Abstract
The structure of a composite staphylococcal cassette chromosome (SCC) carried by a methicillin-resistant Staphylococcus haemolyticus (NW19A) isolated from a bovine milk sample was analyzed. The formation of the circular forms of both single SCC elements and composite SCC elements was detected in NW19A. Twenty heavy metal and antibiotic resistance-related genes coexisted in this composite SCC, suggesting that these genes might be coselected under environmental pressure. The mec gene complex in NW19A, designated type C3, is different from classic C1 or C2 gene complexes structurally and likely evolves differently. Furthermore, results from alignment of the SCC composite island of NW19A with 50 related sequences from different staphylococcal strains provided additional evidence to support the notion that coagulase-negative staphylococci (CoNS) are the original host of heavy metal resistance genes among staphylococci. Given that a SCC composite island could transfer freely among different staphylococcal species from different hosts, more attention should be paid to contamination with heavy metals and antibiotics in dairy farming environments, including wastewater, soil, feces, and feed.
INTRODUCTION
Coresistance, one type of coselection mechanism, occurs when genes specifying resistant phenotypes are located together on the same mobile genetic element (MGE), such as plasmids, transposons, and staphylococcal cassette chromosome (SCC) elements (1). SCC elements are widely distributed in different staphylococci and highly diverse in their structural organization and genetic contents. SCCmec is a type of SCC element. It carries the mec gene complex and has broad-spectrum beta-lactam resistance encoded by the mecA (or mecC) gene. Methicillin-resistant Staphylococcus aureus (MRSA) strains are a leading cause of health care-associated infections and community-associated infections worldwide and can be associated with mastitis in dairy cattle (2, 3). MRSA is generated when methicillin-sensitive Staphylococcus aureus (MSSA) exogenously acquires an SCCmec element, but with an unclear mechanism (4). There is increasing, but largely indirect, evidence suggesting that coagulase-negative staphylococci (CoNS) might act as a source of SCCmec for MRSA. However, very little is known about genetics of SCCmec in CoNS (5).
In China, the average usage of veterinary antibiotics has reached approximately 6,000 tons annually (6), and numerous antibiotic-resistant bacteria have appeared (7). Abundant antibiotics are also used in dairy cattle in China because of the high incidence of mastitis and endometritis (8) and may increase the prevalence of antibiotic-resistant bacteria. In order to investigate antibiotic-resistant staphylococci from mastitis milk, in this study, a methicillin-resistant Staphylococcus haemolyticus (MRSH) strain from a milk sample from a Holstein cow was studied for its SCC element in terms of structural organization and genetic contents. Besides a mecA gene, a tetracycline resistance gene and many heavy metal resistance genes were located together in the SCCs in this strain.
MATERIALS AND METHODS
Bacterial strain and genome sequencing.
Strain NW19A was isolated from a mastitis milk sample from a Holstein cow. It was identified as S. haemolyticus based on 16S rRNA sequencing. Genomic DNA was extracted and the whole-genome sequencing (WGS) was performed by BGI-Shenzhen (Shenzhen, China) using the Solexa paired-end sequencing technology. A genomic DNA library was prepared with an insert size of 500 bp. WGS sequence data were assembled into contigs by using SOAPdenovo (version 1.05). Furthermore, the contigs were joined into scaffolds using paired-end information. The order of the scaffolds was determined by aligning to the genome of S. haemolyticus strain JCSC1435 (GenBank accession no. AP006716) using SOAPaligner (version 2.21).
Assemblage and identification of SCC elements in NW19A.
To map SCC elements, every scaffold of NW19A was subjected to a BLAST search against marker genes, including orfX, the mec gene complex, and the ccr gene complex. Positive scaffolds were aligned against SCCmec V(5C2&5)c of S. aureus P126 (GenBank accession no. KF593809) to confirm their genetic organization. Gaps were closed by PCR followed by sequencing. Gene prediction in NW19A was performed using Glimmer 3.0. The nucleotide sequences of predicted genes and their deduced amino acid sequences were subsequently compared mainly against the nucleotide collection (nr/nt) database and the nonredundant protein database provided by NCBI, separately. A second set of PCRs was also performed for investigating possible SCCmec excision, with extracted genomic DNA from stationary-phase cultures as templates. Primer sequences are listed in Table S1 in the supplemental material.
