent findings have demonstrated that PAS resistance in M. tuberculosis can emerge via multiple mechanisms that include preventing sufficient bioactivation within the folate synthesis pathway, mitigating the impact of target inhibition, and limiting drug accumulation within the bacilli.
Mitigating the impact of target inhibition.
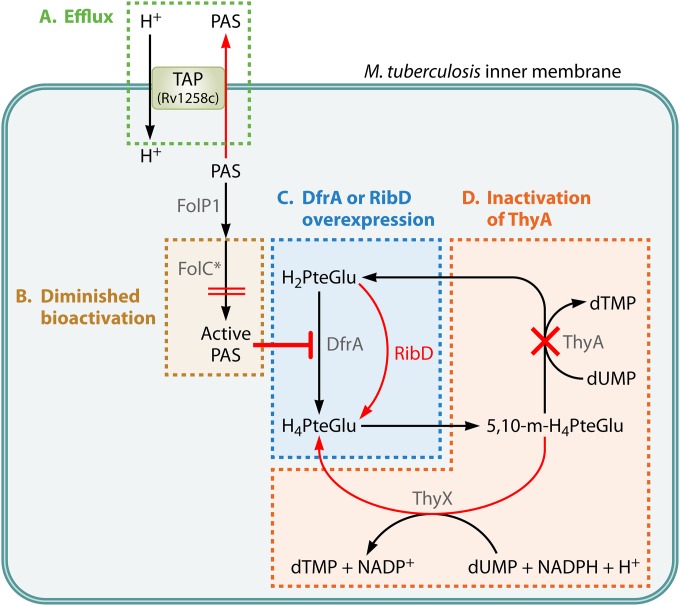
The first genetic evidence of the involvement of folate metabolism in the antitubercular action of PAS was that loss-of-function mutations in one of the genes for thymidylate synthase conferred resistance on M. tuberculosis and M. bovis (83). In biological systems, thymidylate synthase is essential for the 5,10-methylene-H4PteGlu-dependent conversion of dUMP to dTMP. In most organisms, this reaction is performed by a ThyA-type thymidylate synthase that releases H2PteGlu following catalysis. This H2PteGlu must be reduced by DHFR to re-enter folate metabolism (Fig. 3). Some organisms encode a ThyX-type thymidylate synthase. Like most other folate-dependent enzymes, ThyX regenerates H4PteGlu following catalysis (Fig. 3) (112, 113). In contrast to ThyX utilization, ThyA utilization results in an increased demand for DHFR activity to provide sufficient levels of H4PteGlu for one-carbon metabolism and is critical for the susceptibility of many organisms to DHFR inhibitors. Loss-of-function mutations in thyA significantly decrease the demand for DHFR activity and are commonly associated with resistance to DHFR inhibitors (89). In many pathogens, thyA loss-of-function mutations lead to thymine auxotrophy and loss of fitness during infection. Thus, thyA-mediated antifolate drug resistance is rarely clinically relevant (114, 115). In contrast to most bacterial pathogens, M. tuberculosis encodes both ThyA- and ThyX-type thymidylate synthases (99). As bioactivated PAS inhibits DHFR in M. tuberculosis, thyA loss-of-function mutations confer up to 100-fold resistance to PAS (83, 99). Since ThyX can support the cellular dTMP requirement, M. tuberculosis thyA mutants are not attenuated and are associated with clinical resistance to PAS (83, 116–118).
FIG 3.
Mechanisms of PAS resistance. The four characterized mechanisms of PAS resistance are shown. (A) Efflux. Intracellular PAS is excluded by the efflux antiporter TAP of M. tuberculosis encoded by Rv1258c (green). (B) Diminished bioactivation. FolC variants (FolC*) with an altered substrate binding pocket show decreased bioactivation of PAS (gold). (C) DfrA or RibD overexpression. Inhibition of dihydrofolate reduction can be negated by overexpression of the target DfrA or by overexpression of the alternative reductase RibD (blue). (D) Inactivation of the thymidylate synthase ThyA. Loss of ThyA function is tolerated because of the alternate thymidylate synthase ThyX and confers resistance to PAS by decreasing the catalytic demand on DHFR (orange).
In addition to thyA loss-of-function mutations, artificial overexpression of dfrA has been shown to confer PAS resistance on M. tuberculosis (35). This observation is consistent with the idea that the active form of PAS inhibits DfrA. Interestingly, it was also found that a mutation 11 bp upstream of the ribD translational start site increased ribD expression and gave rise to PAS resistance in M. tuberculosis. ribD encodes a riboflavin biosynthesis protein that contains a C-terminal oxidoreductase domain with 43% amino acid sequence similarity to DfrA. It was suggested that overexpression of ribD confers PAS resistance by compensating for the inhibited DfrA function (Fig. 3). While ribD promoter mutations have been identified in PAS-resistant clinical isolates of M. tuberculosis, dfrA mutations have yet to be reported in such isolates (35, 118).
Reduced bioactivation of PAS.
