Abstract
Oxidants were shown to contribute to the lethality of bactericidal antibiotics in different bacterial species, including the laboratory strain Streptococcus pneumoniae R6. Resistance to penicillin among S. pneumoniae R6 mutants was further shown to protect against the induction of oxidants upon exposure to unrelated bactericidal compounds. In the work described here, we expanded on these results by studying the accumulation of reactive oxygen species in the context of antibiotic sensitivity and resistance by including S. pneumoniae clinical isolates. In S. pneumoniae R6, penicillin, ciprofloxacin, and kanamycin but not the bacteriostatic linezolid, erythromycin, or tetracycline induced the accumulation of reactive oxygen species. For the three bactericidal compounds, resistance to a single molecule prevented the accumulation of oxidants upon exposure to unrelated bactericidal antibiotics, and this was accompanied by a reduced lethality. This phenomenon does not involve target site mutations but most likely implicates additional mutations occurring early during the selection of resistance to increase survival while more efficient resistance mechanisms are being selected or acquired. Bactericidal antibiotics also induced oxidants in sensitive S. pneumoniae clinical isolates. The importance of oxidants in the lethality of bactericidal antibiotics was less clear than for S. pneumoniae R6, however, since ciprofloxacin induced oxidants even in ciprofloxacin-resistant S. pneumoniae clinical isolates. Our results provide a clear example of the complex nature of the mode of action of antibiotics. The adaptive approach to oxidative stress of S. pneumoniae is peculiar, and a better understanding of the mechanism implicated in response to oxidative injury should also help clarify the role of oxidants induced by antibiotics.
INTRODUCTION
Streptococcus pneumoniae is an opportunistic colonizer of the nasopharynx and the causative agent of many serious diseases, such as pneumonia, sepsis, meningitis, and otitis media (1, 2). Antimicrobial therapy based on β-lactam antibiotics is the recommended treatment regimen against pneumococcal diseases (3–5). However, resistance is now common in many countries, resulting into a shift toward the use of other molecules, including respiratory fluoroquinolones and third-generation cephalosporins (6–11). While the rates of resistance to these alternative agents remain globally low, some countries are nonetheless experiencing decreased susceptibilities (6, 8, 12–16), and a precise understanding of the mode of action (MOA) of antibiotics and of the cellular response that they induce should prove useful for the prevention of further resistance.
Bactericidal antibiotics have been proposed to contribute to bacterial death through a common mechanism involving reactive oxygen species (ROS) as a common effector (17–24). While generating great enthusiasm regarding novel therapeutic strategies (25, 26), this unified model remains a matter of debate given recent contradictory findings about the role of oxidants in the MOA of bactericidal antibiotics (27–29). While ROS are not the sole arbiters, recent additional work convincingly showed that they contribute to the lethality of antibiotics (30, 31). In the case of S. pneumoniae, the proposed model is difficult to reconcile with the fact that, while being vulnerable to killing by bactericidal antibiotics, S. pneumoniae lacks genes encoding a complete electron transport chain or the tricarboxylic acid cycle (32), which are both central to the production of antibiotic-induced ROS (17, 18, 22, 33). S. pneumoniae is also apparently tolerant to the adverse effects of the Fenton reaction (34) owing to the sequestration of the majority of the Fe2+ (and thus of the reactive OH) away from DNA (35) and to the scarcity of pneumococcal proteins containing iron-sulfur clusters (20, 35). S. pneumoniae produces substantial levels of H2O2 through the activity of its pyruvate oxidase SpxB (36), and it is possible that deregulation in iron homeostasis following exposure to bactericidal antibiotics could feed the Fenton reaction for the production of OH to a point no more sustainable by the cell. Indeed, bactericidal antibiotics but not their bacteriostatic counterparts were previously shown to induce oxidative stress in the laboratory strain S. pneumoniae R6 (37), and in the case of the fluoroquinolone levofloxacin, this was shown to result from an increased expression of iron import genes that contributed to cell death (38). Interestingly, the selection for a nonsense mutation in a putative iron importer in penicillin (PEN)-resistant S. pneumoniae R6 was shown to protect against the triggering of ROS not only by PEN but also upon exposure to unrelated bactericidal molecules, including fluroroquinolones (37). Such cross-tolerance to antibiotic-induced ROS did not translate into substantial cross-resistance, however, suggesting a role for additional mechanisms in antibiotic-induced lethality in S. pneumoniae (37). ROS could also be formed when molecular oxygen collides with flavoenzymes and steals their electrons (reviewed in reference 20). The autoxidation rates of flavoenzymes vary widely, however, depending on the reduction potential of the flavin and the degree to which it is solvent exposed. Flavoenzymes have been described for S. pneumoniae (39, 40), but whether they represent a significant source of oxidant remains unknown.
Due to the peculiarity of S. pneumoniae regarding oxidative stress (34, 35, 41), we sought in this study to determine whether a similar connection between bactericidal antibiotics, ROS, and resistance also occur in resistant clinical isolates, more specifically, if resistance to a single bactericidal antibiotic confers tolerance to oxidative stress induced by unrelated bactericidal molecules. Our results confirmed previous observations obtained with the laboratory strain S. pneumoniae R6 but revealed a more complex situation for clinical isolates, whereby bactericidal antibiotics are potent inducers of ROS in a context of antibiotic sensitivity but also often in a context of antibiotic resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed and described in Table 1. Pneumococci were grown on blood agar containing 5% defibrinated sheep's blood or in brain heart infusion broth (BHI; Difco). Cultures were incubated for 16 to 24 h at 35°C in a 5% CO2 atmosphere. All strains were conserved frozen at −80°C in BHI containing 15% glycerol. The selection of resistant mutants was conducted from independent clones of S. pneumoniae R6 (wild type [WT]), CCRI-14635, CCRI-14703, or CCRI-14598. The selection of resistance was performed on Szybalski plates containing antibiotic concentration gradients. For subculturing, colonies were picked from the area of highest antibiotic concentrations and streaked onto agar plates containing either the same concentration of antibiotic or a gradient of increased antibiotic concentrations. The MIC of the resistant cells isolated from the plates with the highest concentrations of antibiotic was determined to confirm the resistance phenotype.