MIC determinations.
MICs for the antimicrobial agents listed in Table 1 were determined for strain NW19A using an agar dilution method recommended by the CLSI (9). MICs of heavy metal ions were measured as previously described (10). Bacterial suspensions of McFarland 0.5 standard were prepared for the tests. Mueller-Hinton agar plates with different final concentrations of heavy metal ions were prepared [CuSO4 · 5H2O for copper, Cd(NO3)2 · 4H2O for cadmium, NaAsO2 for arsenic, and ZnSO4 · 7H2O for zinc]. The strains S. aureus RN4220 and S. haemolyticus BB02312 served as controls for MIC determination of both antibiotics and heavy metal ions.
TABLE 1.
Results of susceptibility testing against heavy-metal compounds and antibiotics of NW19A
| Strain (species) | MICa (mM) of metal |
MICb (μg/ml) of antimicrobial agent |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CuSO4 | Cd(NO3)2 | ZnSO4 | NaAsO2 | GEN | PEN | OXA | FOX | TET | LIN | CIP | TEC | TMP | SMZ | |
| NW19A (S. haemolyticus) | 8 | 0.4 | 4 | 8 | >256 | 16 | >256 | 32 | >256 | >256 | 32 | 8 | 0.5 | >256 |
| RN4220 (S. aureus) | 4 | 0.015 | 4 | 0.015 | 1 | 1 | 0.5 | 2 | 1 | 1 | 2 | 2 | 1 | >256 |
| BB02312 (S. haemolyticus) | 2 | 0.25 | 1 | 0.25 | 2 | 16 | 1 | 4 | 2 | 128 | 4 | 4 | 2 | >256 |
All MIC determinations were performed in agar dilution assays, and a tentative breakpoint for heavy metal resistance was defined as a MIC of >2 mM.
GEN, gentamicin; PEN, penicillin G; OXA, oxacillin; FOX, cefoxitin; TET, tetracycline; LIN, lincomycin; CIP, ciprofloxacin; TEC, teicoplanin; TMP, trimethoprim; SMZ, sulfamethoxazole.
Bioinformatics analyses.
In order to determine the role of NW19A in the emergence and evolution of MRSA, the NCBI nucleotide BLAST software was used for genome alignments in the nr/nt database with default parameters (accessed on 11 September 2014). The query sequence was the identified nucleotide sequence of CI19A (the SCC composite island of NW19A) with a length of 69.7 kb. The top 50 matched sequences with both a maximum score greater than 10,000 and a total score greater than 40,000 were chosen for detailed analyses. The comparison of different SCC elements was performed using BLASTN with the software Mauve (11), ACT (12), and Easyfig (13). The direct repeat unit (DRU) was analyzed as suggested elsewhere (14).
RESULTS
General features of S. haemolyticus NW19A and the SCC composite island in it.
Strain NW19A was resistant to a range of antimicrobial drugs (Table 1). The MICs of copper, cadmium, arsenic, and zinc ions were 8 mM, 0.4 mM, 8 mM, and 4 mM, respectively. The WGS sequence data of 280 Mb from NW19A, giving approximately 100-fold genome coverage, were generated and assembled into 161 contigs. Subsequently, the contigs were joined into 77 scaffolds, with coverage of 89.99% against the human-origin strain S. haemolyticus JCSC1435. Among the 77 assembled scaffolds, 14 were identified as components of SCC elements. Twenty-one PCRs were performed to close the gaps between/in these scaffolds. Finally, the SCC composite island of NW19A was determined and designated CI19A, with a length of 69.7 kb (GenBank accession no. KM369884). It consisted of one intact SCC element designated SCCmec19A (42.9 kb) and three pseudo-SCC elements designated ΨSCC19A2-ΨSCC19A3-ΨSCCcad/ars/cop, from left to right (Fig. 1a). Among the 71 predicted open reading frames (ORFs) in CI19A, 19 ORFs were resistance-associated genes (2 for antibiotics and 17 for heavy metals) and 12 ORFs were related to DNA transfer (6 insertion sequences and 6 recombination-related genes) (see Table S2 in the supplemental material). CI19A contained 4 integration site sequences (ISS) in addition to the one in orfX. Only one SCC retained the cassette chromosome recombinase (ccr) gene essential for SCC movement. Other SCCs were thus considered remnants of SCCs and designated ΨSCCs.