As described above, PAS is a prodrug that requires bioactivation within the folate biosynthetic pathway by the concerted action of DHPS and DHFS. By selecting for spontaneous PAS-resistant mutant strains of M. tuberculosis H37Rv and H37Ra and M. bovis BCG, it was found that mutations within folC (encoding DHFS) conferred resistance to PAS (35, 36). Mutations in folC were also identified in PAS-resistant M. tuberculosis clinical isolates and were associated with an up to 64-fold increase in PAS resistance (36, 118). A recent study tested clinically isolated PAS-resistant M. tuberculosis strains from northern China and identified folC mutations as the most predominant mutations among the PAS-resistant strains (118). In three recent studies, the reported folC mutations mapped within positions corresponding to substrate binding and nucleoside binding domains that are essential for DHFS activity (35, 36, 118). Indeed, when the DHFS activity of several of these FolC variants was evaluated, reduced conversion of H2Pte to H2PteGlu (10 to 20% of wild-type activity) was observed (36). Importantly, conversion of hydroxy-H2Pte to hydroxy-H2PteGlu was below the limit of detection (36). Further, while a metabolite extract from PAS-treated wild-type M. tuberculosis contained DHFR inhibitory activity, this activity was absent from a metabolite extract from a PAS-treated FolC variant strain of M. tuberculosis (35). These observations suggest that the reported FolC variants confer resistance by precluding sufficient bioactivation of PAS.
Active efflux.
Efflux pumps play a major role in bacterial drug resistance (119). The M. tuberculosis genome encodes at least 46 putative drug efflux systems, and 22 drug efflux pumps have been shown to confer drug resistance (120). Among the M. tuberculosis drug efflux pumps, it was recently found that a major facilitator superfamily drug efflux pump, Tap (Rv1258c), confers resistance to PAS on M. bovis BCG (121). Overexpression of tap in M. bovis BCG increased the MICs of PAS, gentamicin, streptomycin, spectinomycin, tetracycline, triclosan, and vancomycin by more than 4-fold (121). In M. tuberculosis, expression of tap is induced in the presence of rifampin and ofloxacin in vitro (122) and tap expression levels have been found to be elevated in some clinical isolates of M. tuberculosis (123).
CLOSING REMARKS
In this review, we have summarized our current understanding of the basis of the susceptibility and resistance of M. tuberculosis to PAS. After nearly 70 years of clinical use of this drug in treating TB, recent findings clearly establish a role for perturbation of one-carbon metabolism as a major consequence of PAS incorporation in and disruption of folate metabolism. These findings are consistent with the observation that PAS-resistant clinical isolates show alterations in three distinct nodes in folate metabolism. Despite these incontrovertible findings, many standing questions remain regarding the action of this drug against M. tuberculosis. One major question is whether folate disruption is the exclusive antitubercular action of PAS, as there is compelling evidence that this drug disrupts iron assimilation in other species of mycobacteria. Such an effect on iron assimilation is in no way mutually exclusive with the impact on folate metabolism and warrants further investigation. In addition to gaps in our understanding of the mode of action of PAS, it is also clear that additional resistance mechanisms have yet to be described. Studies with cultured bacilli implicate PAS efflux via TAP as a potential resistance mechanism (121), yet a role for TAP in the limitation of PAS efficacy has yet to be established in infection models or in a clinical setting. Further, a PAS inactivation pathway involving the uncharacterized SAM-dependent methyltransferase Rv0560c has been suggested (34, 109, 124, 125) and may be the basis for methionine-linked antagonism of PAS activity (81, 82). Yet, whether this pathway limits the full potential action of PAS remains to be demonstrated. By interfering with pathways that antagonize PAS activity, it may be possible to potentiate the action of this drug and perhaps restore susceptibility in the context of PAS resistance.
ACKNOWLEDGMENTS
This work was supported by a grant from Minnesota Partnership for Biotechnology and Medical Genomics (ML2012, chapter 5, article 1, section 5, subdivision 5e) and by startup funds from the University of Minnesota to A.D.B.
Biographies

Yusuke Minato received his B.S. in Pharmacy and his Ph.D. in Pharmaceutical Sciences from Okayama University. Dr. Minato performed his postdoctoral training at Oregon State University prior to joining the Baughn lab as a postdoctoral associate in the Department of Microbiology at the University of Minnesota Medical School. Dr. Minato has broad training in molecular pathogenesis, bacterial physiology and metabolism, and antimicrobial chemotherapy and resistance. Currently, Dr. Minato is targeting bacterial metabolism to develop novel strategies to treat bacterial infections, with an emphasis on tuberculosis.

Joshua M. Thiede received his B.S. in Biology and Classical Studies from the University of Wisconsin, where he was a research assistant in the Biological and Chemical Engineering Department in the lab of Dr. Brian Pfleger. After graduating, Josh joined the Microbiology, Immunology, and Cancer Biology graduate program at the University of Minnesota, where he is currently pursuing his Ph.D. in Microbiology. He is training in the lab of Dr. Anthony Baughn with a focus on understanding the downstream cellular effects of antifolate treatment in M. tuberculosis.