TABLE 1.
List of strains and transformants used in this study
| Strain | Description | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|---|
| PEN | CIP | KAN | LZD | ERY | TET | ||
| R6 | Wild type | 0.03 | 0.5 | 32 | 0.5 | 0.03 | 0.125 |
| R6M2B-CIP | R6 clone selected for CIP resistance | 0.03 | 128 | 32 | 0.5 | NA | NA |
| R6M2-PEN | R6 clone selected for PEN resistance | 2 | 0.5 | 32 | 0.5 | 0.03 | 0.125 |
| R6M2-KAN | R6 clone selected for KAN resistance | 0.03 | 0.5 | 1,024 | 0.5 | 0.03 | 0.06 |
| R6parCgyrA-R6M2BCIP | R6 transformed with gyrA and parC derived from R6M2B-CIP (gyrA G253A; parC C245T) | 0.03 | 32 | 32 | NA | NA | NA |
| R6parC-R6M2BCIP | R6 transformed with parC derived from R6M2B-CIP (parC C245T) | 0.03 | 2 | 32 | NA | NA | NA |
| CCRI-14635 | Susceptible S. pneumoniae clinical isolate | 0.03 | 0.5 | 32 | 0.5 | NA | NA |
| CCRI-14703 | Susceptible S. pneumoniae clinical isolate | 0.06 | 0.5 | 35 | 0.5 | 0.125 | 0.125 |
| CCRI-14598 | Susceptible S. pneumoniae clinical isolate | 0.12 | 0.5 | 32 | 0.5 | 0.125 | 0.125 |
| CCRI-14635 M2-CIP | CCRI-14635 selected for resistance to CIP (gyrA G253A; parC C245T) | 0.03 | 128 | 32 | 0.5 | 0.125 | 0.125 |
| CCRI-14598 M2-CIP | CCRI-14598 selected for resistance to CIP (gyrA G253A; parC C245T) | 0.12 | 128 | 32 | 0.5 | 0.125 | 0.125 |
| CCRI-14703 M2-CIP | CCRI-14703 selected for resistance to CIP (parC C245T) | 0.06 | 4 | 32 | 0.5 | 0.125 | 0.125 |
| CCRI-45693 | CIP-resistant S. pneumoniae clinical isolate (gyrA G253A; parC C245T) | 0.06 | 128 | 32 | NA | NA | NA |
| CCRI-50154 | CIP-resistant S. pneumoniae clinical isolate (parC C245T) | 0.03 | 4 | 32 | NA | NA | NA |
MICs were determined from three independent biological replicates. NA, not available.
Antibiotic susceptibility determination.
The MICs of all drugs used in this study were determined by microdilution according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The MICs were recorded as the lowest dilution showing no growth. When it was possible, we also used Etest strips (AB bioMérieux, Stockholm, Sweden) on Mueller-Hinton agar plates supplemented with 5% sheep blood according to the manufacturer's instructions. MIC measurements were done at least in triplicate.
Detection of ROS.
Intracellular ROS measurements relied on dichlorodihydrofluorescein diacetate (DCF-DA; Invitrogen, Grand Island, NY), whose fluorescence intensity is proportional to the level of ROS. In a typical experiment, cells were grown to the onset of exponential phase (optical density at 600 nm [OD600], 0.12) before addition of the antibiotic. PEN and ciprofloxacin (CIP) were added at a final concentration corresponding to twice the MIC, and kanamycin (KAN) was added at 13 times the MIC. The bacteriostatic antibiotics linezolid (LZD), erythromycin (ERY), and tetracycline (TET) were added at a final concentration corresponding to 20 times the MIC. One-milliliter culture aliquots were collected at baseline (prior to the addition of antibiotic) and at 1, 2, and 3 h following the addition of antibiotic. The aliquots were washed once with 1× phosphate-buffered saline (PBS; pH 7.2), resuspended in 500 μl of 1× PBS (pH 7.2) containing 5 μM DCF-DA, and incubated at 37°C in the dark for 30 min. The labeled cells were washed once with 1× PBS (pH 7.2) and resuspended in 500 μl of 1× PBS. The fluorescence signal of a 200-μl aliquot was analyzed using a Victor fluorometer (Perkin-Elmer, Waltham, MA) at 485-nm excitation and 535-nm emission wavelengths. Results were normalized according to viable cell counts and are expressed as rates of ROS production in the presence of antibiotics compared to that in a no-drug control. Rates were calculated after 3 h of exposure to drugs. Each experiment was performed with three technical and three biological replicates.
Genetic transformation.
DNA transformation was done as previously described (37, 42, 43). For reconstructing CIP resistance, the CIP-susceptible S. pneumoniae R6 was transformed with PCR fragments covering the quinolone-resistance-determining regions (QRDR) of parC and gyrA derived from the CIP-resistant mutant S. pneumoniae R6M2B (44). When needed, an rpsL+ fragment conferring resistance to streptomycin (Lys57Thr) was cotransformed as a surrogate selection marker along with the DNA fragment of interest (37).