FIG 1.
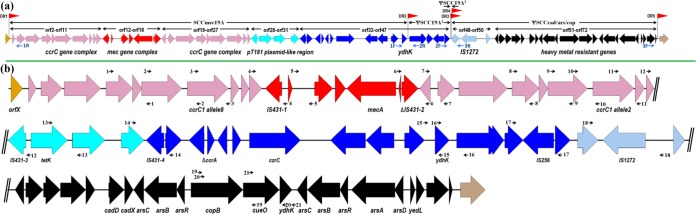
(a) Schematic representation of the SCC composite island (CI19A) in S. haemolyticus NW19A. ORFs are shown as arrows indicating the transcription direction, and different colors represent different fragments. Thin black arrows indicate the regions that are SCCs or fragments. Red arrowheads (DR1, DR2, DR3, DR4, and DR5) indicate the locations of integration site sequences for the SCC. Thin blue arrows indicate the location of PCR primers used to detect excision. (b) Locations of ORFs encoding antibiotic resistance proteins, heavy metal resistance proteins, transposases, or recombinases in the SCC composite island. Thin black arrows indicate the locations of PCR primer pairs used to close the gaps.
PCR were performed to detect putative extrachromosomal circular intermediates of each element situated between direct repeats (DRs). Strongly positive PCR amplicons were detected not only for the primer sets 1R+1F, 2R+2F, and 3R+3F but also for the primers sets 1R+2F, 1R+3F, and 2R+3F (data not shown). These results confirmed the formation of the circular forms in NW19A of both single SCC elements (SCCmec19A, ΨSCC19A2, and ΨSCCcad/ars/cop) and composite SCC elements (CI19A, SCCmec19A-ΨSCC19A2, and ΨSCC19A2-ΨSCC19A3-ΨSCCcad/ars/cop).
SCCmec19A was identified immediately downstream of orfX and bracketed by two DRs (Fig. 1a). SCCmec19A showed 99.1% DNA sequence identity to SCCmec type V (5C2&5)c of S. aureus P126 (GenBank accession no. KF593809) and was classified as the same type. It was a composite of a ccrC1 allele 8 gene complex (orf5 to orf11), a type V SCCmec carrying a class C2-like mec gene complex (orf12 to orf18), and a ccrC1 allele 2 gene complex (orf22 to orf27). The mec gene complex was composed of IS431-1, ΔmvaS, ugpQ, maoC, mecA, ΔmecR1, and ΔIS431-2 genes in the order. Unlike the class C2 mec gene complex, the IS431-2 was truncated in NW19A. A pT181-like fragment was bracketed by two insertion sequences (IS431-3 and IS431-4) and contained a gene encoding a tetracycline resistance protein (tetK) (Fig. 1b). Furthermore, a region of 10 ORFs (orf33 to orf42) located downstream of the pT181-like fragment. The first three genes were related to chromosome recombination (see Table S2 in the supplemental material). The last seven genes might be associated with heavy metal resistance (orf36 to orf38), putative metallo β-lactamase resistance (orf39), and anion transposition (orf40 to orf42) (see Table S2 in the supplemental material).
The ΨSCC19A2 was 4.5 kb long and harbored an IS256 (orf46), which divided a gene into two nonfunctional fragments (orf45 and orf47) (Fig. 1b and 2). ΨSCC19A3 was 0.1 kb long and did not contain any ORF. ΨSCCcad/ars/cop was 22.2 kb long, with 25 ORFs (orf48 to orf72) harboring resistance genes for cadmium, arsenic, and copper (Fig. 1b; also, see Table S2 in the supplemental material). Two ars operons were found in ΨSCCcad/ars/cop as arsC-arsB-arsR and arsC-arsB-arsR-arsA-arsD, separately.
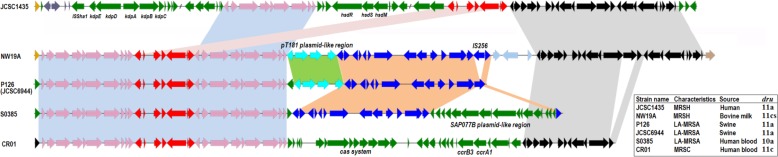
FIG 2.
