Significance
Although antibodies can normally be obtained against a wide variety of antigens, there are still hard targets, including weakly immunogenic epitopes, which are not readily amenable to existing production techniques. In addition, such techniques can be relatively time-consuming and costly, especially if the screening for a specific epitope is required. In this work we describe a rational design method that enables one to obtain antibodies targeting any specific epitope within a disordered protein or disordered region. We show that this method can be used to target three disordered proteins and peptides associated with neurodegenerative and systemic misfolding diseases.
Keywords: protein design, protein aggregation, complementary peptides
Abstract
Antibodies are powerful tools in life sciences research, as well as in diagnostic and therapeutic applications, because of their ability to bind given molecules with high affinity and specificity. Using current methods, however, it is laborious and sometimes difficult to generate antibodies to target specific epitopes within a protein, in particular if these epitopes are not effective antigens. Here we present a method to rationally design antibodies to enable them to bind virtually any chosen disordered epitope in a protein. The procedure consists in the sequence-based design of one or more complementary peptides targeting a selected disordered epitope and the subsequent grafting of such peptides on an antibody scaffold. We illustrate the method by designing six single-domain antibodies to bind different epitopes within three disease-related intrinsically disordered proteins and peptides (α-synuclein, Aβ42, and IAPP). Our results show that all these designed antibodies bind their targets with good affinity and specificity. As an example of an application, we show that one of these antibodies inhibits the aggregation of α-synuclein at substoichiometric concentrations and that binding occurs at the selected epitope. Taken together, these results indicate that the design strategy that we propose makes it possible to obtain antibodies targeting given epitopes in disordered proteins or protein regions.
Antibodies are versatile molecules that are increasingly used in therapeutic and diagnostic applications, as they can be used to treat a wide range of diseases, including cancer and autoimmune disorders (1–5). These molecules can be obtained with well-established methods, such as immunization or phage and associated display methods, against a wide variety of targets (6–11). In some cases, however, these procedures may require significant amounts of time and resources, in particular if one is interested in targeting weakly immunogenic epitopes in protein molecules. In this work, we introduce a computational method of rational design of complementarity determining regions (CDRs) that makes it possible to obtain antibody against virtually any target epitope within intrinsically disordered peptides and proteins or within disordered regions in structured proteins.
Intrinsically disordered proteins, in particular, play major roles in a wide range of biochemical processes in living organisms. A range of recent studies has revealed that the functional diversity provided by disordered regions complements that of ordered regions of proteins, in particular in terms of key cellular functions such as signaling and regulation (12–18). The high flexibility and lack of stable secondary and tertiary structures allow intrinsically disordered proteins to have multiple interactions with multiple partners, often placing them at the hubs of protein–protein interaction networks (19–21). It has also been realized that the failure of the regulatory processes responsible for the correct behavior of intrinsically disordered proteins is associated with a variety of different pathological conditions (22–24). Indeed, intrinsic disorder is often observed in peptides and proteins implicated in a series of human conditions, including cancer, cardiovascular diseases, and neurodegenerative disorders (22–24). It would therefore be very helpful to develop methods to facilitate the generation of antibodies against disordered proteins, a goal that has a great therapeutic potential (25, 26).
Here, we address this problem by introducing a rational design procedure that enables one to obtain antibodies that bind specifically target disordered regions. This procedure is based on the identification of a peptide complementary to a target region and on its grafting on to the CDR of an antibody scaffold. Related methods of altering rationally antibodies have been discussed in the literature, which include the exploration of specificity-enhancing mutations (27, 28), the design of CDRs to bind structured epitopes (28, 29), and the grafting of peptides extracted from aggregation prone proteins (30–32) or from other antibodies (33) in the CDR of an antibody scaffold. Here we show that designed antibodies can be obtained by the method that we present for essentially any disordered epitope. We illustrate the method for the Aβ peptide, α-synuclein, and the islet amyloid polypeptide (IAPP, or amylin peptide), which are respectively involved in Alzheimer’s and Parkinson’s diseases and type II diabetes (24).
Results
In this work, we present a method of rational design of antibodies targeting chosen epitopes within disordered regions of peptides and proteins. We first describe the method and then present the results obtained to test it, which show that the designed antibodies bind with good affinity and specificity their target proteins.
Rational Design of Complementary Peptides.
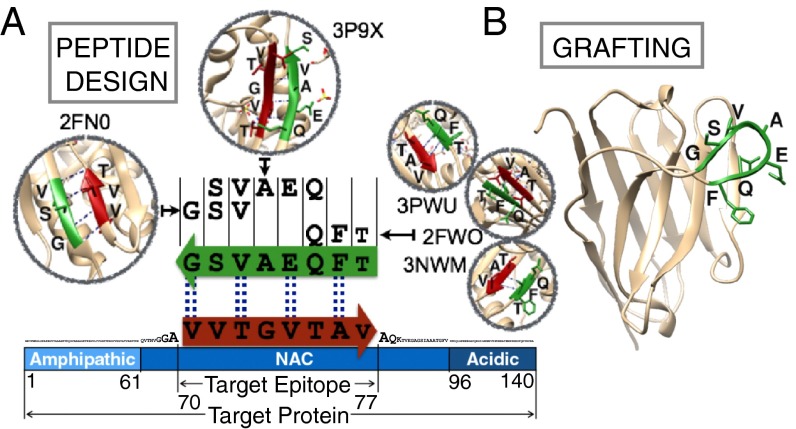
The first step in the rational design of antibodies involves the identification of peptides, called here complementary peptides, that bind with good specificity and affinity target regions of a protein molecule (Fig. 1). The identification of these complementary peptides is based on the analysis of the interactions between amino acid sequences in the Protein Data Bank (PDB). More specifically, we exploit the availability of a large number of protein structures in the PDB to identify potential interaction partners (i.e., the complementary peptides) for any given target sequence. With this choice, the affinity and the specificity of the interactions between the complementary peptides and their targets are already proven in a biological context. The complementary peptides are built through a fragment-and-join procedure (SI Materials and Methods), starting from short peptides found to interact in a β-strand with segments of the target sequence in at least one of the protein structures in the PDB database. The peptide design procedure consists in two steps. First, we collect from the PDB database all protein sequences that face in a β-strand any subsequence of at least three residues of a given target epitope. Second, complementary peptides to the target epitope are built by merging together some of these sequence fragments using a cascade method (Fig. 1A and SI Materials and Methods). In essence, this cascade method starts from one of these fragments and grows it to the length of the target epitope by joining it with some of the others following three rules: (i) all fragments generating the same complementary peptide must come from β-strands of the same type (i.e., parallel or antiparallel), (ii) all fragments must partly overlap with their neighboring fragments, and (iii) the overlapping regions must be identical both in the sequence and in the backbone hydrogen bond pattern (Fig. 1A and Fig. S1). Given this design strategy, the resulting complementary peptides are expected to bind the target epitope by enforcing a β-strand–like conformation. Therefore, such complementary peptides will be particularly effective in binding solvent-exposed regions of protein sequences that do not form persistent hydrogen bonds with other parts of the protein, such as in the case of disordered regions. Alternatively, this method may be used to design complementary peptides against any region of a target protein, including regions in the core of the native state. Such peptides could be used for example for a peptide-based detection in diagnostic, as recently proposed with naturally occurring peptides (34). Once a complementary peptide has been designed, it can be grafted in place of the CDR loop of an antibody scaffold (Fig. 1B). We also note that such a peptide could be used on its own as a drug candidate. However, the grafting on an antibody scaffold offers several advantages over the use of a peptide molecule by itself. As therapeutic molecules, with respect to peptides, antibodies have a longer half-life in vivo (35) and often lower immunogenicity, at least for human scaffolds. Moreover, in both research and diagnostics, antibodies can readily be used in a large number of biochemical and biophysical assays in vitro, including Western blotting, immunoprecipitation, and confocal imaging.
Fig. 1.
Illustration of the method of designing antibodies targeting specific epitopes within disordered proteins. (A) Sequence-based design of complementary peptides. Sequence fragments in β-strand conformations are extracted from the PDB and combined using the cascade method to generate a peptide complementary to the target epitope (SI Materials and Methods). The example shows an antiparallel peptide for an epitope (residues 70–77) in the NAC region of α-synuclein. Dashed lines connect the amino acids predicted to form backbone-backbone hydrogen bonds. (B) The designed peptide is then grafted in place of the CDR loop of an antibody. In this example it is grafted in place of the CDR3 of a human single domain antibody scaffold (SI Materials and Methods). This example corresponds to DesAb-F in Table 1.
Fig. S1.
Example of results from the cascade procedure. The sequences shown are the ones that we have selected for the experimental validation (Figs. 3–5 and Table 1). The first target sequence from the top belongs to the IAPP peptide, the second to Aβ42, and the last three to α-synuclein. In bold is the sequence of the complementary peptide that we actually grafted on the single domain antibody scaffold; a colon marks the residues predicted to participate in backbone-backbone hydrogen bonding with the target sequence.
Generality of the Design Strategy of Complementary Peptides.