RESULTS
Bactericidal antibiotics induce ROS in S. pneumoniae R6.
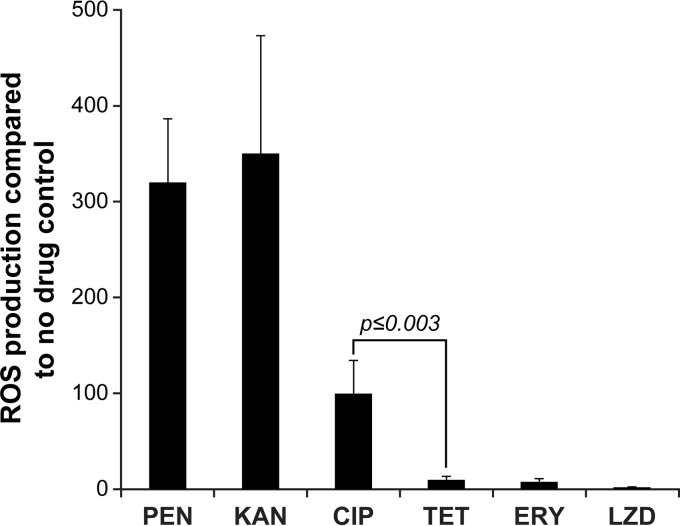
WT S. pneumoniae R6 subjected to a 3-h exposure to PEN, CIP, and KAN at concentrations equivalent to two times the respective MICs exhibited a 100- to 450-fold increase in ROS levels compared to that of the untreated control (Fig. 1). The accumulation of ROS was measured using the dichlorofluorescein diacetate dye, whose fluorescence intensity is indicative of the levels of intracellular ROS (37, 42). In contrast, the bacteriostatic antibiotics ERY, LZD, and TET only minimally increased ROS levels, even when they were added at concentrations equivalent to 20 times their MICs (P ≤ 0.003) (Fig. 1).
FIG 1.
ROS production in WT S. pneumoniae R6 after 3 h of exposure to bactericidal and bacteriostatic antibiotics. Results were normalized according to viable-cell counts and are expressed as the rate of ROS production compared to that in the no-drug control. Results are averages from three independent experiments. The significance of differences in ROS production was confirmed by Student's t test. PEN, penicillin G; KAN, kanamycin; CIP, ciprofloxacin; TET, tetracycline; ERY, erythromycin; LZD, linezolid.
Antibiotic resistance in S. pneumoniae R6 protects against the induction of ROS by unrelated antibiotics.
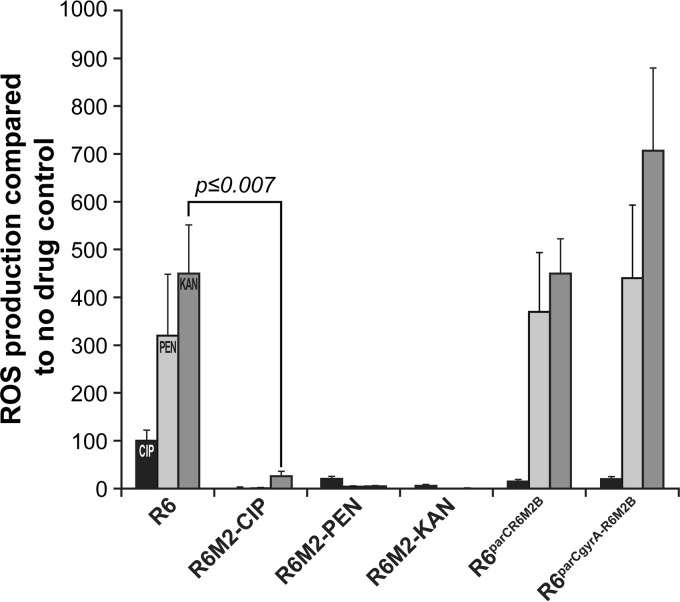
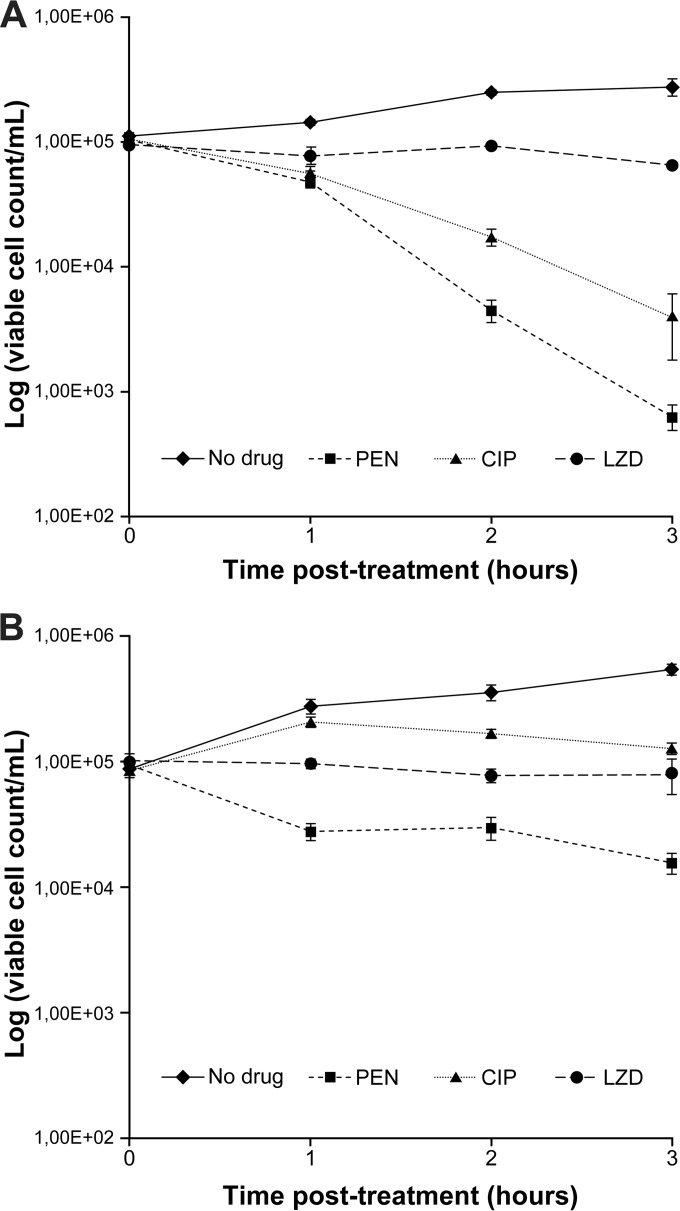
Resistance to PEN and CIP among S. pneumoniae R6 laboratory-derived mutants was previously shown to prevent the accumulation of ROS that these antibiotics usually induce (37, 44). This was confirmed and extended here, whereby the highly resistant mutants S. pneumoniae R6M2-PEN, R6M2B-CIP, and R6M2-KAN (Table 1) failed to produce ROS not only when exposed to the antibiotic defining their resistance but also when challenged with any of the three ROS-inducing antibiotics (Fig. 2). We further studied the consequences of this inability of R6 mutants to produce ROS in the presence of bactericidal antibiotics. Bacterial cells resistant to one bactericidal antibiotic were not cross resistant to the other bactericidal antibiotics (Table 1). While PEN and CIP inhibit the growth of WT S. pneumoniae R6 by killing it (Fig. 3A), it would appear that PEN inhibits the CIP-resistant mutant S. pneumoniae R6M2B-CIP by arresting growth (Fig. 3B). This reduced lethality of PEN for S. pneumoniae R6M2B-CIP was further confirmed by determining the MBC/MIC ratios of PEN, CIP, and LZD as previously described (45–47) (Table 2).
FIG 2.
