Significance
Transcription is a biological procedure in which DNA is transcribed to an RNA molecule. However, only fragments of this RNA are needed for protein synthesis. These fragments are exons that are interrupted by introns. Introns are removed by so-called RNA splicing process. Some exons could be alternatively included or excluded from the final RNA molecule. In this study, we have found that U2 snRNP auxiliary factor 65 kDa (U2AF65), a general splicing regulator, can significantly promote the exclusion of alternative exons. Strikingly, U2AF65 suppresses flanking intron splicing of alternative exons, and even constitutive intron splicing. We deduce that the stimulatory effects of U2AF65 on alternative exon exclusion are induced by the splicing inhibitory effects of U2AF65.
Keywords: U2AF65, pre-mRNA splicing, splicing inhibition, exon exclusion, SMN
Abstract
U2 snRNP auxiliary factor 65 kDa (U2AF65) is a general splicing factor that contacts polypyrimidine (Py) tract and promotes prespliceosome assembly. In this report, we show that U2AF65 stimulates alternative exon skipping in spinal muscular atrophy (SMA)-related survival motor neuron (SMN) pre-mRNA. A stronger 5′ splice-site mutation of alternative exon abolishes the stimulatory effects of U2AF65. U2AF65 overexpression promotes its own binding only on the weaker, not the stronger, Py tract. We further demonstrate that U2AF65 inhibits splicing of flanking introns of alternative exon in both three-exon and two-exon contexts. Similar U2AF65 effects were observed in Fas (Apo-1/CD95) pre-mRNA. Strikingly, we demonstrate that U2AF65 even inhibits general splicing of adenovirus major late (Ad ML) or β-globin pre-mRNA. Thus, we conclude that U2AF65 possesses a splicing Inhibitory function that leads to alternative exon skipping.
Pre-mRNA splicing is a process in which noncoding intron sequences are removed and exon sequences are then ligated together (1, 2). Pre-mRNA splicing is carried out by spliceosome, a large RNA–protein complex that contains five small nuclear ribonucleoproteins (U snRNPs) and more than 100 additional proteins (3). Pre-mRNA splicing occurs in the consensus sequences at the 5′ splice-site, 3′ splice-site, and branch point that are necessary for splicing. The sequence between 3′ AG dinucleotide and branch point is the polypyrimidine (Py) tract that directs spliceosome assembly on the 3′ splice-site. Alternative splicing provides an important regulatory mechanism in higher eukaryotes for multiple proteins produced from a single gene (4, 5).
The U2 snRNP auxiliary factor 65 kDa (U2AF65) exists as a heterodimer with U2AF35 (6). U2AF65 contains three C-terminal RNA recognition motifs (RRMs) and an N-terminal arginine/serine-rich (RS) domain (7, 8). Using U2AF65 depletion/adding back technology with in vitro HeLa nuclear extract, it was demonstrated that U2AF65 is an essential splicing factor (9). Whereas U2AF65 binds to Py tract to promote prespliceosome assembly and branchpoint/U2 snRNA base pairing, U2AF35 plays a role in the 3′ splice-site (10, 11). As U2AF65 prefers high C/U-rich sequences in the Py tract, a stronger interaction between U2AF65 and Py tract promotes prespliceosome assembly (12). U2AF65 is also essential in vertebrate development (13, 14). Its expression level is related to myotonic dystrophy, cystic fibrosis, and cancers (15, 16).