Shannon Lynn Kordus received her B.S. in Microbiology and Biochemistry from the University of Wisconsin—LaCrosse. She earned her M.S. in Molecular Biology, Cellular Biology, and Biochemistry from Boston University studying how the structure of Y-family DNA polymerases enable the bypass of DNA adducts. She is now training for her Ph.D. in Microbiology in the Microbiology, Immunology, and Cancer Biology graduate program at the University of Minnesota under the direction of Dr. Anthony Baughn. Her interests are focused on understanding the biochemical aspects of antimicrobial susceptibility and resistance in M. tuberculosis.

Edward J. McKlveen received his A.B. in Chemistry from Harvard University. As a Herchel Smith Undergraduate Research Fellow and Program for Research in Science and Engineering Research Fellow, Edward designed mononuclear complexes for redox flow battery energy storage in the Theodore Betley Laboratory at Harvard University. Currently, Edward is continuing work on energy storage solutions with the goal of increased utilization of renewable energy.

Breanna J. Turman recently received her B.S. in Microbiology from the University of Minnesota. As an undergraduate researcher in the lab of Dr. Anthony Baughn, Breanna has focused on the identification of PAS resistance mechanisms and the elucidation of the mode of action of PAS in M. tuberculosis. Research in this area will lead to more-rapid diagnosis of PAS-resistant M. tuberculosis infections and aid in the identification of novel antitubercular drug targets.

Anthony D. Baughn received his B.S. in Microbiology from the University of New Hampshire and his Ph.D. in Microbiology and Molecular Biology from Tufts University. As a Merck Fellow of the Helen Hay Whitney Foundation, Dr. Baughn performed his postdoctoral training at the Albert Einstein College of Medicine in the area of mycobacterial metabolism and drug discovery. Currently, Dr. Baughn is an Assistant Professor in the Department of Microbiology at the University of Minnesota Medical School, where his laboratory is focused primarily on understanding genetic and biochemical aspects of antimicrobial susceptibility and resistance in M. tuberculosis. The long-term goal of these studies is to facilitate the rational design of more-effective therapeutic interventions for infectious diseases.
REFERENCES
- 1.World Health Organization. 2013. Global tuberculosis report. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/. [Google Scholar]
- 2.East African-British Medical Research Councils. 1974. Controlled clinical trial of four short-course (6-month) regimens of chemotherapy for treatment of pulmonary tuberculosis. Third report. Lancet ii:237–240. [PubMed] [Google Scholar]
- 3.Yee D, Valiquette C, Pelletier M, Parisien I, Rocher I, Menzies D. 2003. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. Am J Respir Crit Care Med 167:1472–1477. doi: 10.1164/rccm.200206-626OC. [DOI] [PubMed] [Google Scholar]
- 4.Weis SE, Slocum PC, Blais FX, King B, Nunn M, Matney GB, Gomez E, Foresman BH. 1994. The effect of directly observed therapy on the rates of drug resistance and relapse in tuberculosis. N Engl J Med 330:1179–1184. doi: 10.1056/NEJM199404283301702. [DOI] [PubMed] [Google Scholar]
- 5.Ormerod LP. 2005. Multidrug-resistant tuberculosis (MDR-TB): epidemiology, prevention and treatment. Br Med Bull 73-74:17–24. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Xu S, Wang L, Chin DP, Wang S, Jiang G, Xia H, Zhou Y, Li Q, Ou X, Pang Y, Song Y, Zhao B, Zhang H, He G, Guo J, Wang Y. 2012. National survey of drug-resistant tuberculosis in China. N Engl J Med 366:2161–2170. doi: 10.1056/NEJMoa1108789. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J. 2010. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann J. 1946. para-Aminosalicylic acid in the treatment of tuberculosis. Lancet 247:15–16. doi: 10.1016/S0140-6736(46)91185-3. [DOI] [PubMed] [Google Scholar]
- 9.Pfuetze KH, Pyle MM. 1949. Streptomycin in the treatment of tuberculosis. JAMA 139:634–639. doi: 10.1001/jama.1949.02900270018005. [DOI] [PubMed] [Google Scholar]
- 10.Youmans GP, Raleigh GW, Youmans AS. 1947. The tuberculostatic action of para-aminosalicylic acid. J Bacteriol 54:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shane SJ, Laurie JH, Riley C, Boutilier M. 1952. Effect of combined therapy (dihydrostreptomycin and PAS) on the emergence of streptomycin-resistant strains of tubercle bacilli. N Engl J Med 246:132–134. doi: 10.1056/NEJM195201242460404. [DOI] [PubMed] [Google Scholar]
- 12.Graessle OE, Pietrowski JJ. 1949. The in vitro effect of para-aminosalicylic acid (PAS) in preventing acquired resistance to streptomycin by Mycobacterium tuberculosis. J Bacteriol 57:459–464. [PMC free article] [PubMed] [Google Scholar]
