The concept that Ras proteins may function as oligomers was first described almost three decades ago when Santos et al. (1) found evidence for wild-type or mutant H-Ras multimerization using radiation inactivation assays. The idea was largely dropped when the myriad of crystal structures of H-Ras that followed over the ensuing decades suggested that Ras proteins are functional monomers. Inouye et al. (2) briefly resurrected the idea when they reported detecting H-Ras dimers by cross-linking and protein fragmentation complementation. In addition, these authors reported that forced dimerization of nonmembrane-targeted H-Ras activated Raf-1 kinase activity. Also suggestive of signaling from higher-order assemblies of Ras proteins is an extensive literature from Hancock and colleagues (3–5) who analyzed the contribution of nanoclusters of five to eight Ras molecules to signaling through the two canonical Ras effector pathways, Raf–MEK–ERK and PI3K–AKT. More recently, two groups using molecular-dynamic modeling and a variety of biophysical methods have reported dimerization of the G domains of N-Ras (6) and H-Ras (7) when tethered via C-terminal lipid modifications to artificial phospholipid bilayers. Finally, Nussinov and colleagues (8), using similar techniques, very recently reported two different modes of dimerization of bacterially expressed, and therefore unprocessed, K-Ras4B in solution, one involving α-helices 3 and 4 and the other involving β-sheet interactions of the effector binding region. Importantly, both modes of dimerization were dependent on GTP binding. This flurry of activity was fueled in part by the recent discovery that Raf kinases, the Ras effectors that signal down the mitogen-activated protein kinase pathway, function as dimers (9–11). In PNAS, Nan et al. (12) lend another voice to the growing buzz. They use superresolution photoactivated localization microscopy (PALM) to reveal K-Ras4B dimers in living cells.
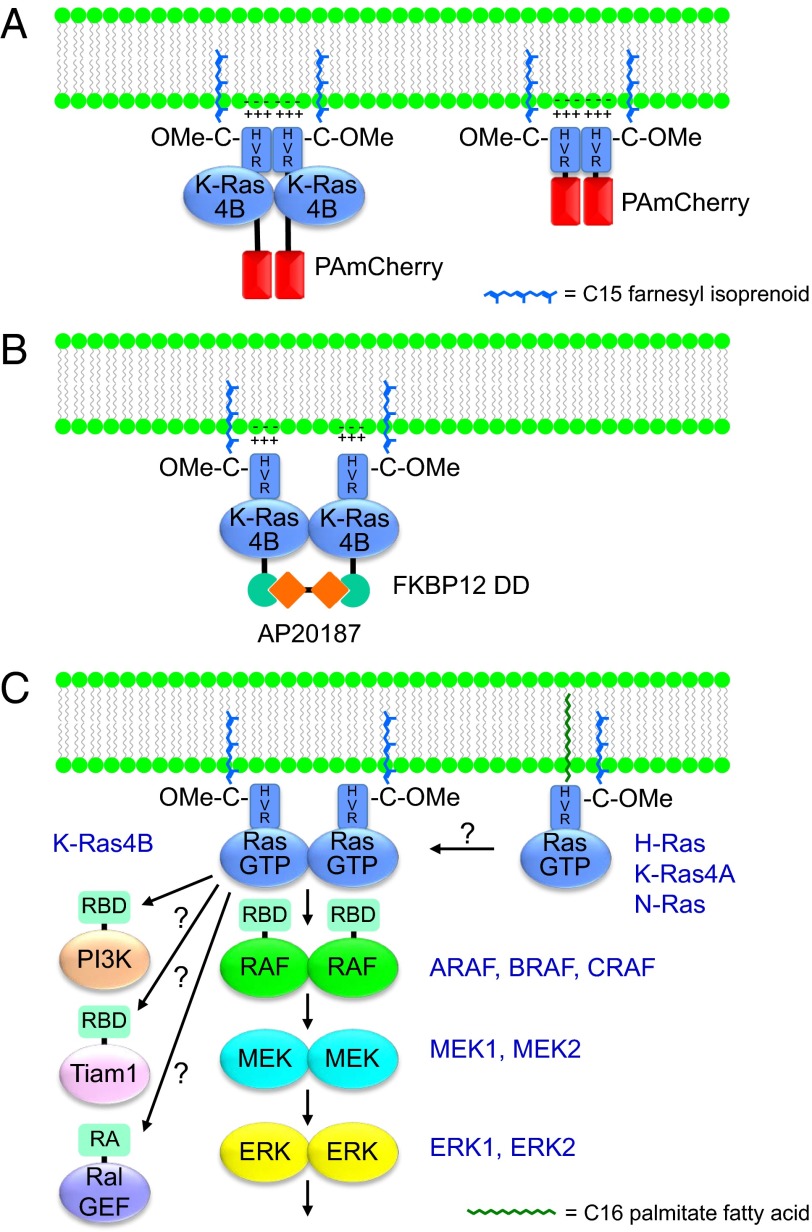
Nan et al. begin by showing that, in cells in which exogenous expression of PAmCherry1-tagged, mutationally activated K-RasG12D is under the control of a doxycycline-inducible promoter such that expression levels can be carefully controlled, there is a threshold effect with regard to activation of ERK. Using PALM and simulation-aided density-based spatial clustering analysis with noise (13), the authors determined that this threshold was associated with K-Ras monomer-to-dimer transition (Fig. 1A). Dimerization was seen at physiologic levels of K-Ras expression, with multimer formation (>4) seen with further overexpression. To confirm K-Ras signaling through dimerization, the authors used a dimerization domain (DD) derived from FKBP12 that is efficiently dimerized in the presence of the symmetrical rapalog AP20187 (Fig. 1B). The authors appended this DD onto the N terminus of K-Ras4B and expressed the construct below the ERK signaling threshold. They observed robust phosphorylation of ERK upon addition of AP20187, concordant with the results of Inouye et al. (2). The authors concluded that K-Ras dimers signal much more efficiently than do monomers and raise the possibility that K-Ras dimers are required for signaling.