Proximal spinal muscular atrophy (SMA) is an autosomal recessive genetic disease (17) and a leading cause of infant mortality. The motor neurons in the anterior horn of spinal cord are severely damaged in patients with type 1 SMA, usually leading to death before age 2 y as a result of a lack of respiratory support (18, 19). In patients with SMA, the SMN1 gene is deleted or mutated, whereas the SMN2 gene, a duplicate of the SMN1 gene, is included (20). SMN2 genomic DNA contains a few nucleotide mutations compared with SMN1 (21, 22). Full-length SMN protein functions in the U snRNP assembly/disassembly, as well as in the β-actin mRNA transport in neurons (23, 24). However, the mutations in SMN2 pre-mRNA cause predominantly skipping of exon 7, which produces SMNΔ7, a truncated and less stable protein with reduced self-oligomerization activity. Alternative exon 7 splicing of SMN pre-mRNA was modulated by orchestrated RNA–protein and protein–protein interactions, secondary structures of RNA, and RNA sequences (25–27). Among the mutations on SMN2 pre-mRNA, the most functionally understood one is the C-to-U point mutation on exon 7, which plays an important role in alternative splicing of exon 7 (25–27). In vitro analysis using HeLa nuclear extract and S100 extract demonstrates that SRSF1 promotes exon 7 inclusion through contacting the enhancer sequence on exon 7 of SMN1 pre-mRNA, and that C-to-U mutation on SMN2 pre-mRNA disrupts SRSF1 binding and then consequently disrupts the enhancer function of SRSF1 (28). However, cell-based analysis shows a different result, indicating that SRSF1 does not play an essential role in SMN exon 7 splicing (29). In contrast, cell transfection analysis demonstrates that heterogeneous nuclear ribonucleoprotein (hnRNP) A1 interacts with the C-to-U mutation on SMN2 pre-mRNA to inhibit exon 7 splicing (29). A possible explanation for these different results is that different analysis systems could provide different conclusions.
Although the roles of U2AF65 in alternative splicing are verified to some extent, the function and mechanism are unclear. The previous reports have shown that U2AF65 roles in alternative splicing are the target of alternative splicing regulatory factors, as demonstrated with increased U2AF65 binding by other splicing regulatory proteins (30, 31). More recently, genome-wide analysis has demonstrated that upstream intronic binding of U2AF65 interferes with the immediate downstream 3′ splice-site of alternative or constitutive exons to cause exon skipping or inclusion (32). In the SMN pre-mRNA, it was demonstrated that U2AF65 interacts more strongly with the SMN1 Py tract than the SMN2 Py tract (33). However, it is unclear how U2AF65 itself regulates alternative splicing.
Here we identified the function of U2AF65 in the alternative splicing. Through siRNA-knockdown and overexpression of U2AF65, we show that U2AF65 promotes alternative exon exclusion of both SMN2 and SMN1 pre-mRNA. Mutations of 5′ splice-site in exon 7 to a higher score sequence abolished the U2AF65 effects. Highly expressed U2AF65 also represses splicing of exon 7, flanking introns in three or two exon contexts. Strikingly, U2AF65 also inhibits intron splicing of adenovirus major late (Ad ML) and β-globin pre-mRNA. In addition, U2AF65 selectively increases its own binding on the weaker Py tract sequence, but not the stronger Py tract. Our results support the conclusion that the U2AF65 activity in promoting alternative exon skipping comes from its own splicing inhibitory activity.
Results
U2AF65 Stimulates Alternative Exon Exclusion.