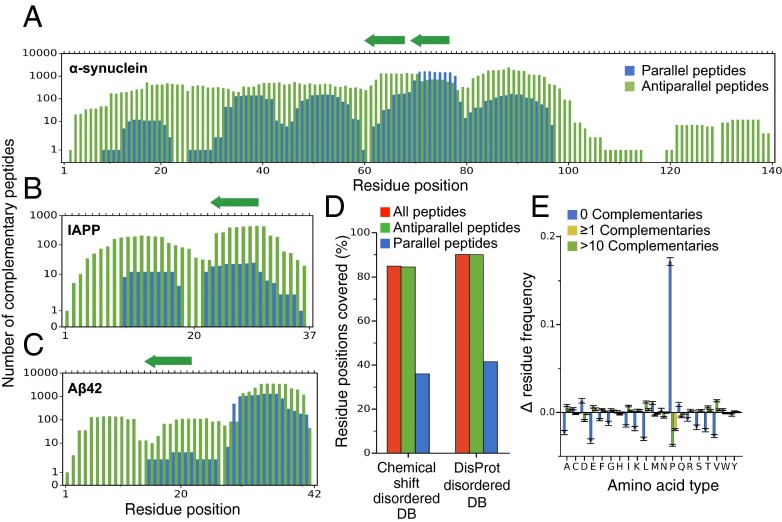
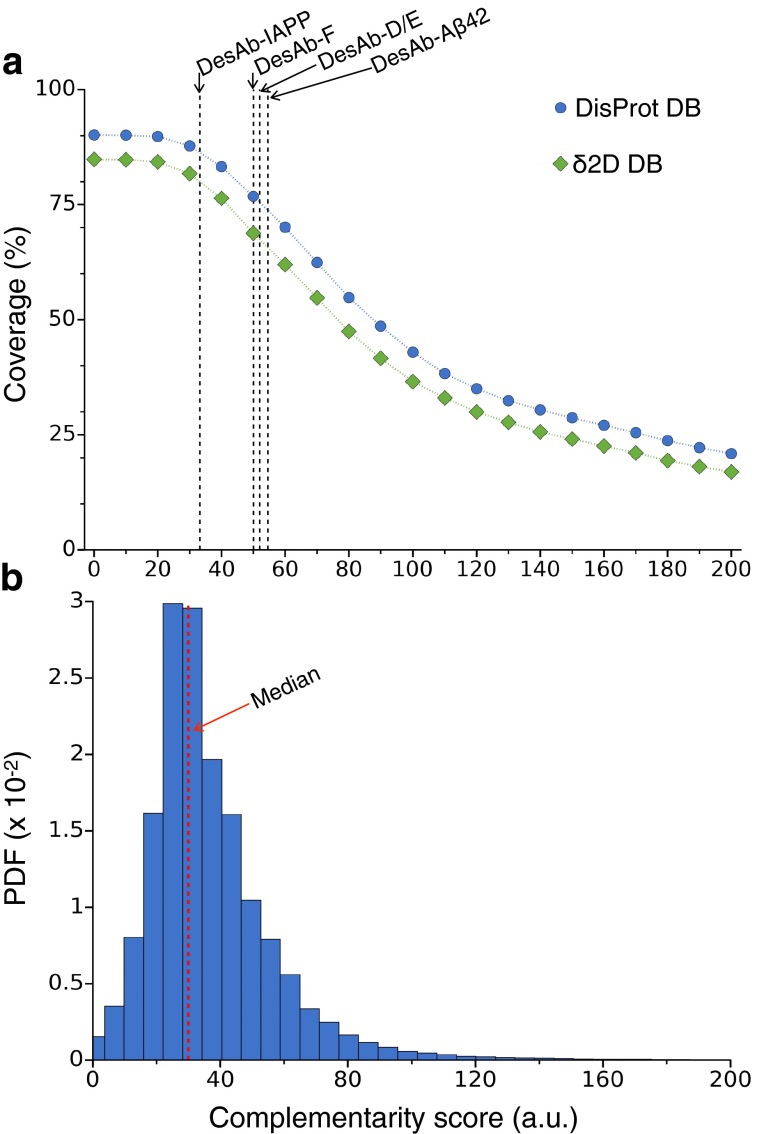
Because the design of complementary peptides depends on the availability of specific sequences facing each other in a β-strand in the protein structures in the PDB, it may not always be possible to construct a complementary peptide for a given epitope. We thus asked how generally applicable our method is by investigating systematically how many complementary peptides can be found for all possible epitopes in a target protein. We ran the cascade method on each possible epitope of eight amino acids for three well-characterized and disease-related intrinsically disordered peptides and proteins: α-synuclein, Aβ42, and IAPP (24). Although other choices are possible, we used eight-residue epitopes because we reasoned that such complementary peptide size should be amenable for grafting in most antibody scaffolds, at least for the longer CDR loops. Consequently, it represents a good epitope size to assess the generality of the cascade method. Furthermore, naturally occurring amyloidogenic eight-residue peptides were found to be specific in recognizing their targets (34), suggesting that this is a convenient length for specific β-strand-like recognition. Our results show that more than 95% of the residue positions in these three proteins can be targeted with at least one peptide. Moreover, typically, the number of different complementary peptides covering one position is much larger than 1. We found that the median number is 200, and the mean is 570 (Fig. 2 A–C). Thus, at least in these three cases, our method can produce several complementary peptides to choose from for most target epitopes. Given these results, one can ask whether a given complementary peptide may have multiple possible target sequences, thus undermining the specificity of the interaction. We investigated this possibility by blasting all of the 15,587 eight-residue peptides shown in Fig. 2 A–C against the human proteome (SI Materials and Methods). The results show that only 0.2% of the designed peptides are actually found in the proteome, suggesting that the great majority of the complementary peptides will specifically interact with their targets, as also shown by the experimental tests below. To estimate the coverage at a proteomic scale, we ran the design method on two databases of disordered proteins. The first consists of all regions annotated as disordered in the DisProt database (36), whereas the second has been constructed by identifying disordered regions from measured NMR chemical shifts (37, 38). The dataset derived from DisProt included 980 different gapless disordered regions, whereas the one derived from the NMR chemical shifts 710. We found that 90% of the residue positions in the DisProt dataset and 85% in the chemical shift dataset are covered by at least one complementary peptide (Fig. 2D). Antiparallel peptides are more frequent than parallel peptides, reflecting the fact that parallel β-strands are less abundant than antiparallel ones in the PDB. An amino acid composition analysis (Fig. 2E) revealed that those positions that are not covered by any complementary peptide are highly enriched in proline residues (), in agreement with the observation that prolines disfavor secondary structure formation (37). Other amino acids preferentially found in regions not covered by complementary peptides, but to a much weaker extent, are aspartic acid () methionine (), and glutamine (). Taken together, these results suggest that our design strategy is general and provides multiple candidates to choose from for most target epitopes (SI Materials and Methods and Fig. S2).
Fig. 2.
Generality of the cascade method. (A–C) Coverage of α-synuclein (A), Aβ42 (B), and IAPP (C). For each residue in the sequence (x axis) we report the number of different complementary 8-residue peptides predicted to bind an epitope containing it. Peptides built from parallel β-strands are in blue and from antiparallel ones in green. The arrows on the top axis mark the positions of the peptides selected for experimental validation (Table 1). (D) Percentage of residues in the disordered regions of the δ2D database (37, 38) (Left) and of the DisProt database (36) (Right) covered by at least one complementary peptide. (E) Difference between the residue frequencies (y axis) observed in three classes of sequence regions within the two databases considered in D and those of the databases themselves. The classes are regions not covered by any complementary peptide (blue), by at least 1 complementary peptide (yellow) and by more than 10 complementary peptides (green).
Fig. S2.
Ranking the complementary peptides. (A) Variation of the complementary peptide coverage reported in Fig. 2D of the disordered regions in the DisProt database (blue) and the δ2D database (green) as a function of the complementarity score described in Ranking the Candidates. The dashed lines are the complementarity scores of the five peptides (Fig. S3) we grafted on the human sdAb scaffold for experimental validation (the one-loop DesAb variants). (B) Probability density function of the complementarity scores of all peptides covering the two databases; the red dashed line is the median value.
A Single Domain Antibody Scaffold for the Grafting of the Complementary Peptides.
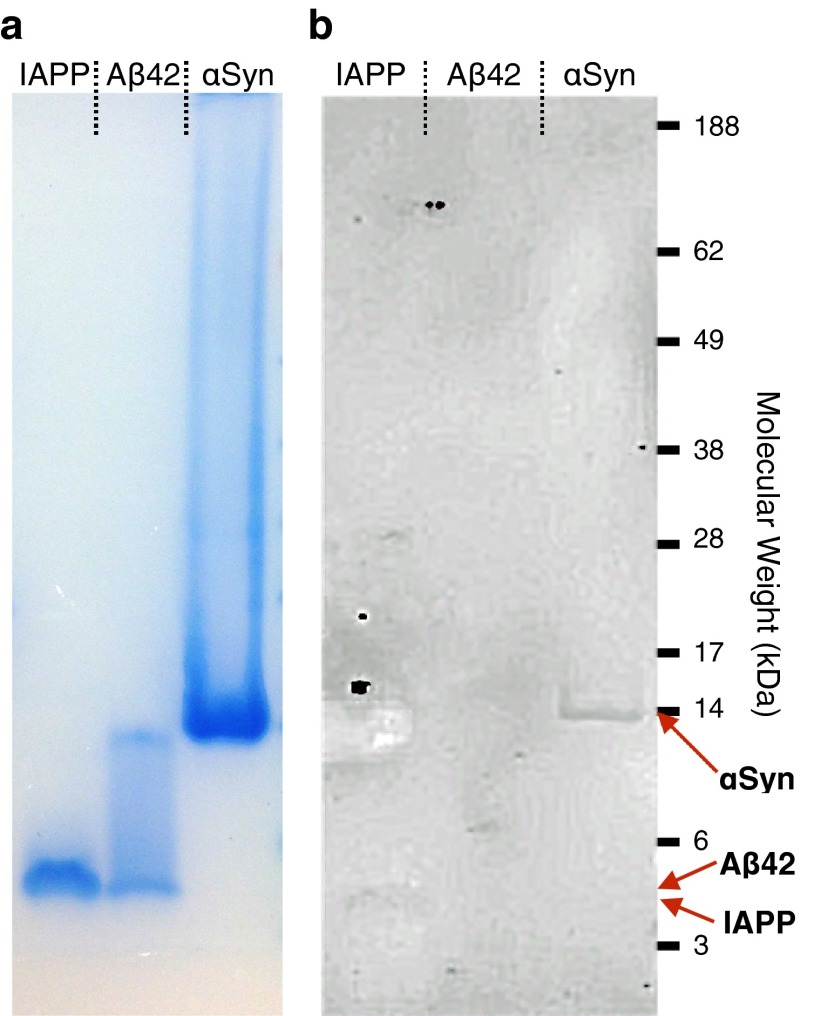
To assess the viability of the design method described above, we rationally designed antibodies targeting disordered proteins. First, we identified a stable antibody scaffold, tolerant to the grafting of peptide segments into one of the CDR loops. We selected a human heavy chain variable (VH) domain that is soluble and stable in the absence of a light chain partner, and whose folding is insensitive to mutations in its third CDR (CDR3) loop (39). Previous studies showed that this single domain antibody scaffold is relatively unaffected by insertions in its CDR3 (31). We found that this antibody is well expressed in bacteria (>5 mg/L), highly pure after a single chromatography step (>95% purity; SI Materials and Methods), and stable in its folded state (40).
Structural Integrity and Binding Capability of the Designed Antibody Variants.
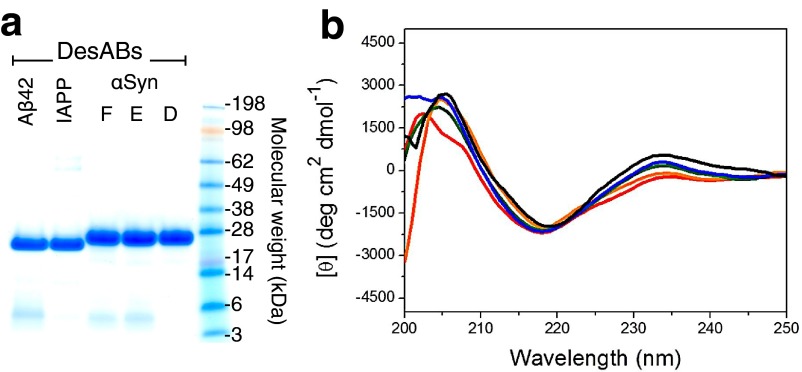
We designed complementary peptides for α-synuclein, Aβ42, and IAPP. The selected epitopes and the corresponding complementary peptides that we grafted in the CDR3 of the single domain antibody scaffolds (Fig. S3) are listed in Table 1. The purity of all of the designed antibodies (DesAb) was characterized by NuPAGE analysis (Fig. S4A) and their structural integrity by far-UV circular dichroism (CD) spectroscopy at 25 °C (SI Materials and Methods and Fig. S4B). All of the grafted variants showed high purity (>95%) and CD spectra compatible with the native-like structure of the single domain antibody scaffold. Therefore, we assessed the viability of the DesAb variants in binding their targets. To this end we used an ELISA test, which uses the basic immunology concept of an antigen binding to its specific antibody (41). We coated the wells with increasing amount of the designed antibodies, and then we incubated in the presence of a fixed amount of target protein (SI Materials and Methods and Fig. S5). All of the designed antibody variants showed a characteristic concentration-dependent curve, which is evidence of antibody–antigen binding (Fig. 3 A–C).