Comparative structure analyses of CI19A against sequences of S. haemolyticus JCSC1435, S. aureus P126, S. aureus JCSC6944, S. aureus S0385, and Staphylococcus capitis CR01. Homologous gene clusters in different strains are shaded in different colors.
Comparative analyses of SCCs in NW19A with those in related bacteria.
Fifty sequences from different staphylococcal strains with the highest similarities with CI19A were chosen for detailed analyses (see Table S3 in the supplemental material). It is noteworthy that 74% of the sequences investigated (37 out of 50) contained defensive systems such as a type I or II restriction-modification system or a CRISPR-associated system (CAS) (see Table S3 in the supplemental material). However, none of the defensive system existed in CI19A (see Table S3 in the supplemental material). Among the 50 strains investigated, the five showing highest scores with CI19A were P126, JCSC6944, S0385, CR01, and JCSC1435.
CI19A shared the region of orf2 to orf27 with P126, JCSC6944, S0385, and CR01. It also shared the regions of orf12 to orf18 and orf19 to orf27 with strain JCSC1435, but with different arrangement of positions (Fig. 2). Interestingly, the IS431-2 gene was intact in JCSC1435 instead of a truncated version in P126, JCSC6944, S0385, and CR01 as well as NW19A. In addition, strains JCSC1435, JCSC6944, and P126 shared DRU type 11a, suggesting that the acquirement of mec gene complexes among them was evolutionarily related (Fig. 2).
The region from orf32 to orf47 in CI19A showed high sequence identities with P126, JCSC6944, and S0385. CI19A, P126, and JCSC6944 contained an integrated pT181-like plasmid (orf28 to orf31), which was absent in the genome of S0385 (Fig. 2). Interestingly, a separate plasmid, pS0385-1, that was nearly identical to orf28 to orf31 was found in S0385 (GenBank accession no. AM990993). Furthermore, similar interruption of IS256 in CI19A was observed in the genome of S0385, where 17 genes were inserted (Fig. 2). These 17 genes showed 98% sequence identity with the ORFs in S. aureus plasmid SAP077B (GenBank accession no. GQ900429). These results suggest the existence of many insertion events in CI19A mediated by plasmids and close evolutional relationships in the region of orf28 to orf47 among these four strains.
The orf28 to orf47 region was absent in CR01 and JCSC1435. Most genes in this region were related to antibiotic resistance (orf29 and orf39), heavy metal resistance (orf36 to orf38), or chromosome transfer (orf28, orf30, orf32, orf33, orf34, orf35, and orf46). The corresponding region in CR01 and JCSC1435 was composed mainly of the CAS system and type I restriction-modification system, respectively.
The region encompassing orf51 to orf72 in CI19A was rich in heavy metal resistance genes, conferring resistance to cadmium, arsenic, and copper. JCSC1435 shared this region with CI19A, but its copB gene (orf61) was truncated; CR01 contains a part of this region. P126 and S0385 did not contain ΨSCC19A3-ΨSCCcad/ars/cop as in NW19A, suggesting that at least two more recombination events occurred in NW19A in comparison with them (Fig. 2).
DISCUSSION
Compared to a single SCCmec, composite SCC elements contain many more resistance genes (15, 16). A novel composite SCC from S. haemolyticus NW19A has been revealed in this study. It is characterized not only by containing a large proportion of heavy-metal resistance genes and antibiotic resistance genes but also by including a surprising number of ISs and recombinases. Several circular intermediates of SCC elements were detected in this study, implying that CI19A could be excised as a whole or in part. It has been reported that soil, water, crops, animal feed, and feces on dairy farms in China were contaminated with heavy metals (17). Hence, the coexistence of both types of determinants in the same genetic element of bacteria may allow antibiotic resistance to be selected upon heavy metal selective pressure in the contaminated environment.
The gene czrC (orf38) is related to zinc and cadmium resistance (18); both CopB (orf61) and CueO (orf62) can transport copper from the cytoplasm to the extracellular area, decreasing the damage of bacteria by copper (19); CadD (orf56) is a putative cadmium transporter, which is regulated by CadX (orf57) (19). These proteins conferred zinc, copper, and cadmium resistance to NW19A, which was determined by heavy metal resistance assay. Two groups of arsenic operons are present in NW19A, giving the strain strong resistance to arsenic (MIC = 8 mM), since the proteins ArsA (orf67) and ArsD (orf68) could greatly enhance the arsenic efflux capacity of the ArsB pump (20).