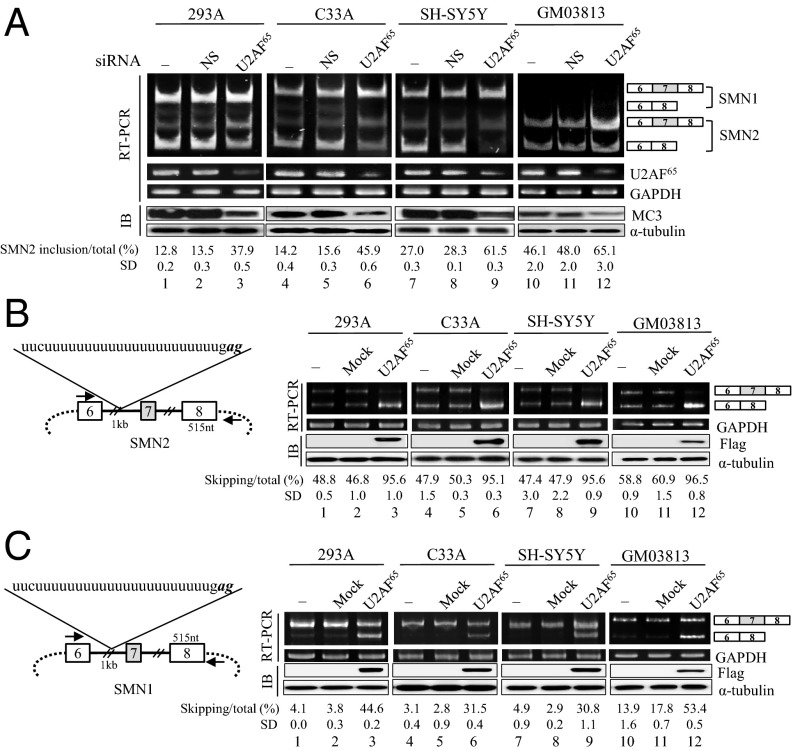
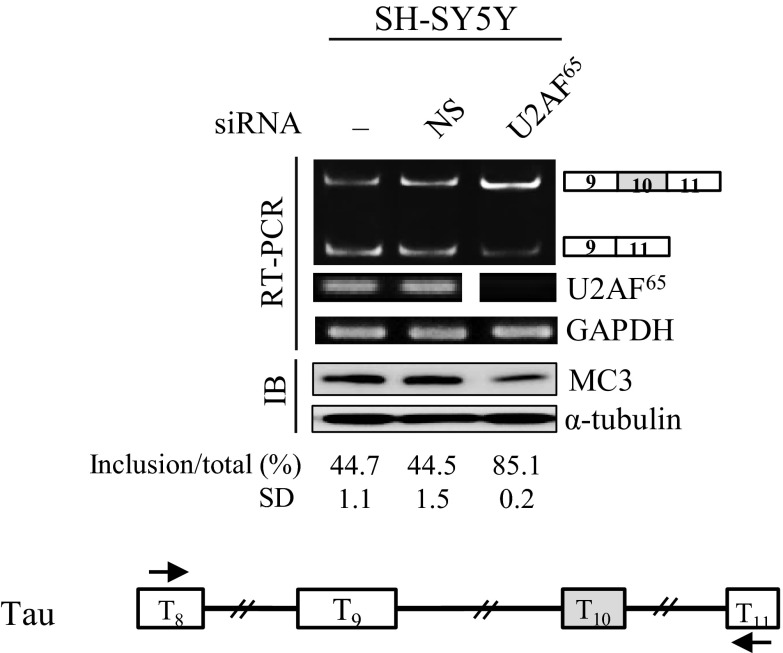
To determine the role of U2AF65 in alternative exon spicing, we first applied siRNA-directed knockdown of U2AF65. We examined U2AF65 effects mostly on alternative exon 7 splicing of SMN pre-mRNA. In the first set of experiments, we obtained the 293A (human embryonic kidney) cells in which U2AF65 expression is reduced by shRNA-virus treatment, and the control cells with nonsilencing shRNA. Immunoblotting analysis with anti-U2AF65 antibody (MC3) and RT-PCR results showed that the treatment with U2AF65-siRNA, but not nonsilencing shRNA, virus significantly decreased the expression of U2AF65 (Fig. 1A). To examine the effects of U2AF65 on endogenous exon 7 splicing of SMN1 and SMN2 pre-mRNA, RT-PCR was performed. We have found that reduced expression of U2AF65 promotes exon 7 inclusion in SMN2 pre-mRNA significantly (∼25%) in 293A cells (lane 3). The promoting activity on alternative exon inclusion was confirmed in C33A (human epithelial carcinoma), SH-SY5Y (human neuroblastoma), and SMA patient cells (GM03813) (∼31%, ∼34%, and ∼13%, respectively; Fig. 1A, lanes 6, 9, and 12). Thus, our results indicate that reduced expression of U2AF65 promotes exon 7 inclusion in endogenous SMN2 pre-mRNA. Interestingly, we also found that effects of U2AF65 knockdown are not limited to SMN pre-mRNA, as we demonstrated that as shown in Fig. S1, reduced U2AF65 expression also promoted exon 10 inclusion of Tau pre-mRNA.