Fig. S3.

Sequence of the single domain antibody scaffold used in this work for all DesAb variants with one loop engineered. The different complementary peptides used in the CDR3 are listed in Table 1.
Table 1.
List of target proteins, target epitopes and their sequences, designed complementary peptides, and designed antibodies (DesAb) used in this work for experimental validation
| Target protein | Target epitope | Complementary peptide | DesAb |
| IAPP | 23FGAILSS29 | RLGVYQR | DesAb-IAPP |
| Aβ42 | 15QKLVFFA21 | FKLSVIT | DesAb-Aβ |
| α-Synuclein | 70VVTGVTA76 | FQEAVSG | DesAb-F |
| α-Synuclein | 61EQVTNVG67 | DILVSYQ | DesAb-D |
| α-Synuclein | 61EQNTNVG67 | EILVSYQ | DesAb-E |
| α-Synuclein | 65NVGGAVV | QEFVAAFSHTE | Two-loop DesAb |
| TGVTAVA79 | +EVFQEAVSGS |
Fig. S4.
(A) SDS/PAGE analysis on the different purified DesAb variants. (B) Far-UV CD structural characterization of the DesAb variants (DesAb-D in blue, DesAb-E in green, DesAb-F in red, DesAb-IAPP in orange, DesAb-Aβ in black).
Fig. S5.
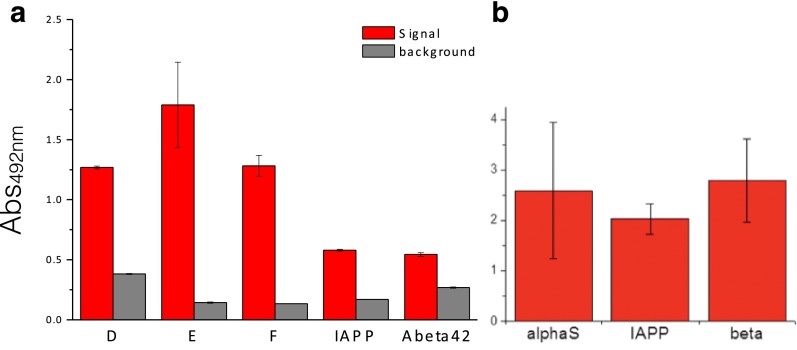
Control experiments for the ELISA tests. (A) Cross-reactivity of primary and secondary antibodies in the ELISA test. The bar plot represents the absorbance at 492 nm of ELISA wells coated with the DesAb variants (x axis; Table 1 and SI Materials and Methods) in the presence (red) and absence (gray) of the antigen protein of the DesAbs. The coated amounts were 10 μg for DesAb D, E, and F, 2.4 μg for DesAb-Aβ, and 7 μg for DesAb-IAPP. (B) Reactivity of the primary antibodies in the ELISA for their specific target.
Fig. 3.
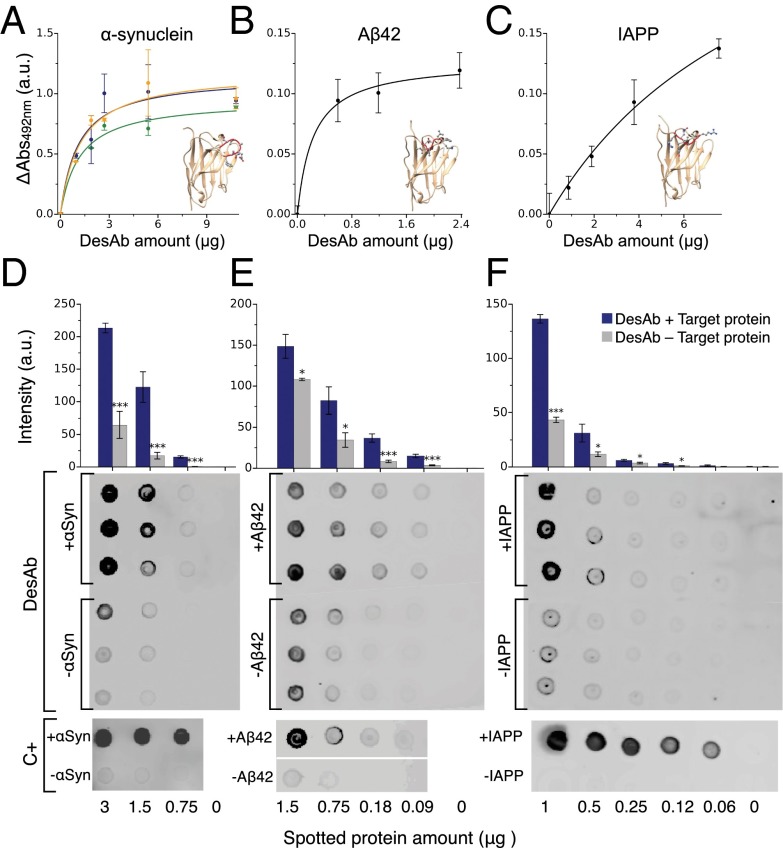
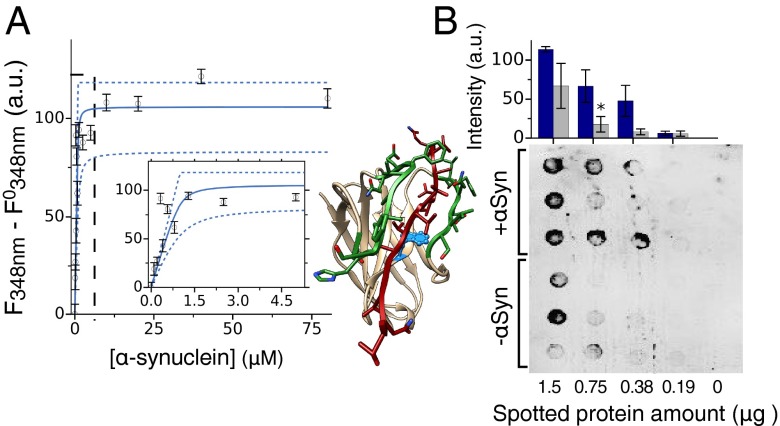
Binding and specificity of the designed antibodies (DesAb). (A–C) ELISA test of the DesAbs in Table 1 with one complementary peptide grafted in the CDR3 that specifically target α-synuclein (A) (DesAb-D in green, DesAb-E in blue, and DesAb-F in orange), Aβ42 (B) (DesAb-Aβ), and IAPP (C) (DesAb-IAPP); the lines are a guide for the eye. Homology models of the structures of the designed antibodies are represented with the grafted complementary peptide in red. (D–F) Dot blot assay performed with three DesAb variants: DesAb-F (D), DesAb-Aβ (E), and DesAb-IAPP (F) and three commercially available antibodies used as a positive control (C+) for the binding to E. coli lysates from cell lines expressing the target protein (dots labeled with +, blue columns) and not expressing it (−, gray column). In the case of DesAb-IAPP, synthetic amylin peptide was mixed to the E. coli lysate (+IAPP) before performing the experiment, as a cell line expressing IAPP was not available. Protein amount is the micrograms of total protein (lysate) spotted on the membrane. The bar plot is a quantification of the intensities of the DesAb dot blots (SI Materials and Methods). Intensities are *>2 away, ** > 3 , and *** > 4 , with and SE being the standard error from the intensities of the three dots.
Specificity of the Designed Antibodies.
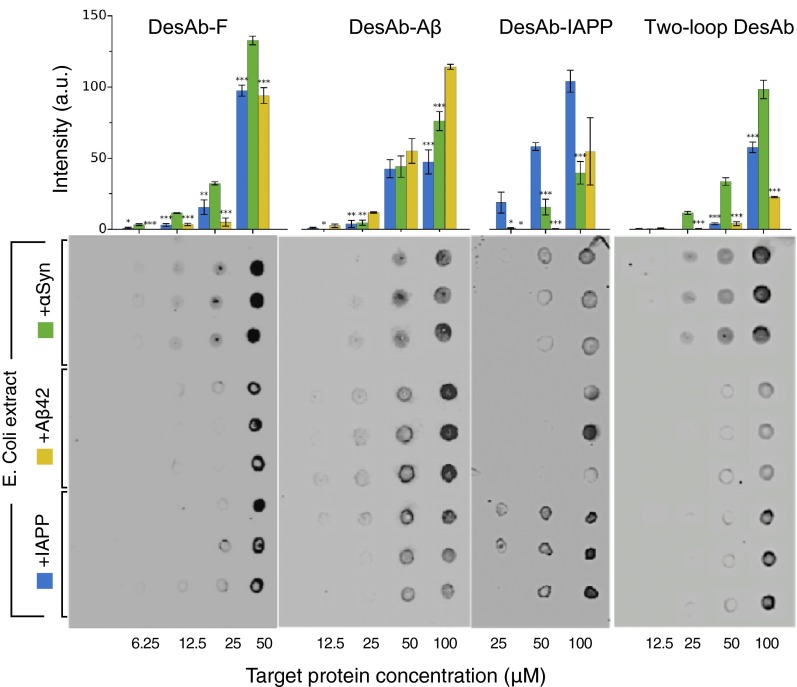
The specificity of the DesAbs was assessed with a dot blot test by spotting different amounts of proteins from Escherichia coli cell lysates on a nitrocellulose membrane (SI Materials and Methods). The binding of three DesAb variants (DesAb-F, DesAb-Aβ, and DesAb-IAPP; Table 1) to lysates from cell lines where the expression of the antigen protein had been induced was compared with that to lysates where the expression had not been induced (Fig. 3 D–F). Because an E. coli cell line expressing IAPP was not available, 100 μM of synthetic IAPP was mixed to the E. coli lysate (+IAPP) before performing the experiment with DesAb-IAPP. The total protein amount of the lysate without IAPP (−IAPP) was adjusted accordingly. The results show that for all tested DesAb variants the intensity of the dots corresponding to cell lysates containing the target protein is always significantly greater than that of dots from lysates not containing it. Moreover, a control experiment performed with commercially available antibodies (C+ in Fig. 3 D–F; SI Materials and Methods) suggests that for α-synuclein and Aβ42, there may be a degree of basal expression of the antigen protein even without induced expression. As an additional control, we tested the cross-reactivity of the DesAb variants by probing with each designed antibody blots prepared with E. coli lysate mixed with equal concentrations of α-synuclein, Aβ, and IAPP, respectively (SI Materials and Methods). A clear trend is observed in this case as well, whereby each DesAb preferentially binds to its target (Fig. S6).