ROS production in WT S. pneumoniae R6 and laboratory-derived resistant mutants and resistant transformants after 3 h of exposure to bactericidal antibiotics. Results were normalized according to viable cell counts and are expressed as the rate of ROS production compared to that in the no-drug control. Results are averages from three independent experiments. The significance of differences in ROS production was confirmed by Student's t test. The last two strains were WT S. pneumoniae R6 transformed with the parC or the parC and gyrA genes derived from R6M2B-CIP.
FIG 3.
Killing efficiency of bactericidal antibiotics in ROS-proficient and ROS-deficient S. pneumoniae backgrounds. The viable cell counts of WT S. pneumoniae R6 (A) and S. pneumoniae R6M2B-CIP (B) were monitored after 3 h of exposure to the bactericidal antibiotics CIP and PEN (at twice the MIC for the strain) and to the bacteriostatic antibiotic LZD (at 20 times the MIC for the strain) by plating in the absence of drugs. Results represent means from three independent experiments.
TABLE 2.
MBC/MIC ratios of PEN, CIP, and LZD for S. pneumoniae R6 and R6M2B-CIP
| Drug | Value fora: |
|||||
|---|---|---|---|---|---|---|
|
S. pneumoniae R6 |
S. pneumoniae R6M2B-CIP |
|||||
| MBC | MIC | MBC/MIC | MBC | MIC | MBC/MIC | |
| PEN | 0.03 | 0.03 | 1 | 0.12 | 0.03 | 4 |
| CIP | 0.5 | 0.5 | 1 | 2,048 | 128 | 16 |
| LZD | 16 | 0.5 | 32 | 16 | 0.5 | 32 |
MBCs and MICs are expressed as micrograms per milliliter and were determined from three independent biological replicates.
In the case of S. pneumoniae R6M2-PEN, the cross-tolerance to ROS accumulation was previously shown to be unrelated to the primary determinants of resistance (i.e., penicillin-binding proteins with a decreased affinity for β-lactams) but instead to result from the selection of a mutation in a putative iron permease (37). We show here that cross-tolerance to ROS accumulation in S. pneumoniae R6M2B-CIP was also independent of target mutations. The CIP resistance of R6M2B-CIP was previously shown to result mostly from the acquisition of mutations in the genes parC and gyrA, coding for DNA topoisomerase IV and DNA gyrase, respectively (44). As expected, the introduction of a parC allele derived from R6M2B-CIP alone and in combination with a mutated gyrA gene into WT S. pneumoniae R6 increased the CIP MIC for the resulting S. pneumoniae transformants 4-fold and 64-fold, respectively (Table 1). However, while the mutations in parC and gyrA were sufficient for preventing CIP from inducing ROS (Fig. 2), these could not prevent the accumulation of ROS triggered by PEN and KAN (Fig. 2). The extended protection against antibiotic-induced ROS should thus also require additional events besides target site mutations in S. pneumoniae.
Bactericidal antibiotics also induce ROS in S. pneumoniae sensitive clinical isolates.