Fig. 1.
U2AF65 stimulates exon 7 exclusion in both SMN2 and SMN1 pre-mRNA. (A) RT-PCR results of both endogenous SMN1 and SMN2 pre-mRNA are shown from untreated, nonsilencing shRNA-treated and U2AF65-shRNA-treated 293A, C33A, SH-SY5Y, and GM03813 cells. The percentage of exon 7 included RNA versus total RNA and its SD are indicated at the bottom. Immunoblotting analysis of these cells using an anti-U2AF65 (MC3) antibody are shown. (B, Left) The scheme of SMN2 minigene is shown with the intron RNA as a thicker line, whereas the vector sequence as a dot line. The RNA sequence of pseudo 3′ splice-site is shown. (Right) Shown is RT-PCR analysis from cells that express SMN2 minigene with overexpression of Flag-U2AF65 plasmid or control plasmid. Immunoblotting analysis with antiflag antibody is illustrated. (C, Left) SMN1 minigene scheme is shown. (Right) Results of RT-PCR analysis of the SMN1 minigene.
Fig. S1.
Reduced expression of U2AF65 promotes exon 10 inclusion of Tau pre-mRNA. (Upper) RT-PCR results of endogenous Tau pre-mRNA are shown from U2AF65-shRNA-treated SH-SY5Y cells. GAPDH was used as a loading control. Quantitation results of SMN2 pre-mRNA splicing are shown. Immunoblotting analysis of these cells using anti-U2AF65 (MC3) antibody are shown, with α-tubulin as a control. (Lower) The scheme of Tau pre-mRNA is shown.
To ask whether increased U2AF65 expression has the opposite effect on SMN2 splicing, we overexpressed U2AF65 in 293A cells that were transiently transfected with the SMN2 minigene. As previously shown, SMN2 minigene harbors exon 6–8 sequences with a deletion of intron 6 (∼1 kb) (Fig. 1B, Left) (34) and produces predominantly exon 7-skipped isoform, with only a small amount of exon 7-included isoform (lanes 1, 4, 7, and 10; Fig. 1B). The results in Fig. 1B (Right) show that U2AF65 overexpression promotes significantly exon 7 skipping of SMN2 pre-mRNA (∼47%; Fig. 1B, lane 3), which is opposite to the effects of U2AF65 siRNA knockdown. Consistently, U2AF65 also supports the increase in the exon 7-skipped form of SMN2 pre-mRNA in C33A, SH-SY5Y, and GM03813 cells (∼48%, ∼48%, and ∼38% independently; lanes 6, 9, and 12; Fig. 1B). Therefore, we conclude that U2AF65 promotes exon 7 skipping of SMN2 pre-mRNA.