Fig. S6.
Dot blot assay for the specificity of the designed antibodies. Dot blot assay for the binding (from left to right) of DesAb-F DesAb-Aβ, DesAb-IAPP, and two-loop DesAb to same concentrations (x axis) of purified αSyn (green bars, top three lines of dots), synthetic Aβ42 (yellow bars, middle three lines of dots), and IAPP (blue bars, lower three lines of dots) mixed to an E. coli lysate. The bar plot is a quantification of the intensities of the DesAb dot blots (SI Materials and Methods). Intensities are *>2 away, ** > 3 , and *** > 4 with , SE being the standard error from the intensities of the three dots.
Detailed Characterization of DesAb-F.
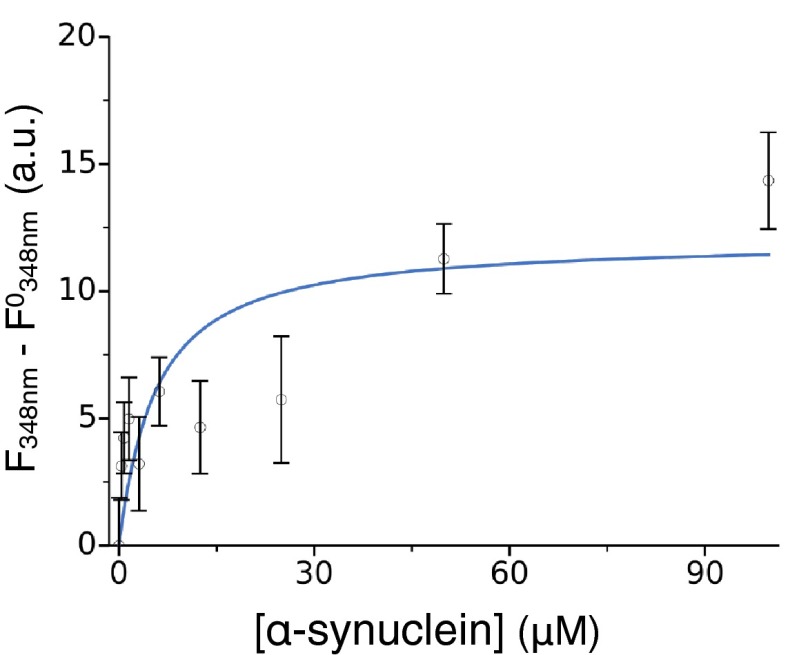
To obtain a more comprehensive characterization on the interaction of the designed antibody variants, we selected one (DesAb-F, with grafted sequence FQEAVSG; Table 1), for which we quantitatively assessed affinity, specificity, and effect on protein aggregation. To characterize the specificity of binding, in addition to the dot-blot test presented in Fig. 3 and Fig. S6, we quantified the reactivity against α-synuclein, Aβ42 peptide, and IAPP. Thus, we performed an ELISA in which we coated the wells of the ELISA plates with a given amount of DesAb-F and then we incubated in the presence of the same amount of the three different antigens (SI Materials and Methods). The amount of α-synuclein, Aβ42, and IAPP bound to DesAb-F was estimated measuring the absorbance at 492 nm after verifying that the primary antibodies exhibited similar reactivity against an equal amount of antigen absorbed to the ELISA well (Fig. S5). We found that DesAb-F clearly shows a preferential binding for α-synuclein than for Aβ42 peptide or IAPP (Fig. 4A). We then characterized in a more quantitative manner the binding constant of the antibody for monomeric α-synuclein. To do so, we assessed the ability of the antibody to bind a labeled variant of α-synuclein carrying the fluorophore dansyl (dansyl-α-synuclein) at position 90. Following a strategy already used for other systems (42, 43), the formation of the complex was studied by titrating increasing quantities of DesAb-F into solutions containing dansyl-α-synuclein and following the fluorescence properties of the dansyl moiety (Fig. 4B). The results of the titration experiments reveal that DesAb-F was able to bind α-synuclein with a Kd of 18 µM, derived assuming a single-site binding model (SI Methods). As the Kd value is highly sensitive to small displacements of the data points, we calculated the 95% CI on the fitting parameters with the bootstrap method (SI Materials and Methods), which placed the Kd between 11 and 27 µM. We note that this affinity, which is within a biologically relevant range but smaller than that of typical antibodies, has been reached by engineering only one loop of the antibody scaffold, whereas standard antibodies generally have more than two loops involved in antigen binding. Furthermore, a relatively high Kd can be effective in affecting protein aggregation (see below), because the antibody can actively interfere with the aggregation process rather than sequestering individual antigen monomers. Finally, to verify that DesAb-F binds specifically the chosen target epitope of α-synuclein, we generated one α-synuclein variant (α-synuclein-P73) with a proline residue inserted in the middle of the target epitope sequence (VVTGPVTA). The reason for this choice is that, if binding indeed occurs at this site, we expect such insertion to cause a significant inhibition of the interaction between the complementary peptide of DesAb-F and α-synuclein. Thus, we performed a florescence competition assay in the presence of 2 μM dansyl-α-synuclein and equimolar concentrations of nonlabeled α-synuclein WT or α-synuclein-P73 (SI Materials and Methods). In the presence of α-synuclein, the percentage of the complex DesAb-F:dansyl-α-synuclein decreased more than 50% in agreement with a competitive reversible inhibition (Fig. 4C). On the contrary, when the mutant variant α-synuclein-P73 was present in solution, no significant decrease was observed (Fig. 4C). The fact that α-synuclein-P73 was not able to compete with dansyl-α-synuclein for the binding to DesAb-F indicates that the proline insertion was able to disrupt the interaction between the designed antibody and α-synuclein, and, therefore, that the complementary peptide of DesAb-F is specifically binding to the region of α-synuclein containing the target epitope.
Fig. 4.
Comprehensive characterization of the designed antibody DesAb-F. (A) The binding of DesAb-F to its target α-synuclein is much stronger than that for Aβ42 and IAPP; in the ELISA, we report the increase in the Abs490nm in the three cases. (B) Fluorescence titration with dansylated α-synuclein in the presence of increasing concentrations of DesAb-F (following the red shift of λmax). The solid blue line represents the best fit (Kd = 18 μM) using a single-binding model, and the broken lines the 95% CI on the fitting parameters (Kd between 11 and 27 μM). (C) Fluorescence competition assay; the y axis report the fraction of complex dansyl-α-synuclein:DesAb-F in the absence (blue) and presence of nonlabeled α-synuclein (red) or α-synuclein-P73 (purple). In A and C, the statistical significance of the difference with the first column was assessed with a Welch's t test (*P < 0.05).
Antiaggregation Activity of DesAb-F.
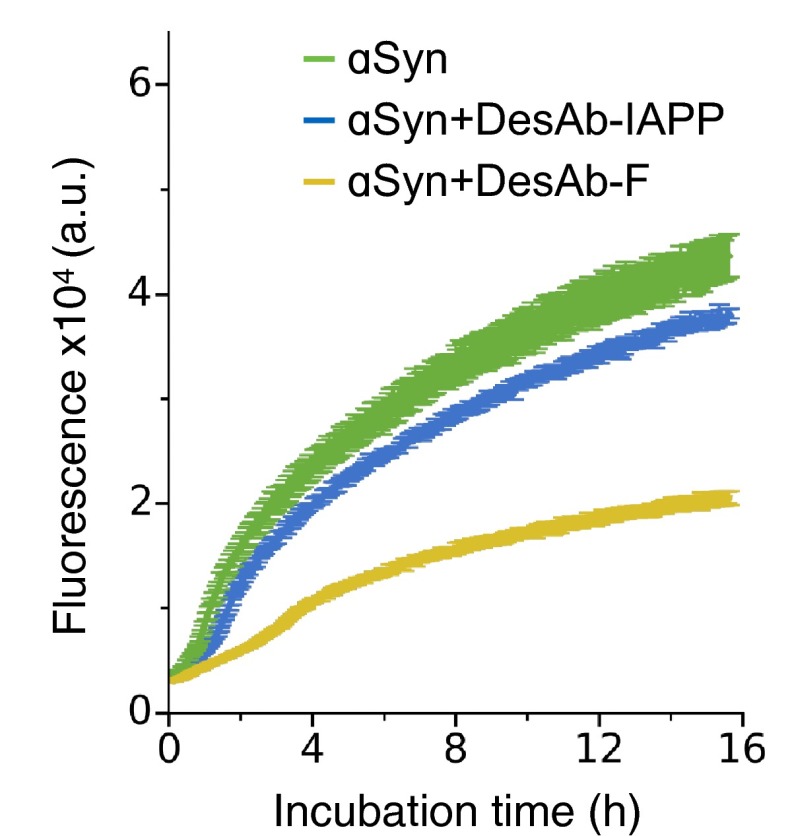
A general feature of amyloid-like aggregates is that they preferentially contain parallel β-sheet conformations (24), which, differently from β-sheets typically found within globular proteins, have one or more β-strands exposed to the solvent (i.e., the fibril elongation sites). Because the designed antibodies contain complementary peptides that enforce a β-strand conformation on their target sequence, we expect that the affinity toward target proteins should be higher when these are found in aggregated species rather than as free monomers in solution, as the entropic cost of binding should be smaller in this case. By monitoring soluble α-synuclein over four-day aggregation (SI Materials and Methods), we found that DesAb-F has a strong inhibitory effect, even at a substoichiometric concentration (1:10) (Fig. 5A). This result suggests that this designed antibody preferentially binds aggregated species rather than to monomeric forms of α-synuclein. To support this conclusion, we performed seeded aggregation assays at increasing concentrations of DesAb-F (SI Materials and Methods). We found a specific concentration-dependent effect of the antibody on the elongation phase of α-synuclein aggregation (Fig. 5 B and C), and we also detected a strong dependence on the concentration of α-synuclein seeds (Fig. 5D). Besides, the fact that DesAb-IAPP only shows a negligible effect on the aggregation of α-synuclein, even at a 1:2 DesAb-monomer ratio (Fig. S7), suggests that the observed inhibition specifically comes from the grafted complementary peptide. Taken together, these data show that DesAb-F is able to reduce α-synuclein aggregation.
Fig. 5.
The designed antibody DesAb-F inhibits α-synuclein aggregation. (A) Analysis on the soluble fraction of α-synuclein during its aggregation in the absence (black line) and presence (red line) of 1:10 molar ratio of DesAb-F:α-synuclein. (B) Seeded aggregation assay (3% seeds) at increasing molar ratios of DesAb-F (reported on the right axis). Different replicates for each condition are reported. (C) Initial growth rates over the molar ratios of single domain antibody scaffold. (D) Initial growth rate over the percentage of seeds in the presence of a fixed ratio of antibody (1:5).
Fig. S7.