- 13.Anonymous. 1952. Prevention of streptomycin resistance by combined chemotherapy; a Medical Research Council investigation. Br Med J 1:1157–1162. doi: 10.1136/bmj.1.4769.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capon AW. 1954. Streptomycin and PAS vs. streptomycin, PAS and isoniazid in the treatment of pulmonary tuberculosis. Can Med Assoc J 70:62–67. [PMC free article] [PubMed] [Google Scholar]
- 15.WHO Collaborating Centre for Tuberculosis Chemotherapy. 1976. A comparative study of daily and twice-weekly continuation regimens of tuberculosis chemotherapy, including a comparison of two durations of sanatorium treatment. Tubercle 57:45–48. doi: 10.1016/0041-3879(76)90016-7. [DOI] [PubMed] [Google Scholar]
- 16.Anonymous. 1973. Co-operative controlled trial of a standard regimen of streptomycin, PAS and isoniazid and three alternative regimens of chemotherapy in Britain. A report from the British Medical Research Council. Tubercle 54:99–129. [DOI] [PubMed] [Google Scholar]
- 17.Ferebee SH, Doster BE, Murray FJ. 1966. Ethambutol: a substitute for para-aminosalicylic acid in regimens for pulmonary tuberculosis. Ann N Y Acad Sci 135:910–920. doi: 10.1111/j.1749-6632.1966.tb45533.x. [DOI] [PubMed] [Google Scholar]
- 18.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 19.Cossins EA. 2000. The fascinating world of folate and one-carbon metabolism. Can J Bot 78:691–708. doi: 10.1139/cjb-78-6-691. [DOI] [Google Scholar]
- 20.Green JG, Matthews RG. 2007. Folate biosynthesis, reduction, and polyglutamylation and the interconversion of folate derivatives. EcoSal Plus doi: 10.1128/ecosalplus.3.6.3.6. [DOI] [PubMed] [Google Scholar]
- 21.Bermingham A, Derrick JP. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 22.Li R, Sirawaraporn R, Chitnumsub P, Sirawaraporn W, Wooden J, Athappilly F, Turley S, Hol WGJ. 2000. Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs. J Mol Biol 295:307–323. doi: 10.1006/jmbi.1999.3328. [DOI] [PubMed] [Google Scholar]
- 23.Brown GM. 1962. The biosynthesis of folic acid. II. Inhibition by sulfonamides. J Biol Chem 237:536–540. [PubMed] [Google Scholar]
- 24.Roland S, Ferone R, Harvey RJ, Styles VL, Morrison RW. 1979. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J Biol Chem 254:10337–10345. [PubMed] [Google Scholar]
- 25.Smilack JD. 1999. Trimethoprim-sulfamethoxazole. Mayo Clin Proc 74:730–734. doi: 10.4065/74.7.730. [DOI] [PubMed] [Google Scholar]
- 26.Wormser G, Keusch G, Heel R. 1982. Co-trimoxazole (trimethoprim-sulfamethoxazole). Drugs 24:459–518. doi: 10.2165/00003495-198224060-00002. [DOI] [PubMed] [Google Scholar]
- 27.Ellman P, Lawrence JS, Cumings JN. 1941. Investigation in the value of sulphapyridine in the treatment of pulmonary tuberculosis. Tubercle 22:296–302. doi: 10.1016/S0041-3879(41)80057-9. [DOI] [Google Scholar]
- 28.Freilich EB, Coe GC, Wien NA. 1939. The use of sulfanilamide in pulmonary tuberculosis: preliminary report. Ann Intern Med 13:1042–1045. doi: 10.7326/0003-4819-13-6-1042. [DOI] [Google Scholar]
- 29.Forgacs P, Wengenack NL, Hall L, Zimmerman SK, Silverman ML, Roberts GD. 2009. Tuberculosis and trimethoprim-sulfamethoxazole. Antimicrob Agents Chemother 53:4789–4793. doi: 10.1128/AAC.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameen SM, Drancourt M. 2013. In vitro susceptibility of Mycobacterium tuberculosis to trimethoprim and sulfonamides in France. Antimicrob Agents Chemother 57:6370–6371. doi: 10.1128/AAC.01683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilchèze C, Jacobs WR. 2012. The combination of sulfamethoxazole, trimethoprim, and isoniazid or rifampin is bactericidal and prevents the emergence of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:5142–5148. doi: 10.1128/AAC.00832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alsaad N, van Altena R, Pranger AD, van Soolingen D, de Lange WCM, van der Werf TS, Kosterink JGW, Alffenaar J-WC. 2013. Evaluation of co-trimoxazole in the treatment of multidrug-resistant tuberculosis. Eur Respir J 42:504–512. doi: 10.1183/09031936.00114812. [DOI] [PubMed] [Google Scholar]
- 33.Davies Forsman L, Schön T, Simonsson USH, Bruchfeld J, Larsson M, Juréen P, Sturegård E, Giske CG, Ängeby K. 2014. Intra- and extracellular activities of trimethoprim-sulfamethoxazole against susceptible and multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:7557–7559. doi: 10.1128/AAC.02995-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty S, Gruber T, Barry CE, Boshoff HI, Rhee KY. 2013. para-Aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339:88–91. doi: 10.1126/science.1228980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Rubin EJ, Bifani P, Mathys V, Lim V, Au M, Jang J, Nam J, Dick T, Walker JR, Pethe K, Camacho LR. 2013. para-Aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis. J Biol Chem 288:23447–23456. doi: 10.1074/jbc.M113.475798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao F, Wang XD, Erber LN, Luo M, Guo AZ, Yang SS, Gu J, Turman BJ, Gao YR, Li DF, Cui ZQ, Zhang ZP, Bi LJ, Baughn AD, Zhang XE, Deng JY. 2014. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:1479–1487. doi: 10.1128/AAC.01775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerum AB, Ulmer JE, Jacobus DP, Jensen NP, Sherman DR, Sibley CH. 2002. Novel Saccharomyces cerevisiae screen identifies WR99210 analogues that inhibit Mycobacterium tuberculosis dihydrofolate reductase. Antimicrob Agents Chemother 46:3362–3369. doi: 10.1128/AAC.46.11.3362-3369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixon MR, Saionz KW, Koo MS, Szymonifka MJ, Jung H, Roberts JP, Nandakumar M, Kumar A, Liao R, Rustad T, Sacchettini JC, Rhee KY, Freundlich JS, Sherman DR. 2014. Folate pathway disruption leads to critical disruption of methionine derivatives in Mycobacterium tuberculosis. Chem Biol 21:819–830. doi: 10.1016/j.chembiol.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Ye QZ, Liu J, Walsh CT. 1990. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc Natl Acad Sci U S A 87:9391–9395. doi: 10.1073/pnas.87.23.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 41.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 43.Slock J, Stahly DP, Han CY, Six EW, Crawford IP. 1990. An apparent Bacillus subtilis folic acid biosynthetic operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid, and the dihydropteroate synthase gene. J Bacteriol 172:7211–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf WA, Brown GM. 1969. The biosynthesis of folic acid. X. Evidence for an Amadori rearrangement in the enzymatic formation of dihydroneopterin triphosphate from GTP. Biochim Biophys Acta 192:468–478. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki Y, Brown GM. 1974. The biosynthesis of folic acid. XII. Purification and properties of dihydroneopterin triphosphate pyrophosphohydrolase. J Biol Chem 249:2405–2410. [PubMed] [Google Scholar]
- 46.Mathis JB, Brown GM. 1970. The biosynthesis of folic acid. XI. Purification and properties of dihydroneopterin aldolase. J Biol Chem 245:3015–3025. [PubMed] [Google Scholar]
- 47.Goulding CW, Apostol MI, Sawaya MR, Phillips M, Parseghian A, Eisenberg D. 2005. Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme. J Mol Biol 349:61–72. doi: 10.1016/j.jmb.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 48.Richey DP, Brown GM. 1969. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J Biol Chem 244:1582–1592. [PubMed] [Google Scholar]
- 49.Shiota T, Baugh C, Jackson R, Dillard R. 1969. The enzymatic synthesis of hydroxymethyldihydropteridine pyrophosphate and dihydrofolate. Biochemistry 8:5022–5028. doi: 10.1021/bi00840a052. [DOI] [PubMed] [Google Scholar]
- 50.Roback P, Beard J, Baumann D, Gille C, Henry K, Krohn S, Wiste H, Voskuil MI, Rainville C, Rutherford R. 2007. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res 35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, Walk TC, Zhang P, Karp PD. 2008. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 36(Database issue):D623–D631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gengenbacher M, Xu T, Niyomrattanakit P, Spraggon G, Dick T. 2008. Biochemical and structural characterization of the putative dihydropteroate synthase ortholog Rv1207 of Mycobacterium tuberculosis. FEMS Microbiol Lett 287:128–135. doi: 10.1111/j.1574-6968.2008.01302.x. [DOI] [PubMed] [Google Scholar]
- 53.Baca AM, Sirawaraporn R, Turley S, Sirawaraporn W, Hol WGJ. 2000. Crystal structure of Mycobacterium tuberculosis 6-hydroxymethyl-7,8-dihydropteroate synthase in complex with pterin monophosphate: new insight into the enzymatic mechanism and sulfa-drug action. J Mol Biol 302:1193–1212. doi: 10.1006/jmbi.2000.4094. [DOI] [PubMed] [Google Scholar]
- 54.Young PG, Smith CA, Metcalf P, Baker EN. 2008. Structures of Mycobacterium tuberculosis folylpolyglutamate synthase complexed with ADP and AMPPCP. Acta Crystallogr D Biol Crystallogr D64(Pt 7):745–753. doi: 10.1107/S0907444908012262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bognar AL, Shane B. 1986. Bacterial folylpoly(gamma-glutamate) synthase-dihydrofolate synthase. Methods Enzymol 122:349–359. doi: 10.1016/0076-6879(86)22193-X. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence SA, Titus SA, Ferguson J, Heineman AL, Taylor SM, Moran RG. 2014. Mammalian mitochondrial and cytosolic folylpolyglutamate synthetase maintain the subcellular compartmentalization of folates. J Biol Chem 289:29386–29396. doi: 10.1074/jbc.M114.593244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitfield CD, Steers EJ, Weissbach H. 1970. Purification and properties of 5-methyltetrahydropteroyltriglutamate-homocysteine transmethylase. J Biol Chem 245:390–401. [PubMed] [Google Scholar]
- 58.Blakley RL, MacDougall BM. 1961. Dihydrofolic reductase from Streptococcus faecalis R. J Biol Chem 236:1163–1167. [Google Scholar]