Fig. 1.
Ras dimerization and effector signaling. (A) K-Ras4B membrane association is mediated by the 21-aa C-terminal hypervariable region (HVR) that terminates in a CAAX tetrapeptide sequence. The CAAX sequence is modified such that the C is farnesylated and, after the AAX amino acids are proteolytically removed, is also methylesterified (–OMe) to create a hydrophobic C terminus. The modified CAAX sequence acts in conjunction with a polybasic sequence within the HVR to anchor K-Ras4B to the inner leaflet of the plasma membrane. Fusion of PAmCherry to the N terminus of K-Ras4B (Left) revealed dimers when cells were examined by PALM. Similar dimers were observed when PAmCherry was extended only with the K-Ras4B HVR (Right), suggesting that it is the HVR that mediated dimerization. (B) Fusion of the protein dimerization domain (DD) of FKBP12 to the N terminus of K-Ras4B allowed for forced dimerization by addition of the homobifunctional rapalog AP20187. Forced dimerization was associated with enhanced signaling. (C) Regulation by dimerization of Ras and downstream elements of the Raf–MEK–ERK protein kinase cascade. In addition to the three Raf isoforms, the activities of the highly related MEK1/MEK2 and ERK1/ERK2 isoforms are also regulated by homodimerization and/or heterodimerization. Whether other Ras isoforms also form homodimers and/or heterodimers and whether K-Ras dimerization is also required for activation of other Ras-binding (RBD) or Ras association (RA) domain-containing effectors remain to be determined.
The major advance of the study by Nan et al. is the observation of Ras dimers in living cells and at physiologic levels of expression. Previous reports are based on either modeling, expression of recombinant Ras proteins on artificial bilayers, or ex vivo analysis of membrane sheets or cell lysates. Although the techniques used by Nan et al. require ectopic expression of chimeric proteins, the tetracycline-inducible system allowed for tight control of expression levels and therefore diminished the chances of artifact.
Perhaps the most surprising aspect of the study by Nan et al. is that the authors observed exactly the same dimerization signal by PALM when mCherry was extended with either full-length K-Ras or the C-terminal 21 aa that constitute the hypervariable region (HVR) that contains all of the membrane-targeting information (Fig. 1A). The authors argue that the monomeric form of Cherry fluorescent protein could not mediate oligomerization and therefore conclude that it is the HVR that mediates dimerization of K-Ras. This is unexpected for two reasons. First, the HVR is disordered and has a charge of +8 at physiological pH, making the mechanism of dimerization difficult to understand. Second, all of the recent biophysical studies that show dimerization reveal the dimerization interface to be in the catalytic G domain (6–8).