α-Synuclein aggregation with a control DesAb. Seeded aggregation assay (3% seeds) of 70 μM α-synuclein alone (green) and in the presence of DesAb-F (yellow) and DesAb-IAPP (blue). The DesAb:α-synuclein monomer ratio is 1:2 for both DesAb variants. Error bars are SEs over three replicates.
Affinity Increase by Grafting Two Complementary Peptides.
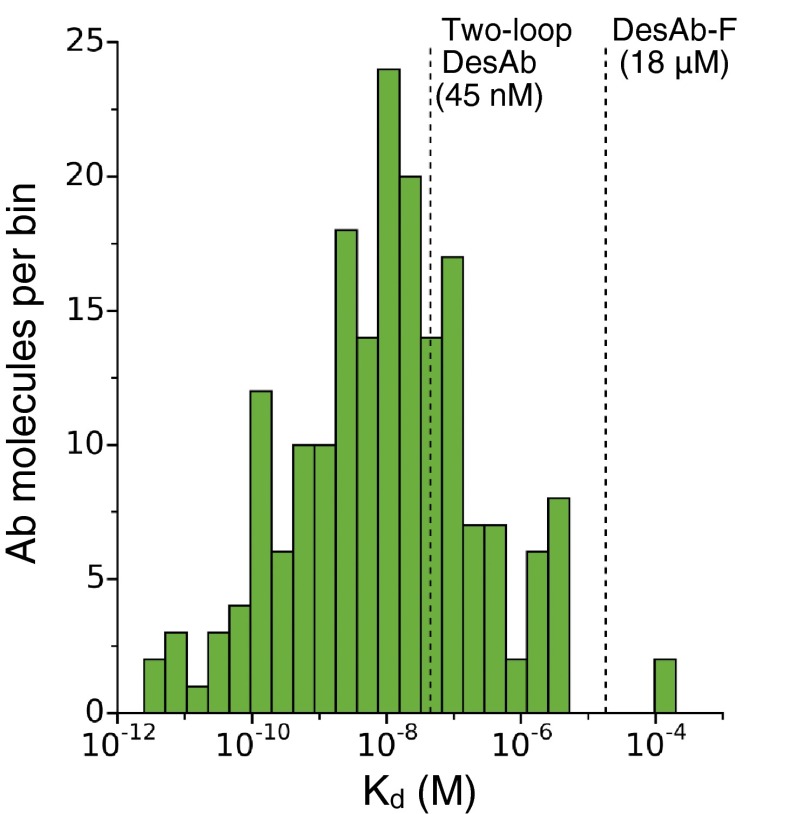
Although the affinity of DesAb-F for monomeric α-synuclein (Kd ∼ 20 μM) is probably ideal for inhibiting protein aggregation (see previous sections), it is still far from that of typical antibodies obtained with standard techniques. Because antibodies usually bind their antigens with more than one CDR loop, we decided to design an additional DesAb variant targeting α-synuclein with two loops engineered (two-loop DesAb in Table 1). These loops contain two complementary peptides predicted to cooperatively bind to the target epitope (SI Materials and Methods and Fig. S8). To add a second loop to our scaffold, we replaced 6 amino acids in the region of the CDR2 with 12 amino acids containing a complementary peptide (modeled structure in Fig. 6 and SI Materials and Methods). Thus, in the attempt of compensating for the impact on the domain stability, we changed the expression system to an E. coli strain that enables the formation of the intrachain disulphide bond (SI Materials and Methods), and we changed the purification protocol by eluting the protein with imidazole rather than at low pH. With this strategy, we were able to successfully purify the protein and confirm its structural integrity with far-UV CD (Fig. S8E). However, as we envisaged, this human single VH domain with two extended loops is quite unstable, and for instance, it starts to precipitate at about pH 6. An advantage of this construct is that the binding site is now located between the CDR3 and CDR2, which is in close vicinity of residue Trp-47 on the scaffold (Fig. 6). This feature allows the binding to be measured in a label-free way by monitoring the change in the intensity of the intrinsic fluorescence of the DesAb at 348 nm, with varying concentration of WT α-synuclein, which does not contain Trp residues (SI Materials and Methods). The titration curve in Fig. 6A is best fitted with a Kd of 45 nM, and the 95% CI analysis places its upper value at 185 nM. For comparison, we performed the same type of experiment with the one-loop DesAb-F. In this case, the change in fluorescence is much weaker, probably because the binding site is further away from the fluorescent Trp on the DesAb (Fig. S9). The fitting of the titration curve gives a Kd of 5 μM, in agreement with the more accurate dansyl fluorescence estimate (Fig. 4B). In addition, we assessed the specificity of the two-loop DesAb with a dot-blot experiment as performed for the one-loop DesAb variants (SI Materials and Methods). The preferential binding for the cell lysate containing α-synuclein is apparent at a qualitative level (Fig. 6B and Fig. S6). Finally, we successfully employed the two-loop DesAb in the Western blot detection of its antigen protein (SI Materials and Methods and Fig. S10). No signal, however, was observed when probing the Western blot with the one-loop DesAb variants, probably because of the relatively low affinity of these variants and the fact that in a SDS/PAGE, the monomers preferentially populate elongated conformations, which may further weaken the interactions with the grafted complementary peptide. Because of its instability, the two-loop DesAb cannot be considered a viable antibody for most applications, but it represents a proof of principle that it is possible to greatly improve the affinity (by two or three orders of magnitude) in a rational way using our design method, by engineering two binding loops.
Fig. S8.

Design of the two-loop DesAb variant. (A–D) structures of three nanobodies (A–C) and the HEL4 human single domain antibody (D) used for the design of the two-loop construct; loops containing the CDR3 and CDR2 are colored in dark gray, and disulphide bridges stabilizing the CDR3 are highlighted in cyan when present. (A) Structure of a nanobody (1RI8) with the ligand that binds between the CDR3 and the CDR2 shown in transparent orange. (B) Structure of a nanobody (2X6M) bound to the C-terminal peptide of α-synuclein (orange), Lys105 and Asn52 are colored in magenta, and their hydrogen bond network with the backbone of the CDR3 loop is drawn (formed hydrogen bonds are in dark blue, possible ones in light blue). (C) Structure of a nanobody (4KRP) showing Asn52 in magenta, Ala33 in yellow and Tyr122-Asp123-Tyr124 on the stem of the CDR3 in green. (E) Structural integrity of the two-loop DesAb assessed with far-UV CD. (F) Alignment of the four template sequences with the two-loop DesAb sequence (named 2Loops) with the grafted CDR sequences underlined and the complementary peptides colored in green; the sequence of the one-loop DesAb scaffold is also reported (1Loop), and the residues in the two-loop DesAb sequence that differ from those in the one-loop DesAb outside the CDR loops are highlighted in yellow. These residues are also those colored in the corresponding template structures (A–C) with the exception of Glu130, which was selected from ref. 40. (G) Representation of the pincer-like binding to α-synuclein of the complementary peptides grafted in the CDR2 and CDR3 loops of the two-loop DesAb construct; equal signs mark residues predicted to be involved in backbone-backbone hydrogen bonding and arrows denote the N to C terminus direction. (H) Results from the cascade method for the two peptides grafted in the two-loop DesAb scaffold; grafted sequences are shown in bold.
Fig. 6.
Binding and specificity of the 2-loop DesAb. (A) Intrinsic fluorescence (Trp) titration assay performed at a constant concentration of two-loop DesAb (1 μM) and increasing concentration of α-synuclein (x axis). The solid blue line represents the best fit (Kd = 45 nM) and the broken lines the 95% CI on the fitting parameters (Kd up to 185 nM). (Inset) Zoom of the region highlighted by the dashed-black line. (B) Dot blot assay for the binding of the two-loop DesAb variant to an E. coli lysate from a cell line expressing α-synuclein (top three rows, blue column) and not expressing it (bottom three rows, gray columns). The structure shown is a coarse-grained model to illustrate the concept of two loops grafted into the sdAb scaffold, the complementary peptides are in green and the α-synuclein epitope (residues 64–80) in red, and Trp-47 is shown in light blue.
Fig. S9.
DesAb-F intrinsic fluorescence. Intrinsic fluorescence (Trp) titration assay performed at a constant concentration of DesAb-F (1 μM) and increasing concentration of α-synuclein (x axis). The solid blue line represents the best fit (Kd = 5 μM).
Fig. S10.
Western blot analysis of the reactivity of the two-loop DesAb for α-synuclein, Aβ42, and IAPP. SDS/PAGE (A) and corresponding Western blot (B) of samples at the same concentration (200 μM) of α-synuclein (αSyn), Aβ42, and IAPP probed with two-loop DesAb as a primary antibody. Different intensities in A are caused by differences in protein sizes.
SI Materials and Methods
Creation of the β-Strand Database.
To create the β-strand (BS) database, which is used to design the complementary peptides, we first downloaded from the PDB database all protein structures with a 90% sequence identity threshold. This threshold was used to avoid double counting resulting from homomers, close homologs, or multiple structures of the same protein. We thus obtained the PDB90 database. We then used the DSSP program (44) to identify the β-sheet regions in each protein structure in the PDB90 database. These regions were used to generate the BS database, which consists of all of the β-strands in the PDB90 database. More specifically, the BS database contains pairs of amino acid sequence fragments that are found facing each other in β-strand conformations (β-pairs). Only fragments of three residues or more were considered, and β-pairs were separated into parallel and antiparallel. In addition, we also associated to each β-pair the hydrogen-bonding pattern formed by the two backbone chains, as this is particularly important when merging two fragments.
Target-Specific Scoring of the β-Strands in the BS Database.
We assigned a score (the count score) to every β-pair in the BS database on the basis of the number of times the two fragments are found facing each other in a β-strand with the same hydrogen-bonding pattern. If different hydrogen-bonding patterns were found for the same pair, the two β-strands were saved as two different β-pairs in the BS database with two different scores. To simplify the construction of the complementary peptides, instead of using the BS database directly, we subdivided it into separate libraries of fragments of fixed lengths (the BSn libraries). We considered fragments of length n, with n ranging from 3 to 8. For each n, the BSn libraries was constructed by extracting from the BS database all possible β-pairs of length n, including subfragments belonging to β-pairs of length m > n. The count score of the complementary β-strand partners is updated accordingly so that it always reflects the number of times that the two fragments of length n are found facing each other in β-strands forming the same hydrogen-bonding pattern in the structures that we initially analyzed. For every length n, we built two separate BSn libraries, one with the parallel complementary stretches and one with the antiparallel ones. Next, for every β-pair in the BSn libraries we computed the promiscuity score, to quantify the specificity of the interaction between the two fragments in the β-pair. Such a score is understood by considering the definition of specificity: two binding partners interact specifically if they interact only with one another and not with anyone else. The greater the number of possible binding partners, the lower is the specificity. As a consequence, we looked at the number of β-strand partners of each fragment in the libraries. This number is readily extracted from the BSn libraries themselves, by counting the number of β-pairs where the fragment appears. Then, to correctly assign the promiscuity score to each β-pair, one must avoid counting the partner corresponding to the β-pair under scrutiny and should also count only β-strand partners of the right kind (parallel or antiparallel) and with the same hydrogen bond pattern of the β-pair under scrutiny.