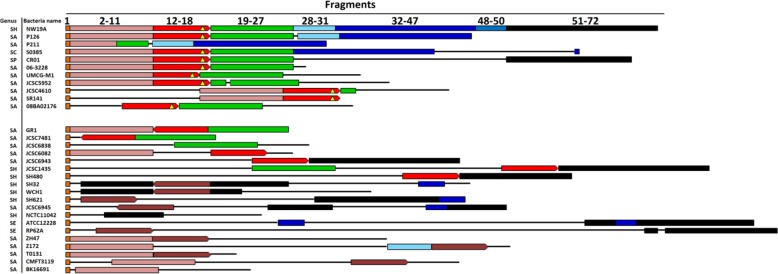
In this study, bioinformatics analyses were performed in order to reveal potential sequences for formation of CI19A. As shown in Fig. 3 and in Table S4 in the supplemental material, CI19A can be divided into 3 constitutive regions (orf2 to orf27, orf28 to orf50, and orf51 to orf72). The first region is comprised of three independent units: the ccrC1 allele 8 gene complex, the mec gene complex, and the ccrC1 allele 2 gene complex. The second region might be formed by three recombination events: entry of orf28 to orf31 by the pT181-like plasmid, entry of orf32 to orf47 mediated by CcrAB recombinases, and entry of orf48 to orf50 probably by IS1272. The third region possesses many heavy metal resistance genes. Eleven investigated strains as well as NW19A contain all or parts of this region. It is noteworthy that 10 out of the 12 strains are CoNS (Fig. 3; also, see Table S4 in the supplemental material), suggesting that this heavy metal multiresistant region may originate from CoNS. In summary, we hypothesize that the formation of CI19A might be conducted with the help of S. aureus P126, JCSC6944, or S0385 for the first two constitutive regions and the help of S. haemolyticus JCSC1435 for the third constitutive region.
FIG 3.
Comparative analysis of CI19A with the 50 bacteria investigated. Various fragments are differentiated by different colors. The red and brown boxes with arrows represent mec gene complexes and their directions. The red boxes with and without a yellow triangle represent type C3 and C2 mec gene complexes, respectively. The brown boxes represent other mec complex types. The thin black line indicates sequences with no identities among different SCC elements. SH, SA, SC, SP, and SE represent S. haemolyticus, S. aureus, S. capitis, Staphylococcus pseudintermedius, and Staphylococcus epidermidis, respectively. More detailed comparisons and descriptions of strains are given in Table S4 in the supplemental material.
There were several types of mec gene complexes in the strains investigated. A large proportion of strains contain a truncated IS431-2 (Fig. 3). IS431-2 has been proposed to be involved in the formation of the mec gene complex (21). Therefore, a truncated IS431-2 could eliminate the mobility of the mec gene complex. Furthermore, the IS-mediated mecA deletion occurs at much higher rates than the ccr-mediated complete excision of SCCmec (4, 22), so a truncated IS431-2 probably could decrease the frequency of losing the mec gene complex. We confirmed this hypothesis by the finding that the ΔIS431-contained class C2 mec gene complexes were always interlocked with surrounding sequences (Fig. 3), suggesting the presence of a new evolutional tendency of the mec gene complex. There are two distinct class C mec gene complexes (C1 and C2) differentiated by the orientation of their IS431-2. They have likely evolved independently (23). Based on this, we suggest classifying this mec gene complex as a class C3 complex.
It seems that the chromosome of NW19A is very plastic and might have encountered various types of environmental pressure. The CAS and restriction-modification system enable bacteria to degenerate exogenous DNA and protect the stability of chromosome, so the lack of these systems in NW19A may facilitate acquisition of exogenous DNA and consequently speed up the evolutional process of this bacterium. The emergence of livestock-associated MRSA infection constitutes an additional obstacle to control MRSA in humans (24, 25) and to treat mastitis in dairy cattle. A study has provided evidence for a horizontal transfer of a type V SCCmec from MRSH to a MSSA to create a new clone of MRSA (26). Considering the fact that abundant heavy metal-resistant genes and antibiotic resistance genes coexist in a mobile SCC element and could be coselected, we suggest reducing the use of antibiotics and heavy metals and improving animal management systems to limit the selection of resistant bacteria and the emergence of MRSA in dairy cattle.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from National Science Foundation of China (grant number 31372282), the China Thousand Talents program and two University Scientific Research Fund projects (fund number Z111021305 and fund number Z109021431).