The data presented by Nan are compelling with regard to enhanced efficiency of signaling from Ras dimers. The result is an attractive one given the recent understanding of the importance of dimerization in the regulation of Raf kinases. The idea that Ras homodimers could nucleate the formation of Raf dimers as part and parcel of the Raf–MEK–ERK activation mechanism is intriguing (Fig. 1C). However, for a K-Ras homodimer to function in this capacity, some spatial constraints must be overcome. If the Nussinov model (8) is correct such that the effector binding domains of K-Ras can be oriented on opposite sides of the dimer, the Raf kinase must be extended and flexible enough for the dimerization interface of the kinase domain to be brought together under the K-Ras homodimer. How this would allow the cysteine-rich domain of Raf that is adjacent to the Ras-binding domain (RBD) to simultaneously interact with the plasma membrane (14) presents a conundrum. If Nan et al. are correct that it is the HVR that represents the dimerization domain of K-Ras, then the two effector binding domains might have a greater degree of freedom and thereby allow for an orientation that permits the Raf kinase to dimerize while simultaneously binding a K-Ras dimer, the plasma membrane, and a substrate (e.g., MEK). Perhaps the answer will be found in future cryo-electron microscopy studies, which are rapidly supplanting crystallography in analyzing protein complexes and have recently achieved resolutions of 2.2 Å (15).
Although the results reported by Nan et al. represent an advance in the emerging role for dimerization as a key regulator of Ras effector signaling, the story is incomplete. The fact that K-Ras can signal as a forced dimer does not mean that K-Ras must dimerize to signal, and further studies will be required to establish the physiologic relevance of the finding. The answer to the important question of whether monomeric K-Ras can signal will be revealed in future studies that analyze the signaling properties of GTP-loaded K-Ras that is restricted to a monomeric state. An analogy can be drawn from the field of G-protein–coupled receptors (GPCRs). The idea that GPCRs must oligomerize to signal gained traction a decade ago. Using nanodisc technology, Sunahara and colleagues (16) showed that an individual β2-adrenergic receptor could activate adenylcyclase in a ligand-dependent fashion, providing strong evidence for monomeric signaling of GPCRs. Nanodisc technology has been successfully applied to Ras proteins (17) such that the means for a direct analysis of monomeric K-Ras signaling by titrating the K-Ras content of each nanodisc to an average of one molecule should be feasible.
Other important questions raised by the work of Nan et al. and the other reports of Ras dimers include whether H-Ras and N-Ras, with divergent HVR sequences (8% sequence identity), also undergo dimerization, whether different Ras isoforms can heterodimerize, and whether dimerization with other CAAX-terminating proteins can occur (Fig. 1C). Additionally, whether K-Ras dimerization regulates other Ras effectors such as PI3K, Ral exchange factors, Tiam1, and PLCε will be important to establish to predict the biological effects of K-Ras dimerization. Given the importance of K-Ras in human cancer and the recently intensified quest to develop anti–K-Ras therapeutics (18, 19), the question of the physiologic significance of K-Ras dimerization will be of great interest to both basic cell biologists and translational cancer biologists because K-Ras dimerization may offer a previously unappreciated mode of therapeutic intervention if agents can be developed that inhibit dimerization.
Footnotes
The authors declare no conflict of interest.
See companion article on page 7996 in issue 26 of volume 112.
References
- 1.Santos E, Nebreda AR, Bryan T, Kempner ES. Oligomeric structure of p21 ras proteins as determined by radiation inactivation. J Biol Chem. 1988;263(20):9853–9858. [PubMed] [Google Scholar]
- 2.Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J Biol Chem. 2000;275(6):3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- 3.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160(2):165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian T, et al. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9(8):905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 5.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102(43):15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Güldenhaupt J, et al. N-Ras forms dimers at POPC membranes. Biophys J. 2012;103(7):1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin WC, et al. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc Natl Acad Sci USA. 2014;111(8):2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muratcioglu S, et al. GTP-dependent K-ras dimerization. Structure. 2015;23(7):1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61(9):3595–3598. [PubMed] [Google Scholar]
- 10.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461(7263):542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 12.Nan X, et al. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci USA. 2015;112(26):7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nan X, et al. Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling. Proc Natl Acad Sci USA. 2013;110(46):18519–18524. doi: 10.1073/pnas.1318188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bondeva T, Balla A, Várnai P, Balla T. Structural determinants of Ras-Raf interaction analyzed in live cells. Mol Biol Cell. 2002;13(7):2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartesaghi A, et al. Electron microscopy. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348(6239):1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA. 2007;104(18):7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazhab-Jafari MT, et al. Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci USA. 2015;112(21):6625–6630. doi: 10.1073/pnas.1419895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]