The Cascade Method of Constructing Complementary Peptides.
For a target sequence of N amino acids, we start by considering the longest length n ≤ N available in the BSn library. We then slide a window of size n on the target sequence, storing the β-strand partners facing the sequence window under scrutiny, together with their scores and hydrogen-bonding patterns. We repeat this procedure for every length n (down to 3 amino acids) so that we end up with a list of all possible complementary fragments of different length. This list is composed by groups of β-strand partners of the same length, and, within each group, we sort them first according to decreasing count score and then to increasing promiscuity score. In this way, fragments that are found more times as β-strand partners of the corresponding bit of the target sequence appear first, and—within fragments with the same count score—low promiscuity score fragments appear first.
Starting from the top, we select the β-strand partner with longest length and highest count score, which we refer to as cascade initializer. There are now three important aspects that are taken into account. The first is whether this fragment was part of a parallel or of an antiparallel β-sheet, because we consider only fragments of the same type until we initiate a new cascade (to look for another complementary peptide candidate). The second is the position of the initializer with respect to the target sequence. If it is a β-strand partner of the beginning of the sequence, we only have to grow the peptide to the right, and if of the end only to the left, and otherwise in both directions. The third is its hydrogen-bonding pattern, as this must be conserved while we grow the peptide.
To better understand the cascade procedure, let’s consider an initializer C1 and assume that it is a β-strand partner of the region of the target sequence that goes from position i to position j < N. In the following we consider the case in which we grow the complementary peptide to the right, hence for increasing values of j. C1 was selected at the top of the list, which is sorted according to the lengths and scores of the β-strand partners, and now, with position j fixed, we go down the list until we find another β-strand partner that meets three criteria. First, it has to be of the same type (parallel or antiparallel) as C1. Second, it must be a β-strand partner of a bit of the target sequence that includes at least position j and j + 1. This criterion creates an overlap between this new fragment and C1, i.e., there is a part of the target sequence (which could be only residue j) that is a β-partner of both a region in C1 and a region in the new fragment. The third criterion is that these two regions on the two fragments are identical, both in sequence and in hydrogen-bonding pattern. When the three criteria are all met, the new fragment is merged with C1 to generate a longer complementary candidate C2, which covers the target sequence from position i to position k > j. A new cascade step is carried out until k = N, and the same is done toward the left until i = 0.
At every step after the first one, we begin by looking at new fragments from the top of the list so that we always use the longest/highest-count-score fragment. Moreover, it could be that there are multiple fragments that meet the three criteria and also have the same length and count score of the one we are looking at. In this case, we spilt the cascade and we generate multiple candidates. Then we use them all to grow different complementary peptides. This fact is true also when we select the initializer, as there could exist multiple fragments of maximum length and count score that are β-strand partner of different bits (or of the same bit) of the target sequence. In this case we initiate multiple cascades that will generate different complementary peptide candidates.
At every step of the cascade there could be more than one suitable candidate that results in a forking of the cascade path and in the generation of different complementary peptides. However, the opposite can also be true: the convergence of the cascade—i.e., the fact that we can reach i = 0 and k = N—is not always granted. In other words, it is not always the case that there exists a complementary fragment that will meet all there criteria at every cascade step. Generally, we simply discard those paths that get stuck and we only keep the ones that allow for the generation of complementary peptides of length N. Nevertheless, if no such path exists, the algorithm looks for different, lower score or shorter length, initializers, until it finds some complete complementary peptide candidate.
Fig. S1 shows the cascade paths that generated the complementary peptides we grafted on the single domain antibody scaffold.
Ranking the Candidates.
The cascade method usually generates more than one complementary peptide for a given target sequence. A choice therefore needs to be made for the best candidate at binding the target sequence with high affinity and specificity. For this choice we exploit the fact that our fragment database provides also useful statistics to rank the candidates. Each of the complementary peptides has been built combining fragments from the BSn libraries with the cascade method. We thus know the length score, the count score, and the promiscuity score of each of the fragments that compose the complementary peptide (Fig. S1).
We regard the count score as a measure of affinity: if nature has used many times the same pair of fragments as β-strand partners in native proteins, it means that they delivered optimal stability to the structure and speed to the folding of the protein. The length score is defined as the number of residues that make up the complementary fragment. Because chances that a long fragment has a large number of possible β-strand partners are very low, we regard this as a measure of specificity, and this is why we begin each step of the cascade by looking at the longest possible fragments. Finally the promiscuity score is a more quantitative indicator of the specificity than the length of the fragment alone.
To rigorously rank the complementary candidates these different values (there are three scores for each fragment that compose the complementary peptide) should be combined in an overall complementarity score. However, there are many possible way to do this and, in the absence of a large number of experimentally measured binding constants and specificity assays to help us guide the choice, we preferred to report the values for every single fragment in each complementary peptide (Fig. S1), favoring the candidates that are made by longer fragments, with higher count score and lower promiscuity score.
Nevertheless, to assess how the coverage results reported in Fig. 2 change when putting a threshold on the quality (i.e., a complementarity score) of the complementary peptides we used a working score defined as , where n is the number of fragments building the complementary peptide, l the length, c the count score, and p the promiscuity score. This choice of C is arbitrary and it simply reflects our rule of favoring fragments of larger length, then of larger count score, and within those of same length and count score of smaller promiscuity score. Fig. S2 reports the coverage of the two database of disordered regions as a function of a threshold on the complementarity score C.
Finally, there exists another important aspect when choosing among different complementary candidates for a target sequence, which is the amino acid composition of the peptide itself. Given the fact that the fragments that make up the complementary peptides are found in β-strands, they tend to be enriched in hydrophobic amino acids and are in general poorly soluble. Thus, some complementary peptide will show a tendency to aggregate and possibly interact nonspecifically with other hydrophobic stretches in different proteins. To control for the solubility of the complementary peptide candidates, we run the sequence-based solubility calculation of CamSol method (40), and we favor those peptides predicted to be more soluble. We also note that the VH scaffold contains a solubility tag consisting in a triple Glu inserted next to the grafting site in the CDR3 (Fig. S3), so that the solubility of the grafted antibody is effectively greater than that of the peptide alone. The effect of the triple Glu insertion was previously assessed (40), where it was shown to act as a powerful gatekeeper that significantly increased the solubility of the VH domain.
The δ2D Database of Intrinsic Disorder.
The δ2D database was previously described (38) and contains protein sequences with assigned NMR chemical shifts measured at about physiological conditions and in the absence of compounds that may affect the solution behavior of the protein (e.g., micelles, denaturants, TFE,…). The chemical shifts were then used to calculate the corresponding secondary structure populations using the δ2D method (37). In the present work a region is defined disordered if 20 or more consecutive amino acids satisfy , where is the secondary structure population of α-helices and of β-strands.
Blasting of Short Peptides on the Human Proteome.
The blast search for the complementary peptides was carried out with the pblast program within the BLAST+ package (45) against the human reference proteome as downloaded from the UniProt website. Given the short size of the peptides (eight amino acids), the blast search was carried out with the command “blastp –query peptide_sequence -db human_proteome -task ‘blastp-short’ -word_size 2 -seg ‘no’ -evalue 20000” to maximize the number of hits.
Design of the Two-Loop DesAb.
Given the fact that natural antibodies generally bind to their antigens by using more than one CDR loop, we investigated whether it was possible to increase the affinity of our one-loop DesAb variants by grafting a second complementary peptide on another binding loop. This second complementary peptide is engineered to bind the target epitope cooperatively with the first complementary peptide.
This strategy, however, presents a number of challenges, especially in the case of the human single domain antibody scaffold that we use. First, both the CDR2 and the CDR1 in this scaffold are too short to accommodate a solvent-exposed complementary peptide and thus one needs to be extended with unavoidable consequences on the stability of the construct. Second, the two complementary peptides must carefully be placed in the loops, so that their cooperative binding to the target epitope is geometrically favored and does not require major conformational changes. These aspects also mean that the two peptides must be designed to bind to essentially the same epitope sequence (because the scaffold is small) without competing with each other for the binding. This result can be accomplished by selecting complementary peptides predicted to bind at either side of the epitope sequence, sandwiching it in a pincer-like way (Fig. S8G and Fig. 6).
One advantage of the human VH single domain that we used is that its stability is essentially independent from the CDR3 sequence (39), which can also be greatly extended; it was for example reported to accommodate a staggering 16-residue insertion (46). On the contrary, however, the sequence of the CDR1 has been shown to be crucial for the scaffold stability and refolding ability (47), which leaves only the relatively short CDR2 as a possible grafting site.
Thus, three results need to be accomplished: (i) to optimize the CDR3 and CDR2 geometry so that the two complementary peptides are allowed to face each other to promote a pincer-like binding, and the antigen epitope can fit between the loops without requiring large conformational changes, (ii) to extend the CDR2 to accommodate a fairly mobile, solvent-exposed complementary peptide, and (iii) to select complementary peptide sequences targeting appropriate regions of the target protein with the appropriate hydrogen-bonding pattern so that the two grafted peptides do not compete with each other for the binding to the antigen (Fig. S8G).
To accomplish i, we used a structure-based design strategy. By examining the structures of three camelid VH antibodies (1RI8, 2X6M, and 4KRP) we noticed that the backbone of the CDR3 loop runs parallel to that of the CDR2 loop for about five residues (Fig. S8 A–C). This feature is ideal to accomplish i, but it does not seem to be present in the crystal structure of the HEL4 human VH single domain (which essentially is our scaffold; Fig. S8 D and F). Its absence may be the sole consequence of the shorter CDR3 of the HEL4 construct, yet the fact that camelid CDR3 contributes to stability by packing against the framework (48) suggests that this could be a feature specific to camelid VH domains.
We thus sought to implement this feature in our human sdAb scaffold. The CDR3 loops of the nanobodies in Fig. S8 A and B appear to be stabilized by a disulphide bond with the scaffold (cyan), but this is not the case for 4KRP in Fig. S8C. Consequently we analyzed the interactions of the CDR3 of 4KRP with the scaffold. We found that the β-bridge between the two stems of the loop (dark green in Fig. S8C) is more twisted than that of the HEL4 crystal structure (Fig. S8D) and that Asn52 in the scaffold (magenta in Fig. S8C), which is not present in HEL4, forms a hydrogen bond with the backbone of the CDR3, possibly stabilizing its conformation. Other interactions involving the side-chains of the CDR3 were neglected in this analysis because we will replace the loop with a complementary peptide. Consequently, we decided to modify our scaffold adding the C terminus stem of the loop of 4KRP (Tyr-Asp-Tyr at position 122 in Fig. S8F) and Asn52; the latter is present in 2X6M as well, where it forms a similar hydrogen bond with the CDR3. From the structure 4KRP we also performed the mutation Y33A, as this is the position that forms the disulphide bond in 1RI8 and 2X6M, and a Tyr residue there may cause a displacement of the CDR3 loop because of its larger side-chain. Moreover, the same type of analysis performed on the structure 2X6M revealed a Lys residue at position 105 (Fig. S8 B and F) that forms a hydrogen bond with the backbone of the CDR3. As the corresponding CDR3 region in our scaffold consists in three repeated Glu residues, we hypothesized that this insertion, in addition to a potential hydrogen bond, would produce an electrostatic attraction between the stems of the loop that may benefit the stability of the domain. Finally, we also performed the mutation L130E, because it was shown to increase the solubility (40).