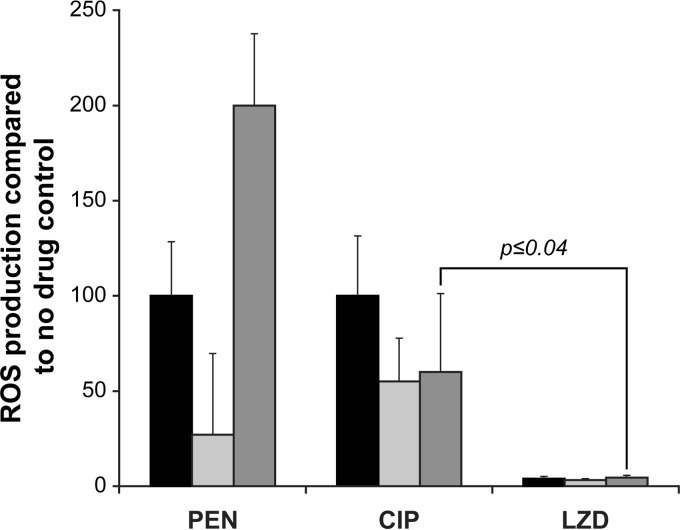
The antibiotic-sensitive clinical isolates CCRI-14635, CCRI-14703, and CCRI-14598 (Table 1) exhibited a 50- to 200-fold increase in ROS levels when exposed for 3 h to PEN and CIP at concentrations equivalent to two times their MICs (Fig. 4). Variations in ROS levels were noted between the isolates, however, with PEN, producing a 10-fold difference in ROS between clinical isolates (Fig. 4). Similar to the case with WT S. pneumoniae R6, the bacteriostatic antibiotic LZD induced only a minimal increase in ROS levels in the three antibiotic-susceptible clinical isolates (P ≤ 0.04), even when added at a concentration equivalent to 20 times the MIC (Fig. 4).
FIG 4.
ROS production in antibiotic-susceptible S. pneumoniae clinical isolates after 3 h of exposure to bactericidal and bacteriostatic antibiotics. Results were normalized according to viable cell counts and are expressed as the rate of ROS production compared to that of the no-drug control. Results are averages from three independent experiments. The significance of differences in ROS production was confirmed by Student's t test. Black bars, CCRI-14635; light gray bars, CCRI-14703; dark gray bars, CCRI-14598.
Complex association between ROS-inducing antibiotics and resistance in clinical isolates of S. pneumoniae.
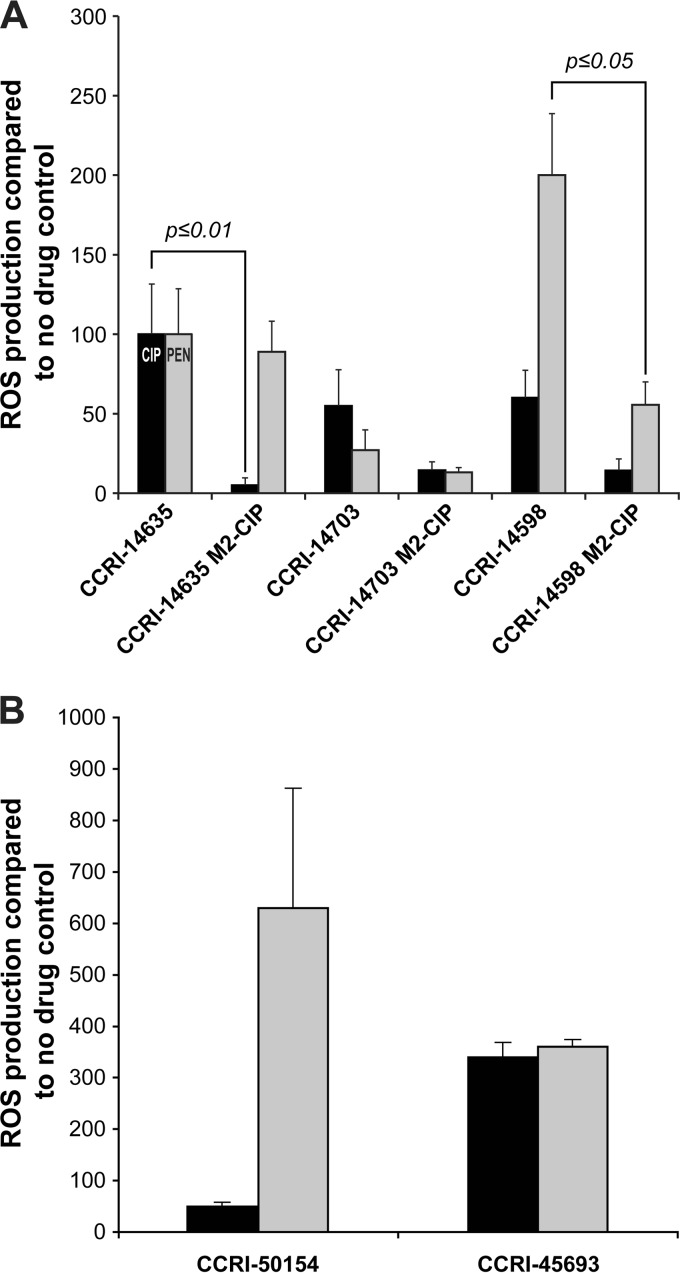
CIP induced ROS in the sensitive isolates S. pneumoniae CCRI-14635 and CCRI-14598 (Fig. 4). These two clinical isolates were then selected in vitro for resistance to CIP until they reached a 250-fold increase in resistance (CCRI-14635 M2-CIP and CCRI-14598 M2-CIP in Table 1) and both mutants had mutations in parC and gyrA (Table 1). Resistance to CIP was also selected in CCRI-14703, but we could not achieve high-level resistance in this strain (CCRI-14703 M2-CIP in Table 1). The CCRI-14635 M2-CIP and CCRI-14598 M2-CIP resistant mutants exhibited a significantly reduced ROS production (P ≤ 0.01) upon exposure to CIP in comparison to that of their sensitive parents (Fig. 5A). Similar to the case with R6, we did observe a cross-protection against the induction of ROS by PEN for CCRI-14598 M2-CIP (P ≤ 0.05) (Fig. 5A). However, ROS levels monitored in the CCRI-14635M2-CIP mutant and its CCRI-14635 parent after a 3-h exposure to PEN were almost identical (Fig. 5A). For mutant CCRI-14703 M2-CIP, differences in ROS levels were not significant with either PEN or CIP compared to its CCRI-14703 parent (Fig. 5A), a phenomenon that might be explained by the low level of resistance of this mutant (Table 1).