- 59.Dias MVB, Tyrakis P, Domingues RR, Leme AFP, Blundell TL. 2014. Mycobacterium tuberculosis dihydrofolate reductase reveals two conformational states and a possible low affinity mechanism to antifolate drugs. Structure 22:94–103. doi: 10.1016/j.str.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 60.Nopponpunth V, Sirawaraporn W, Greene PJ, Santi DV. 1999. Cloning and expression of Mycobacterium tuberculosis and Mycobacterium leprae dihydropteroate synthase in Escherichia coli. J Bacteriol 181:6814–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCullough JL, Maren TH. 1973. Inhibition of dihydropteroate synthetase from Escherichia coli by sulfones and sulfonamides. Antimicrob Agents Chemother 3:665–669. doi: 10.1128/AAC.3.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allegra CJ, Boarman D, Kovacs JA, Morrison P, Beaver J, Chabner BA, Masur H. 1990. Interaction of sulfonamide and sulfone compounds with Toxoplasma gondii dihydropteroate synthase. J Clin Invest 85:371–379. doi: 10.1172/JCI114448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Triglia T, Menting JGT, Wilson C, Cowman AF. 1997. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A 94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasekarn W, Sirawaraporn R, Chahomchuen T, Cowman AF, Sirawaraporn W. 2004. Molecular characterization of bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase from Plasmodium falciparum. Mol Biochem Parasitol 137:43–53. doi: 10.1016/j.molbiopara.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 65.Voeller D, Kovacs J, Andrawis V, Chu E, Masur H, Allegra C. 1994. Interaction of Pneumocystis carinii dihydropteroate synthase with sulfonamides and diaminodiphenyl sulfone (dapsone). J Infect Dis 169:456–459. doi: 10.1093/infdis/169.2.456. [DOI] [PubMed] [Google Scholar]
- 66.Wallace RJ, Nash DR, Steele LC, Steingrube V. 1986. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J Clin Microbiol 24:976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Britton WJ, Lockwood DN. 2004. Leprosy. Lancet 363:1209–1219. doi: 10.1016/S0140-6736(04)15952-7. [DOI] [PubMed] [Google Scholar]
- 68.Poe M, Breeze AS, Wu JK, Short CR, Hoogsteen K. 1979. Dihydrofolate reductase from trimethoprim-resistant Escherichia coli MB 3746 and MB 3747. Purification, amino acid composition, and some kinetic properties. J Biol Chem 254:1799–1805. [PubMed] [Google Scholar]
- 69.White EL, Ross LJ, Cunningham A, Escuyer V. 2004. Cloning, expression, and characterization of Mycobacterium tuberculosis dihydrofolate reductase. FEMS Microbiol Lett 232:101–105. doi: 10.1016/S0378-1097(04)00038-2. [DOI] [PubMed] [Google Scholar]
- 70.Huang T-S, Kunin CM, Yan B-S, Chen Y-S, Lee SS-J, Syu W. 2012. Susceptibility of Mycobacterium tuberculosis to sulfamethoxazole, trimethoprim and their combination over a 12 year period in Taiwan. J Antimicrob Chemother 67:633–637. doi: 10.1093/jac/dkr501. [DOI] [PubMed] [Google Scholar]
- 71.Suling WJ, Reynolds RC, Barrow EW, Wilson LN, Piper JR, Barrow WW. 1998. Susceptibilities of Mycobacterium tuberculosis and Mycobacterium avium complex to lipophilic deazapteridine derivatives, inhibitors of dihydrofolate reductase. J Antimicrob Chemother 42:811–815. doi: 10.1093/jac/42.6.811. [DOI] [PubMed] [Google Scholar]
- 72.Ogwang S, Nguyen HT, Sherman M, Bajaksouzian S, Jacobs MR, Boom WH, Zhang G-F, Nguyen L. 2011. Bacterial conversion of folinic acid is required for antifolate resistance. J Biol Chem 286:15377–15390. doi: 10.1074/jbc.M111.231076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolff KA, Nguyen L. 2012. Strategies for potentiation of ethionamide and folate antagonists against Mycobacterium tuberculosis. Expert Rev Anti Infect Ther 10:971–981. doi: 10.1586/eri.12.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.El-Hamamsy MHRI, Smith AW, Thompson AS, Threadgill MD. 2007. Structure-based design, synthesis and preliminary evaluation of selective inhibitors of dihydrofolate reductase from Mycobacterium tuberculosis. Bioorg Med Chem 15:4552–4576. doi: 10.1016/j.bmc.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 75.Kumar A, Zhang M, Zhu L, Liao RP, Mutai C, Hafsat S, Sherman DR, Wang M-W. 2012. High-throughput screening and sensitized bacteria identify an M. tuberculosis dihydrofolate reductase inhibitor with whole cell activity. PLoS One 7:e39961. doi: 10.1371/journal.pone.0039961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Childs G, Lambros C. 1986. Analogues of N-benzyloxydihydrotriazines: in vitro antimalarial activity against Plasmodium falciparum. Ann Trop Med Parasitol 80:177–181. [DOI] [PubMed] [Google Scholar]