Then, to accomplish ii, we performed homology modeling of a number of combinations of different lengths of the two CDR loops and different complementary regions on the selected α-synuclein epitope (residues 64–80; Fig. S8E), using the scaffold sequence labeled as 2Loops in Fig. S8F, and the four crystal structures as templates (Fig. S8 A–D). Homology modeling was performed as described previously (40), forcing the binding to the selected α-synuclein epitope by imposing distance restraints between the backbone atoms N and O of the CDRs and those of the epitope (Fig. 6). The best combination of CDR2/CDR3 lengths was selected with the PROCHECK-NMR program (49), as the one of the model that scored lower in equivalent resolution and in number of residues in disallowed regions of the Ramachandran maps. This procedure not only provided us with a guess at the length of the loops to be grafted, but also with the ideal offset between the two complementary peptides on the α-synuclein epitope. The offset is better understood by looking at Fig. S8G: we defined it as the number of residues of α-synuclein that separate the first residue complementary to the CDR2 peptide from the first residue complementary to the CDR3 peptide. The analysis yielded an offset of 3 residues with a CDR3 loop that is four amino acids longer than that of the one-loop DesAb variants (Fig. S8F) and a CDR2 of 12 residues, obtained by replacing 6 existing residues and effectively adding 6 (Fig. S8F).
Finally, to accomplish iii, we ran the cascade method on the α-synuclein sequences in Fig. S8H, selecting two candidates with the appropriate hydrogen-bonding pattern (i.e., that of Fig. S8G). Because most likely the termini regions of both complementary peptides will not be able to bind to their target (as it would require an excessive stretching of the loops), we selected the complementary fragments in these regions (Fig. S8H) according to their contribution to the solubility, as assessed with the CamSol method (40) (e.g., presence of negatively charged residues), rather than on their count and promiscuity scores.
Given the overall approach that we took, we do not expect our homology models to be very accurate. Nevertheless, probably aided by the structure-based optimization of the scaffold described above (point i), we were able to confirm the structural integrity of the purified two-loop DesAb variant with far UV circular dichroism (Fig. S8E).
Protein Preparation.
The different complementary peptides were grafted into the CDR3 loop of the single domain antibody scaffold by means of mutagenic PCR with phosphorylated oligonucleotides. The different DesAbs were then expressed in fusion with a N-terminal PelB leader sequence from pET17b in E. coli BL21 (DE3)-pLysS strain (Agilent Technologies) and purified from the growth medium by nickel-affinity chromatography at pH 3 (40).
The two-loop DesAb construct was expressed and purified from pRSET-b vector in E. coli Rosetta Gami 2 (DE3) (Merck Millipore). Cells were grown for 15 h at 30 °C using Overnight Express Instant TB Medium (Merck Millipore) supplemented with ampicillin (100 g/mL). Cells were then harvested by centrifugation, resuspended in PBS and one EDTA-Free Complete Protease Inhibitor Mixture tablet (Roche), and lysed using sonication; cell debris was removed using centrifugation at 15,000 rpm (JA-25.50 Rotor; Beckman Coulter). The cleared lysate was loaded onto a Ni2+-NTA Superflow column (Qiagen), previously equilibrated with PBS containing 10 mM imidazole. After washing with PBS containing 20 mM imidazole, the His-tagged two-loop DesAb was eluted with PBS containing 200 mM imidazole. The protein was then dialyzed extensively against PBS. For all these protein variants, protein concentration was determined by absorbance measurement at 280 nm, using theoretical extinction coefficients calculated with Expasy ProtParam (50).
Human WT α-synuclein (gi:80475099) and the mutants α-synuclein-P73 and A90C were purified as early reported (51). The mutations into α-synuclein sequence were introduced by mutagenic PCR with the QuikChange XLII kit (Qiagen). Protein concentration was determined by absorbance measurement at 275 nm using an extinction coefficient of 5,600 M−1⋅cm−1. Protein purity always exceeded 98% as determined by SDS/PAGE analysis. Aβ42 (Cambridge Biosciences) and IAPP (LKT Laboratories) peptides were chemical synthetized.
CD.
Far-UV CD spectra for all protein variants were recorded using a Jasco J-810 spectropolarimeter equipped with a Peltier holder, using a 0.1-cm-pathlength cuvette. Typically, samples contained 10 μM protein in PBS. The far-UV CD spectra of all of the variants were recorded from 200 to 250 nm at 25 °C, and the spectrum of the buffer was systematically subtracted from the spectra of all protein samples.
ELISA Test.
The wells of the ELISA plates (Thermo Scientific) were coated (at 37 °C for 1 h under constant shaking) with increasing amounts (from 1 to 10 µg) of purified DesAbs and blocked with 5% (wt/vol) BSA in PBS (BSA/PBS). The coating was verified to be higher than 60% and homogeneous for all DesAb amounts; 60-µL samples of 2.5 µM ligand protein (i.e., α-synuclein, Aβ42, and IAPP) were then incubated in the coated wells. Primary antibodies reactive with the client proteins and the appropriate horseradish peroxidase-conjugated secondary antibody were diluted in BSA/PBS and added to the wells following the manufacturer's instructions. All incubations were performed at room temperature for 1 h under shaking and followed by six consecutive washes with PBS. After two final washes with BSA/PBS and PBS, ortho-phenylenediamine at 2.5 mg/mL in 50 mM citric acid and 100 mM Na2HPO4, pH 5, was added. The absorbance at 492 nm (Abs492nm) was measured with a CLARIOstar platereader (BMG Labtech). Nonspecific binding of primary and secondary antibody to the different DesAbs was assessed (Fig. S5A). The primary antibodies used were rabbit monoclonal anti-α-synuclein N terminus antibody (clone EP1646Y; Merck Millipore), mouse monoclonal anti-amyloid β clone W0-2 antibody (Merck Millipore), and monoclonal mouse anti-human IAPP (R10/99) antibody (LifeSpan BioSciences).
Dot-Blot and Western Blot Analyses.
Different amounts of crude extracts from E. coli BL21 (DE3) gold strain (Agilent Technologies) expressing α-synuclein or Aβ42 were spotted (2.5 μL) on a Immobilon-P membrane (GE Healthcare). As negative controls, the same amounts from crude extracts from the same cells in which target protein expression was not induced were used; 100 μM of synthetic peptide was mixed with the E. coli lysate. The total protein amount of the lysate without IAPP was adjusted accordingly. As a further control of specificity (Fig. S6), to verify the reactivity of each of the designed antibodies for the three target substrates (α-synuclein, Aβ42, and IAPP) in a crowded environment, different amounts of solutions containing 100 μM of pure α-synuclein, Aβ42, and IAPPin 50% (vol/vol) DMSO were mixed with 3 μg of an E. coli protein lysate and were spotted as described above. The blots were blocked 1 h (BSA/PBS) and probed overnight at 4 °C with each antibody (at a concentration between 4 and 7 μM). Blots with bound DesAbs were then probed with 6x-His Epitope Tag Antibody (1:5,000 dilution; Life Technologies). Positive controls (C+) were performed with a mouse monoclonal anti-αSyn antibody (Transduction Laboratories), mouse monoclonal anti-amyloid β clone W0-2 antibody (Merck Millipore), and mouse monoclonal anti-human IAPP (R10/99) antibody (LifeSpan BioSciences). All blots were probed with goat anti-mouse IgG (H+L) secondary antibody and Alexa Fluor 488 conjugate (1:5,000 dilution; Life Technologies). Dot-blot signals were quantified by densitometry analysis using ImageJ software (National Institutes of Health).
For the Western blot analyses, 20-μL samples of 200 μM of protein concentration of pure α-synuclein, Aβ42, and IAPP, supplemented with 50% DMSO, were loaded on 4–12% Bis-Tris NuPAGE gels (Life Technologies). Proteins were then transferred on a PVDF membrane using the iBlot Dry Blotting System (Life Technologies). The blots were probed using the same protocol applied for the dot-blots.
Labeling Reaction.
The A90C α-synuclein variant was labeled with DANSYL-MTS (Toronto Research) via the cysteine thiol moiety. The protein was incubated in the presence of 5 molar equivalent excess of the dye in PBS for 3 h at room temperature in the dark. The labeled protein was then purified from the excess of free dye by Zeba Desalt Spin Columns (Thermo Scientific) and further size exclusion chromatography step using a Superdex 75 10/300 GL (GE Healthcare). The labeling efficiency was more than 70% as estimated by MS. The labeled protein concentration was estimated by absorbance measurement at 335 nm using the extinction coefficient of the free dye (4,100 M−1⋅cm−1) (52).
Kd Determination Through Fluorescence Titration.
Dissociation constants (Kd) for the DesAb-F and two-loop DesAb variants were determined with fluorescence titration experiments by fitting the equation
where y is the fluorescence observable (y = y[x] − y0 where y0 is the value at [x] = 0), [F] is the concentration of the fluorescent protein, which is held constant during the titration, and [x] is the concentration of the protein that is titrated. ymax is the other fitting parameter and it equals the plateau. The 95% CI on the fitting parameters was calculated with 105 bootstrap cycles of data resampling, a technique that does not assume independent and identically distributed residuals.
Fluorescence Titration with Monomeric Dansyl-α-Synuclein.
In the case of DesAb-F fluorescence titration experiments were carried out by incubating 2 μM dansyl-α-synuclein in PBS for 30 min at 25 °C in the presence of different concentrations of the designed antibody. Fluorescence emissions spectra from 400 to 630 nm were recorded as the average of 10 spectra, following an excitation at 330 nm (42). Shifts of the maximum emission wavelength were plotted as a function of the antibody concentration and analyzed assuming a one site binding model (see above), where y is the red shift of the fluorescence emission spectrum of dansyl-α-synuclein at a given concentration of DesAb (λx − λ0, where λx is the maximum wavelength at the given concentration [x] of DesAb and λ0 is the maximum wavelength in the absence of DesAb, i.e., 547.5 nm). Fluorescence competition assays were performed in solutions of 200 μM DesAb-F samples in PBS in the presence of 2 μM dansyl-α-synuclein and equimolar concentrations of nonlabeled α-synuclein or α-synuclein-P73. The percentage of the complex dansyl-α-synuclein:DeAb-F formed was derived assuming a competitive reversible inhibition.
Intrinsic Trp Fluorescence Spectroscopy.
The interaction between the DesAb-F and two-loop DesAb with α-synuclein was studied in PBS using intrinsic (Trp) fluorescence spectroscopy by titrating different concentrations of α-synuclein into a solution containing 1 µM DesAb after incubating the mixtures for 1 h at 25 °C. Steady-state Trp fluorescence emission spectra were recorded from 300 to 400 nm, selectively exciting tryptophan residues at 295 nm, using a Cary Eclipse spectrofluorimeter (Varian). Excitation and emission band passes were set to 5 nm each.