FIG 5.
ROS production in antibiotic-resistant S. pneumoniae clinical isolates after 3 h of exposure to bactericidal antibiotics. S. pneumoniae clinical isolates either were selected for resistance to CIP in vitro (A) or were CIP-resistant clinical isolates (B). Results were normalized according to viable cell counts and expressed as the rate of ROS production compared to no drug control. Results are the average of three independent experiments. The significance of differences in ROS production was confirmed by Student's t test.
We tested ROS production in clinical isolates for which resistance was induced in vitro. We also tested for antibiotic-induced ROS production in clinical CIP-resistant isolates. The resistant isolates CCRI-50154 and CCRI-45693 display low- and high-level resistance to CIP, respectively, owing to mutations in parC alone (CCRI-50154) or in combination with mutations in gyrA (CCRI-45693) (Table 1). Despite their resistance, these isolates did not exhibit ROS protection against either CIP (i.e., the drug defining their resistance) or PEN (Fig. 5B). The ROS levels monitored in CCRI-50154 after exposure to CIP were indeed not significantly different from those induced in the CIP-sensitive clinical isolates CCRI-14703 and CCRI-14598 (Fig. 5B). The ROS levels induced in the highly CIP-resistant clinical isolate CCRI-45693 were higher than in any other strain included in this study (Fig. 5B).
DISCUSSION
The bactericidal antibiotics PEN and CIP, but not the bacteriostatic TET, chloramphenicol, or LZD, had previously been shown to induce oxidative stress in the laboratory strain S. pneumoniae R6 (37, 44). Resistance to either PEN or CIP also protected S. pneumoniae R6 mutants against the accumulation of ROS upon exposure to the antibiotic defining their resistance (37, 44). In the case of S. pneumoniae R6 PEN-resistant mutants, such protection against antibiotic-induced ROS was further shown to extend to unrelated bactericidal antibiotics (37). In this study, we confirmed that this phenomenon most likely generalizes to any bactericidal antibiotics in the case of S. pneumoniae R6 mutants, with cross-tolerance to ROS production also occurring among mutants resistant to either of the unrelated bactericidal antibiotics CIP and KAN. These cross-protections do not translate into cross-resistance (Table 1), but we observed a switch from a clean bactericidal MOA to a phenomenon reminiscent of the bacteriostatic MOA, as indicated by the survival growth curves (Fig. 3) and the higher MBC/MIC ratio (Table 2). This is in line with the connection between ROS production and antibiotic-induced lethality extensively discussed in the literature (17, 18, 22, 23, 26, 30, 48). The acquisition of mutations in a given antibiotic's target is not enough for conferring protection against ROS induced by unrelated bactericidal counterparts, as indicated for PEN previously (37) or with the parC and gyrA genes for CIP in this study (Fig. 2). Mutations preventing ROS accumulation are likely to be selected early during the acquisition of resistance (37, 44). Combined with the altered lethality of bactericidal antibiotics in the absence of ROS, this suggests an early survival benefit to endure antibiotic pressure while more efficient mechanisms of resistance are being selected or acquired.
Our analysis of further linkages between bactericidal antibiotics and ROS production in the context of antibiotic sensitivity and resistance indicated that our findings for the lab strain R6 are not as easily extrapolated to clinical isolates. ROS-producing antibiotics were also competent in producing ROS in clinical isolates (Fig. 4 and 5). Interestingly, we found that resistance to one bactericidal antibiotic in R6 led to cross-protection to antibiotic-induced ROS (Fig. 2). A similar ROS cross-protection was observed with the drug-resistant protozoan parasite Leishmania (49). This phenomenon of resistance-mediated absence of ROS production was observed with one clinical isolate selected for CIP resistance in vitro (CCRI-14598M2-CIP) but not in CCRI-14635 (Fig. 5A). In the S. pneumoniae R6 resistant mutants, cross-tolerance to ROS induction implicated mutations other than target site mutations (see the resistant transformants in Fig. 2A), and it is possible that such mutations have not been selected in the resistant mutants derived from CCRI-14635. Neither R6 (Fig. 2) nor clinical isolates (Fig. 5A) selected in vitro for CIP resistance produced ROS in the presence of CIP. CIP resistance generated by DNA transformation in R6 will also lead to the absence of ROS production in the presence of CIP (Fig. 2). However, this is not observed with clinical CIP-resistant isolates for which ROS is produced when these cells are incubated with CIP (Fig. 5B). Thus, resistance per se is not sufficient to decrease antibiotic-induced ROS.