- 77.Meyer SC, Majumder SK, Cynamon MH. 1995. In vitro activities of PS-15, a new dihydrofolate reductase inhibitor, and its cyclic metabolite against Mycobacterium avium complex. Antimicrob Agents Chemother 39:1862–1863. doi: 10.1128/AAC.39.8.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shah LM, DeStefano MS, Cynamon MH. 1996. Enhanced in vitro activity of WR99210 in combination with dapsone against Mycobacterium avium complex. Antimicrob Agents Chemother 40:2644–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernheim F. 1940. The effect of salicylate on the oxygen uptake of the tubercle bacillus. Science 92:204. doi: 10.1126/science.92.2383.204-a. [DOI] [PubMed] [Google Scholar]
- 80.Bernheim F. 1941. The effect of various substances on the oxygen uptake of the tubercle bacillus. J Bacteriol 41:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hedgecock LW. 1956. Antagonism of the inhibitory action of aminosaliylic acid on Mycobacterium tuberculosis by methionine, biotin and certain fatty acids, amino acids, and purines. J Bacteriol 72:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hedgecock LW. 1958. Antagonism of methionine in aminosalicylate-inhibition of Mycobacterium tuberculosis. J Bacteriol 75:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. 2004. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol 53:275–282. doi: 10.1111/j.1365-2958.2004.04120.x. [DOI] [PubMed] [Google Scholar]
- 84.Yegian D, Long RT. 1951. The specific resistance of tubercle bacilli to para-aminosalicylic acid and sulfonamides. J Bacteriol 61:747–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie Z, Siddiqi N, Rubin EJ. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 49:4778–4780. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hawser S, Lociuro S, Islam K. 2006. Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol 71:941–948. doi: 10.1016/j.bcp.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 87.Cohen SS, Barner HD. 1954. Studies on unbalanced growth in Escherichia coli. Proc Natl Acad Sci U S A 40:885–893. doi: 10.1073/pnas.40.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Then R, Angehrn P. 1973. Sulphonamide-induced ‘thymineless death’ in Escherichia coli. J Gen Microbiol 76:255–263. doi: 10.1099/00221287-76-2-255. [DOI] [PubMed] [Google Scholar]
- 89.Ahmad SI, Kirk SH, Eisenstark A. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol 52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 90.Barclay B, Little JG. 1978. Genetic damage during thymidylate starvation in Saccharomyces cerevisiae. Mol Gen Genet 160:33–40. [DOI] [PubMed] [Google Scholar]
- 91.Yamao F, Nagai Y, Kaneda S, Yoshida S, Seno T. 1993. Conditional resistance to thymineless death predominantly selects DNA synthesis-deficient mutants of mammalian cells. Mutat Res 289:83–89. doi: 10.1016/0027-5107(93)90133-Z. [DOI] [PubMed] [Google Scholar]
- 92.Breitman TR, Maury PB, Toal JN. 1972. Loss of deoxyribonucleic acid-thymine during thymine starvation of Escherichia coli. J Bacteriol 112:646–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Freifelder D, Katz G. 1971. Persistence of small fragments of newly synthesized DNA in bacteria following thymidine starvation. J Mol Biol 57:351–354. doi: 10.1016/0022-2836(71)90351-2. [DOI] [PubMed] [Google Scholar]
- 94.Yoshinaga K. 1973. Double-strand scission of DNA involved in thymineless death of Escherichia coli 15 TAU. Biochim Biophys Acta 294:204–213. doi: 10.1016/0005-2787(73)90293-1. [DOI] [PubMed] [Google Scholar]
- 95.Parveen N, Cornell KA. 2011. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahman A, Srivastava S, Sneh A, Ahmed N, Krishnasastry M. 2010. Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase. BMC Biochem 11:35. doi: 10.1186/1471-2091-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boissier F, Bardou F, Guillet V, Uttenweiler-Joseph S, Daffé M, Quémard A, Mourey L. 2006. Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis. J Biol Chem 281:4434–4445. doi: 10.1074/jbc.M510250200. [DOI] [PubMed] [Google Scholar]
- 98.Cornell KA, Primus S, Martinez JA, Parveen N. 2009. Assessment of methylthioadenosine/S-adenosylhomocysteine nucleosidases of Borrelia burgdorferi as targets for novel antimicrobials using a novel high-throughput method. J Antimicrob Chemother 63:1163–1172. doi: 10.1093/jac/dkp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fivian-Hughes AS, Houghton J, Davis EO. 2012. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology 158:308–318. doi: 10.1099/mic.0.053983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen L, Thompson CJ. 2006. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol 14:304–312. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 101.Nagachar N, Ratledge C. 2010. Knocking out salicylate biosynthesis genes in Mycobacterium smegmatis induces hypersensitivity to p-aminosalicylate (PAS). FEMS Microbiol Lett 311:193–199. doi: 10.1111/j.1574-6968.2010.02091.x. [DOI] [PubMed] [Google Scholar]
- 102.Ratledge C, Brown KA. 1972. Inhibition of mycobactin formation in Mycobacterium smegmatis by p-aminosalicylate. A new proposal for the mode of action of p-aminosalicylate. Am Rev Respir Dis 106:774–776. [DOI] [PubMed] [Google Scholar]