Aggregation Assays.
α-Synuclein (70 μM) alone or together with 7 μM DesAb (i.e., 1:10 molar ratio) was incubated in 600 μL PBS (with 0.01% NaN3 to prevent bacterial growth) at 37 °C under constant shaking at 200 rpm in a Innova 4330 shaker (New Brunswick Scientific). At specific incubation times, 10-μL aliquots were centrifuged at 16,000 × g, and the supernatant was analyzed using 4–12% Bis-Tris NuPAGE gels (Life Technologies). Protein amount was estimated by densitometry using ImageJ software. Aggregation data are represented as insoluble protein fraction, normalizing them with the value corresponding to initial soluble protein amount and plotting reciprocal values.
Seeded Aggregation Experiments.
α-Synuclein fibrils were obtained as described above. Fibrils were then centrifuged for 15 min at 16,000 × g in a bench centrifuge and resuspended in the same volume of aggregation buffer to remove any monomer in solution; 70 μM monomeric α-synuclein alone or with increasing concentrations of DesAb-F was then incubated in the presence of different amount of preformed fibrils (if not differently specified) and 20 μM ThT at 37 °C in quiescence. The ThT fluorescence of the samples was continuously monitored using a FLUOstar OPTIMA plate reader (BMG Labtech Ltd.) with excitation and emission wavelengths of 440 and 480 nm (slit widths of 10 nm), respectively.
Conclusions
In this work, we have presented a method of rational design of antibodies, which works through a complementary peptide design and grafting procedure, to target specific epitopes within intrinsically disordered proteins. We have shown that this method generates antibodies that can bind with good specificity and affinity target regions in three disordered peptides and proteins associated with protein misfolding diseases and that they can be effective in reducing their aggregation. Compared with antibodies obtained with standard experimental techniques, however, our designed antibodies exhibit some limitations. The one-loop DesAb variants have relatively low affinity and selectivity, which may undermine their usefulness for some applications (e.g., Western blot detection). To improve on these aspects, we have shown that the simultaneous grafting of two complementary peptides can bring the affinity in the range of that of standard antibodies (Fig. S11) and lead to antigen detection in a Western blot, although this procedure also reduced the stability of the domain scaffold that we used here. We anticipate that our design strategy, and in particular the grafting of multiple loops, will be applicable to scaffolds that are intrinsically more stable than the human VH domain that we used and can better tolerate loop insertions. Also, this rational design approach can be combined with existing in vitro affinity maturation techniques, such as error-prone PCR and phage display. We also suggest that the complementary peptide design strategy that we presented may be applied to rationally engineer interactions of other classes of proteins of biomedical and biotechnological interest.
Fig. S11.
Distribution of Kd values in the Structural Antibody Database (SAbDab). Histogram of 211 Kd values deposited in the SAbDab (53). The distribution includes Kd values from a wide range of antibody types (from full length to nanobodies) obtained with a variety of methods (from immunogenization to in vitro affinity maturation). The vertical dashed lines mark our estimates for the Kd values of DesAb-F (18 μM) and two-loop DesAb (45 nM).
Materials and Methods
The method of identifying complementary peptides and of grafting them on a single domain antibody scaffold is described in SI Materials and Methods. The method of protein expression is also described SI Materials and Methods. The details of all experimental assays are reported in SI Materials and Methods.
Acknowledgments
We thank Dr. Peter Tessier for sending us the plasmid of the WT human heavy chain variable domain that we used in this study as scaffold for grafting the designed complementary peptides. We thank Dr. Paolo Arosio and Dr. Stefano Gianni for useful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.M.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422401112/-/DCSupplemental.
References
- 1.Carter PJ. Introduction to current and future protein therapeutics: A protein engineering perspective. Exp Cell Res. 2011;317(9):1261–1269. doi: 10.1016/j.yexcr.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Elvin JG, Couston RG, van der Walle CF. Therapeutic antibodies: Market considerations, disease targets and bioprocessing. Int J Pharm. 2013;440(1):83–98. doi: 10.1016/j.ijpharm.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 3.Goodman M. Market watch: Sales of biologics to show robust growth through to 2013. Nat Rev Drug Discov. 2009;8(11):837–837. doi: 10.1038/nrd3040. [DOI] [PubMed] [Google Scholar]
- 4.Leader B, Baca QJ, Golan DE. Protein therapeutics: A summary and pharmacological classification. Nat Rev Drug Discov. 2008;7(1):21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 5.Pavlou AK, Reichert JM. Recombinant protein therapeutics: Success rates, market trends and values to 2010. Nat Biotechnol. 2004;22(12):1513–1519. doi: 10.1038/nbt1204-1513. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury AR, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: The power of in vitro display technologies. Nat Biotechnol. 2011;29(3):245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23(9):1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 8.Lee CC, Perchiacca JM, Tessier PM. Toward aggregation-resistant antibodies by design. Trends Biotechnol. 2013;31(11):612–620. doi: 10.1016/j.tibtech.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11(6):610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- 10.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 11.Miersch S, Sidhu SS. Synthetic antibodies: Concepts, potential and practical considerations. Methods. 2012;57(4):486–498. doi: 10.1016/j.ymeth.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337(6101):1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 13.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 14.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 15.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 16.Tompa P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem Sci. 2012;37(12):509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Uversky VN. Intrinsically disordered proteins and novel strategies for drug discovery. Expert Opin Drug Discov. 2012;7(6):475–488. doi: 10.1517/17460441.2012.686489. [DOI] [PubMed] [Google Scholar]
- 18.Varadi M, et al. pE-DB: A database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Res. 2014;42(Database issue):D326–D335. doi: 10.1093/nar/gkt960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 20.Malaney P, Pathak RR, Xue B, Uversky VN, Davé V. Intrinsic disorder in PTEN and its interactome confers structural plasticity and functional versatility. Sci Rep. 2013;3:2035. doi: 10.1038/srep02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mészáros B, Simon I, Dosztányi Z. The expanding view of protein-protein interactions: Complexes involving intrinsically disordered proteins. Phys Biol. 2011;8(3):035003. doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- 22.Uversky VN. Intrinsic Disorder in Proteins Associated with Neurodegenerative Diseases. Protein Folding and Misfolding. Vol 7. Springer; New York: 2009. pp. 21–75. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 24.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 25.Schenk D, et al. Immunization with amyloid-β attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400(6740):173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 26.Näsström T, et al. Antibodies against alpha-synuclein reduce oligomerization in living cells. PLoS One. 2011;6(10):e27230. doi: 10.1371/journal.pone.0027230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantazes RJ, Maranas CD. OptCDR: A general computational method for the design of antibody complementarity determining regions for targeted epitope binding. Protein Eng Des Sel. 2010;23(11):849–858. doi: 10.1093/protein/gzq061. [DOI] [PubMed] [Google Scholar]
- 28.Li T, Pantazes RJ, Maranas CD. OptMAVEn: A new framework for the de novo design of antibody variable region models targeting specific antigen epitopes. PLoS One. 2014;9(8):e105954. doi: 10.1371/journal.pone.0105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu C-M, et al. Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface. PLoS One. 2012;7(3):e33340. doi: 10.1371/journal.pone.0033340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladiwala ARA, et al. Rational design of potent domain antibody inhibitors of amyloid fibril assembly. Proc Natl Acad Sci USA. 2012;109(49):19965–19970. doi: 10.1073/pnas.1208797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perchiacca JM, Ladiwala ARA, Bhattacharya M, Tessier PM. Structure-based design of conformation- and sequence-specific antibodies against amyloid β. Proc Natl Acad Sci USA. 2012;109(1):84–89. doi: 10.1073/pnas.1111232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perchiacca JM, Tessier PM. Engineering aggregation-resistant antibodies. Annu Rev Chem Biomol Eng. 2012;3:263–286. doi: 10.1146/annurev-chembioeng-062011-081052. [DOI] [PubMed] [Google Scholar]
- 33.Kashmiri SV, De Pascalis R, Gonzales NR, Schlom J. SDR grafting–a new approach to antibody humanization. Methods. 2005;36(1):25–34. doi: 10.1016/j.ymeth.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Ganesan A, et al. Selectivity of aggregation-determining interactions. J Mol Biol. 2015;427(2):236–247. doi: 10.1016/j.jmb.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Zalevsky J, et al. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2010;28(2):157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sickmeier M, et al. Disprot: The database of disordered proteins. Nucleic Acids Res. 2007;35(Database issue):D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51(11):2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 38.Sormanni P, Camilloni C, Fariselli P, Vendruscolo M. The s2D method: Simultaneous sequence-based prediction of the statistical populations of ordered and disordered regions in proteins. J Mol Biol. 2015;427(4):982–996. doi: 10.1016/j.jmb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Barthelemy PA, et al. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J Biol Chem. 2008;283(6):3639–3654. doi: 10.1074/jbc.M708536200. [DOI] [PubMed] [Google Scholar]
- 40.Sormanni P, Aprile FA, Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427(2):478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77(2):305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 42.Roodveldt C, et al. Chaperone proteostasis in Parkinson’s disease: Stabilization of the Hsp70/α-synuclein complex by Hip. EMBO J. 2009;28(23):3758–3770. doi: 10.1038/emboj.2009.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aprile FA, et al. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS One. 2013;8(6):e67961. doi: 10.1371/journal.pone.0067961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 45.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perchiacca JM, Ladiwala ARA, Bhattacharya M, Tessier PM. Aggregation-resistant domain antibodies engineered with charged mutations near the edges of the complementarity-determining regions. Protein Eng Des Sel. 2012;25(10):591–601. doi: 10.1093/protein/gzs042. [DOI] [PubMed] [Google Scholar]
- 47.Jespers L, Schon O, James LC, Veprintsev D, Winter G. Crystal structure of HEL4, a soluble, refoldable human V(H) single domain with a germ-line scaffold. J Mol Biol. 2004;337(4):893–903. doi: 10.1016/j.jmb.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Bond CJ, Marsters JC, Sidhu SS. Contributions of CDR3 to V H H domain stability and the design of monobody scaffolds for naive antibody libraries. J Mol Biol. 2003;332(3):643–655. doi: 10.1016/s0022-2836(03)00967-7. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 50.Gasteiger E, et al. The Proteomics Protocols Handbook. Springer; New York: 2005. pp. 571–607. [Google Scholar]
- 51.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149(5):1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasymov OK, Abduragimov AR, Glasgow BJ. Excited protein states of human tear lipocalin for low- and high-affinity ligand binding revealed by functional AB loop motion. Biophys Chem. 2010;149(1-2):47–57. doi: 10.1016/j.bpc.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunbar J, et al. SAbDab: The structural antibody database. Nucleic Acids Res. 2014;42(Database issue):D1140–D1146. doi: 10.1093/nar/gkt1043. [DOI] [PMC free article] [PubMed] [Google Scholar]