Recent work highlighted the complex nature of antibiotics action and reinforced the notion that ROS contribute causatively to drug lethality in addition to the lethal cellular damage induced by the inhibition of target-specific processes (30). S. pneumoniae provides a clear example of such complexity, with the role of ROS differing between laboratory-derived and naturally selected antibiotic-resistant mutants. S. pneumoniae is adapted to survive in the presence of high concentrations of endogenously generated oxidants (H2O2), but it is not entirely immune to their adverse effects (50). It is also peculiar in lacking canonical enzymes to detoxify oxygen radicals or homologues of oxidative stress response regulators (reviewed in reference 34). Still, pneumococcal enzymes implicated in the removal of ROS have been described, which include NADH oxidase, superoxide dismutase, thiol peroxidase, and alkyl hydroperoxidase (reviewed in reference 34), and differences in expression of such detoxifying enzymes or pathways could provide protection against antibiotic-induced oxidants, as recently reported (30). For example, the pneumococcal population was shown to display high genetic diversity, and differences in the ability to survive oxidative stress have been noted between strains (51). We also have previously shown that polymorphism in a putative NAD(P)H-dependent glycerol-3-phosphate dehydrogenase can protect S. pneumoniae against ciprofloxacin-mediated ROS (44). It is therefore possible that depending on their genetic background relative to oxidative stress defense, some clinical isolates do not require additional mutations to thrive in the presence of oxidants induced by antibiotics.
In conclusion, our results suggest that tolerance to antibiotic-induced oxidative stress in S. pneumoniae is not universal and varies according to the genetic background of the strains. Monitoring for new mutations providing ROS protection and the chronology of their appearance under various bactericidal conditions could increase our understanding of the response of S. pneumoniae against bactericidal antibiotics.
ACKNOWLEDGMENTS
This work was supported by CIHR grant (MOP-81266) to M.O. A.L. received a studentship from program de bourses de leadership et developpement durable de l'Université Laval, and M.O. holds the Canada Research Chair in Antimicrobial Resistance.
The authors have no conflict of interest to declare.
REFERENCES
- 1.Klugman KP, Feldman C. 2001. Streptococcus pneumoniae respiratory tract infections. Curr Opin Infect Dis 14:173–179. doi: 10.1097/00001432-200104000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Levine OS, O'Brien KL, Knoll M, Adegbola RA, Black S, Cherian T, Dagan R, Goldblatt D, Grange A, Greenwood B, Hennessy T, Klugman KP, Madhi SA, Mulholland K, Nohynek H, Santosham M, Saha SK, Scott JA, Sow S, Whitney CG, Cutts F. 2006. Pneumococcal vaccination in developing countries. Lancet 367:1880–1882. doi: 10.1016/S0140-6736(06)68703-5. [DOI] [PubMed] [Google Scholar]
- 3.Liñares J, Ardanuy C, Pallares R, Fenoll A. 2010. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect 16:402–410. doi: 10.1111/j.1469-0691.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhanel GG, Karlowsky JA, Palatnick L, Vercaigne L, Low DE, Hoban DJ. 1999. Prevalence of antimicrobial resistance in respiratory tract isolates of Streptococcus pneumoniae: results of a Canadian national surveillance study. The Canadian Respiratory Infection Study Group. Antimicrob Agents Chemother 43:2504–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan SL, Mason EO Jr. 1998. Management of infections due to antibiotic-resistant Streptococcus pneumoniae. Clin Microbiol Rev 11:628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SN, McGeer A, Melano R, Tyrrell GJ, Green K, Pillai DR, Low DE. 2011. Susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Antimicrob Agents Chemother 55:3703–3708. doi: 10.1128/AAC.00237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhanel GG, Ennis K, Vercaigne L, Walkty A, Gin AS, Embil J, Smith H, Hoban DJ. 2002. A critical review of the fluoroquinolones: focus on respiratory infections. Drugs 62:13–59. doi: 10.2165/00003495-200262010-00002. [DOI] [PubMed] [Google Scholar]
- 8.Adam HJ, Hoban DJ, Gin AS, Zhanel GG. 2009. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997–2006. Int J Antimicrob Agents 34:82–85. doi: 10.1016/j.ijantimicag.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Bhavnani SM, Hammel JP, Jones RN, Ambrose PG. 2005. Relationship between increased levofloxacin use and decreased susceptibility of Streptococcus pneumoniae in the United States. Diagn Microbiol Infect Dis 51:31–37. doi: 10.1016/j.diagmicrobio.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Imai S, Ito Y, Ishida T, Hirai T, Ito I, Maekawa K, Takakura S, Iinuma Y, Ichiyama S, Mishima M. 2009. High prevalence of multidrug-resistant pneumococcal molecular epidemiology network clones among Streptococcus pneumoniae isolates from adult patients with community-acquired pneumonia in Japan. Clin Microbiol Infect 15:1039–1045. doi: 10.1111/j.1469-0691.2009.02935.x. [DOI] [PubMed] [Google Scholar]
- 11.Hakenbeck R, Grebe T, Zahner D, Stock JB. 1999. Beta-lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol Microbiol 33:673–678. doi: 10.1046/j.1365-2958.1999.01521.x. [DOI] [PubMed] [Google Scholar]