- 103.Adilakshmi T, Ayling PD, Ratledge C. 2000. Mutational analysis of a role for salicylic acid in iron metabolism of Mycobacterium smegmatis. J Bacteriol 182:264–271. doi: 10.1128/JB.182.2.264-271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snow GA. 1965. Isolation and structure of mycobactin T, a growth factor from Mycobacterium tuberculosis. Biochem J 97:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lane SJ, Marshall PS, Upton RJ, Ratledge C, Ewing M. 1995. Novel extracellular mycobactins, the carboxymycobactins from Mycobacterium avium. Tetrahedron Lett 36:4129–4132. doi: 10.1016/0040-4039(95)00676-4. [DOI] [Google Scholar]
- 106.Gobin J, Moore CH, Reeve JR, Wong DK, Gibson BW, Horwitz MA. 1995. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci U S A 92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown KA, Ratledge C. 1975. The effect of p-aminosalicyclic acid on iron transport and assimilation in mycobacteria. Biochim Biophys Acta 385:207–220. doi: 10.1016/0304-4165(75)90349-9. [DOI] [PubMed] [Google Scholar]
- 108.Kopinc R, Lapanje A. 2012. Antibiotic susceptibility profile of Mycobacterium avium subspecies hominissuis is altered in low-iron conditions. J Antimicrob Chemother 67:2903–2907. doi: 10.1093/jac/dks313. [DOI] [PubMed] [Google Scholar]
- 109.Schaller A, Sun Z, Yang Y, Somoskovi A, Zhang Y. 2002. Salicylate reduces susceptibility of Mycobacterium tuberculosis to multiple antituberculosis drugs. Antimicrob Agents Chemother 46:2636–2639. doi: 10.1128/AAC.46.8.2636-2639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A 97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Turnbull FW, Wallace AT, Stewart S, Crofton JW. 1953. Streptomycin resistance in patients with pulmonary tuberculosis previously treated with PAS alone. Br Med J 1:1244–1246. doi: 10.1136/bmj.1.4822.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, Liebl U. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 113.Chernyshev A, Fleischmann T, Kohen A. 2007. Thymidyl biosynthesis enzymes as antibiotic targets. Appl Microbiol Biotechnol 74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 114.Kok M, Bühlmann E, Pechère JC. 2001. Salmonella typhimurium thyA mutants fail to grow intracellularly in vitro and are attenuated in mice. Microbiology 147(Pt 3):727–733. [DOI] [PubMed] [Google Scholar]
- 115.Cersini A, Salvia AM, Bernardini ML. 1998. Intracellular multiplication and virulence of Shigella flexneri auxotrophic mutants. Infect Immun 66:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mathys V, Wintjens R, Lefevre P, Bertout J, Singhal A, Kiass M, Kurepina N, Wang XM, Mathema B, Baulard A, Kreiswirth BN, Bifani P. 2009. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 53:2100–2109. doi: 10.1128/AAC.01197-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leung KL, Yip CW, Yeung YL, Wong KL, Chan WY, Chan MY, Kam KM. 2010. Usefulness of resistant gene markers for predicting treatment outcome on second-line anti-tuberculosis drugs. J Appl Microbiol 109:2087–2094. doi: 10.1111/j.1365-2672.2010.04840.x. [DOI] [PubMed] [Google Scholar]
- 118.Zhang X, Liu L, Zhang Y, Dai G, Huang H, Jin Q. 2015. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob Agents Chemother 59:1320–1324. doi: 10.1128/AAC.03695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: an update. Drugs 69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Black PA, Warren RM, Louw GE, van Helden PD, Victor TC, Kana BD. 2014. Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2491–2503. doi: 10.1128/AAC.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ramón-García S, Mick V, Dainese E, Martín C, Thompson CJ, De Rossi E, Manganelli R, Aínsa JA. 2012. Functional and genetic characterization of the tap efflux pump in Mycobacterium bovis BCG. Antimicrob Agents Chemother 56:2074–2083. doi: 10.1128/AAC.05946-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. 2004. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a Tap-like efflux pump. Infection 32:109–111. doi: 10.1007/s15010-004-3097-x. [DOI] [PubMed] [Google Scholar]
- 123.Calgin MK, Sahin F, Turegun B, Gerceker D, Atasever M, Koksal D, Karasartova D, Kiyan M. 2013. Expression analysis of efflux pump genes among drug-susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates and reference strains. Diagn Microbiol Infect Dis 76:291–297. doi: 10.1016/j.diagmicrobio.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 124.Schuessler DL, Parish T. 2012. The promoter of Rv0560c is induced by salicylate and structurally-related compounds in Mycobacterium tuberculosis. PLoS One 7:e34471. doi: 10.1371/journal.pone.0034471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Z, Cheng S-J, Zhang H, Zhang Y. 2001. Salicylate uniquely induces a 27-kDa protein in tubercle bacillus. FEMS Microbiol Lett 203:211–216. doi: 10.1111/j.1574-6968.2001.tb10843.x. [DOI] [PubMed] [Google Scholar]