We have no conflicts of interests to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04831-14.
REFERENCES
- 1.Chapman JS. 2003. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegradation 51:271–276. doi: 10.1016/S0964-8305(03)00044-1. [DOI] [Google Scholar]
- 2.Chatterjee SS, Otto M. 2013. Improved understanding of factors driving methicillin-resistant Staphylococcus aureus epidemic waves. Clin Epidemiol 5:205–217. doi: 10.2147/CLEP.S37071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes MA, Zadoks RN. 2011. Methicillin resistant S. aureus in human and bovine mastitis. J Mammary Gland Biol Neoplasia 16:373–382. doi: 10.1007/s10911-011-9237-x. [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Katayama Y, Hiramatsu K. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother 43:1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbier F, Ruppe E, Hernandez D, Lebeaux D, Francois P, Felix B, Desprez A, Maiga A, Woerther PL, Gaillard K, Jeanrot C, Wolff M, Schrenzel J, Andremont A, Ruimy R. 2010. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis 202:270–281. doi: 10.1086/653483. [DOI] [PubMed] [Google Scholar]
- 6.Zhao L, Dong YH, Wang H. 2010. Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci Total Environ 408:1069–1075. doi: 10.1016/j.scitotenv.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L-J, Ying G-G, Liu S, Zhang R-Q, Lai H-J, Chen Z-F, Pan C-G. 2013. Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China. Sci Total Environ 444:183–195. doi: 10.1016/j.scitotenv.2012.11.087. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2004. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Informational supplement M31-S1 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Sowmya M, Rejula MP, Rejith PG, Mohan M, Karuppiah M, Hatha AAM. 2014. Heavy metal tolerant halophilic bacteria from Vembanad Lake as possible source for bioremediation of lead and cadmium. J Environ Biol 35:655–660. [PubMed] [Google Scholar]
- 11.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryffel C, Bucher R, Kayser F, Berger-Bächi B. 1991. The Staphylococcus aureus mec determinant comprises an unusual cluster of direct repeats and codes for a gene product similar to the Escherichia coli sn-glycerophosphoryl diester phosphodiesterase. J Bacteriol 173:7416–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui LZ, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang LJ, Lu XW, Lei K, Zhai YX, Huang J. 2011. Heavy metal pollution in surface sediment of Wei River (Baoji), China. J Agro Environ Sci 30:334–340. [Google Scholar]
- 18.Cavaco LM, Hasman H, Stegger M, Andersen PS, Skov R, Fluit AC, Ito T, Aarestrup FM. 2010. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob Agents Chemother 54:3605–3608. doi: 10.1128/AAC.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silver S, Phung LT. 1996. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay R, Rosen BP, Pung LT, Silver S. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26:311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 21.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother 54:4352–4359. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noto MJ, Fox PM, Archer GL. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 52:1221–1229. doi: 10.1128/AAC.01164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Hiramatsu K, Oliveira DC, de Lencastre H, Zhang KY, Westh H, O'Brien F, Giffard PM, Coleman D, Tenover FC, Boyle-Vavra S, Skov RL, Enright MC, Kreiswirth B, Ko KS, Grundmann H, Laurent F, Sollid JE, Kearns AM, Goering R, John JF, Daum R, Soderquist B, International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 25.Kluytmans-VandenBergh MFQ, Kluytmans J. 2006. Community-acquired methicillin-resistant Staphylococcus aureus: current perspectives. Clin Microbiol Infect 12:9–15. doi: 10.1111/j.1469-0691.2006.01341.x. [DOI] [PubMed] [Google Scholar]
- 26.Berglund C, Soderquist B. 2008. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden-possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin Microbiol Infect 14(Suppl s1):1048–1056. doi: 10.1111/j.1469-0691.2008.02090.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.