- 12.Weigel LM, Anderson GJ, Facklam RR, Tenover FC. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 45:3517–3523. doi: 10.1128/AAC.45.12.3517-3523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baylay AJ, Piddock LJ. 18 November 2014. Clinically relevant fluoroquinolone resistance due to constitutive overexpression of the PatAB ABC transporter in Streptococcus pneumoniae is conferred by disruption of a transcriptional attenuator. J Antimicrob Chemother doi: 10.1093/jac/dku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-David D, Schwaber MJ, Adler A, Masarwa S, Edgar R, Navon-Venezia S, Schwartz D, Porat N, Kotlovsky T, Polivkin N, Weinberg I, Lazary A, Ohana N, Dagan R. 2014. Persistence and complex evolution of fluoroquinolone-resistant Streptococcus pneumoniae clone. Emerg Infect Dis 20:799–805. doi: 10.3201/eid2005.130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domenech A, Tirado-Velez JM, Fenoll A, Ardanuy C, Yuste J, Linares J, de la Campa AG. 2014. Fluoroquinolone-resistant pneumococci: dynamics of serotypes and clones in Spain in 2012 compared with those from 2002 and 2006. Antimicrob Agents Chemother 58:2393–2399. doi: 10.1128/AAC.02669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandevelde NM, Tulkens PM, Diaz Iglesias Y, Verhaegen J, Rodriguez-Villalobos H, Philippart I, Cadrobbi J, Coppens N, Boel A, Van Vaerenbergh K, Francart H, Vanhoof R, Liistro G, Jordens P, d'Odemont JP, Valcke Y, Verschuren F, Van Bambeke F. 2014. Characterisation of a collection of Streptococcus pneumoniae isolates from patients suffering from acute exacerbations of chronic bronchitis: in vitro susceptibility to antibiotics and biofilm formation in relation to antibiotic efflux and serotypes/serogroups. Int J Antimicrob Agents 44:209–217. doi: 10.1016/j.ijantimicag.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. 2012. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell 46:561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang FC. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 20.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA, Collins JJ. 2008. Rewiring bacteria, two components at a time. Cell 133:947–948. doi: 10.1016/j.cell.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 22.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 24.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Halfawy OM, Valvano MA. 2012. Non-genetic mechanisms communicating antibiotic resistance: rethinking strategies for antimicrobial drug design. Expert Opin Drug Discov 7:923–933. doi: 10.1517/17460441.2012.712512. [DOI] [PubMed] [Google Scholar]
- 26.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. 2013. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol 31:160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 30.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dwyer DJ, Collins JJ, Walker GC. 2015. Unraveling the physiological complexities of antibiotic lethality. Annu Rev Pharmacol Toxicol 55:313–332. doi: 10.1146/annurev-pharmtox-010814-124712. [DOI] [PubMed] [Google Scholar]
- 32.Hoskins J, Alborn WE Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 34.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. 2013. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol 21:187–195. doi: 10.1016/j.tim.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185:6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 37.Fani F, Leprohon P, Legare D, Ouellette M. 2011. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome Biol 12:R115. doi: 10.1186/gb-2011-12-11-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrándiz MJ, de la Campa AG. 2014. The fluoroquinolone levofloxacin triggers the transcriptional activation of iron transport genes that contribute to cell death in Streptococcus pneumoniae. Antimicrob Agents Chemother 58:247–257. doi: 10.1128/AAC.01706-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Ruyck J, Janczak MW, Neti SS, Rothman SC, Schubert HL, Cornish RM, Matagne A, Wouters J, Poulter CD. 2014. Determination of kinetics and the crystal structure of a novel type 2 isopentenyl diphosphate: dimethylallyl diphosphate isomerase from Streptococcus pneumoniae. Chembiochem 15:1452–1458. doi: 10.1002/cbic.201402046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrakchi H, Dewolf WE Jr, Quinn C, West J, Polizzi BJ, So CY, Holmes DJ, Reed SL, Heath RJ, Payne DJ, Rock CO, Wallis NG. 2003. Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK). Biochem J 370:1055–1062. doi: 10.1042/BJ20021699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng J, Lupien A, Gingras H, Wasserscheid J, Dewar K, Legare D, Ouellette M. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res 19:1214–1223. doi: 10.1101/gr.089342.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billal DS, Feng J, Leprohon P, Legare D, Ouellette M. 2011. Whole genome analysis of linezolid resistance in Streptococcus pneumoniae reveals resistance and compensatory mutations. BMC Genomics 12:512. doi: 10.1186/1471-2164-12-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lupien A, Billal DS, Fani F, Soualhine H, Zhanel GG, Leprohon P, Ouellette M. 2013. Genomic characterization of ciprofloxacin resistance in a laboratory-derived mutant and a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother 57:4911–4919. doi: 10.1128/AAC.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahal JJ Jr, Simberkoff MS. 1979. Bactericidal and bacteriostatic action of chloramphenicol against meningeal pathogens. Antimicrob Agents Chemother 16:13–18. doi: 10.1128/AAC.16.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson RD, Steigbigel RT, Davis HT, Chapman SW. 1980. Method of reliable determination of minimal lethal antibiotic concentrations. Antimicrob Agents Chemother 18:699–708. doi: 10.1128/AAC.18.5.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor PC, Schoenknecht FD, Sherris JC, Linner EC. 1983. Determination of minimum bactericidal concentrations of oxacillin for Staphylococcus aureus: influence and significance of technical factors. Antimicrob Agents Chemother 23:142–150. doi: 10.1128/AAC.23.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belenky P, Collins JJ. 2011. Microbiology. Antioxidant strategies to tolerate antibiotics. Science 334:915–916. [DOI] [PubMed] [Google Scholar]
- 49.Moreira W, Leprohon P, Ouellette M. 2011. Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis 2:e201. doi: 10.1038/cddis.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J Bacteriol 189:6532–6539. doi: 10.1128/JB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andisi VF, Hinojosa CA, de Jong A, Kuipers OP, Orihuela CJ, Bijlsma JJ. 2012. Pneumococcal gene complex involved in resistance to extracellular oxidative stress. Infect Immun 80:1037–1049. doi: 10.1128/IAI.05563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]