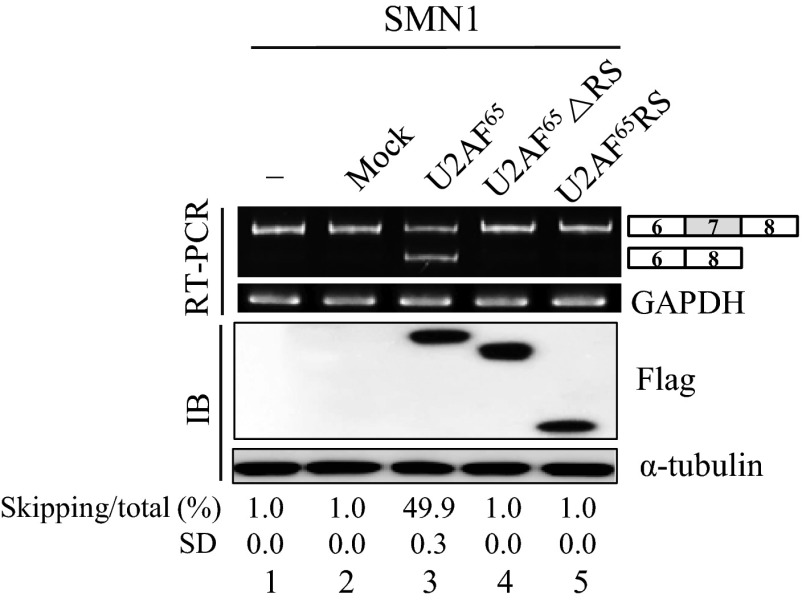
We next wondered whether U2AF65 affects SMN1 splicing. Because the endogenous SMN1 predominantly produced the exon 7-included isoform (Fig. 1A), a further decrease of exon 7 skipping in SMN1 pre-mRNA would be hard to be detected. Nonetheless, similar to the U2AF65 effects on SMN2 pre-mRNA splicing, the reduction of exon 7-skipped isoform of SMN1 pre-mRNA was still observed in the U2AF65-siRNA-treated 293A and C33A cells (Fig. 1A, lanes 3 and 6). We further asked whether increased U2AF65 expression also promotes exon 7 splicing of SMN1 pre-mRNA. As shown in Fig. 1C, in the SMN1 minigene, U2AF65 overexpression promotes exon 7 skipping of SMN1 pre-mRNA significantly in 293A, C33A, and SH-SY5Y cells (∼40%, ∼28%, ∼26%, and ∼40%, respectively; Fig. 1C). Therefore, we conclude that U2AF65 promotes exon 7 skipping of SMN1 pre-mRNA. To analyze the functional requirement for U2AF65 in alternative splicing, we applied U2AF65 mutations with either the RS domain (U2AF65ΔRS) or the RRM domain deleted (U2AF65RS). Our results in Fig. S2 show that neither U2AF65ΔRS nor U2AF65RS was able to support exon 7 skipping (lanes 4 and 5). Thus, we conclude that both the RS domain and the RRM domain of U2AF65 are required for increasing the exon 7-skipped isoform. Taken together, we conclude that U2AF65 promotes exon 7 skipping of both SMN1 and SMN2 pre-mRNA, and therefore, U2AF65 effects are not related to the point mutations on SMN2 pre-mRNA.
Fig. S2.
Both RS domain and RRM domain are required for the role of U2AF65 in exon 7 skipping of SMN1 pre-mRNA. RT-PCR analysis of SMN1 minigene in 293A cells that overexpressed Flag-U2AF65 (or Flag-U2AF65 ΔRS or Flag-U2AF65 RS) plasmid or control plasmid are shown.
A Stronger 5′ Splice Site on Exon 7 of SMN Pre-mRNA Prevents U2AF65 Effects on Exon 7 Splicing.
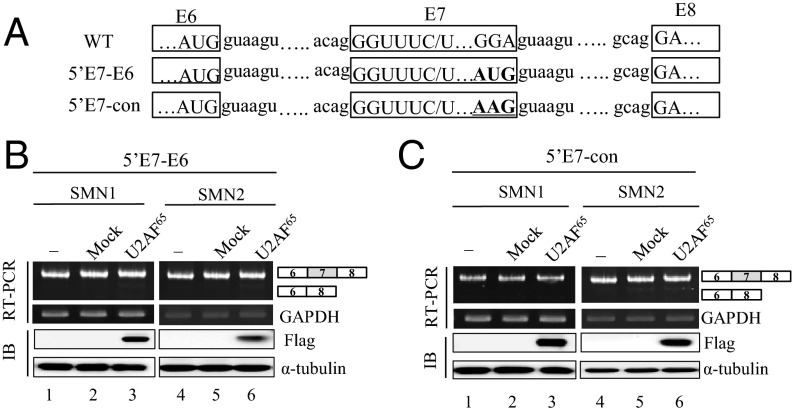
Using a bioinformatics splice-site score calculation program, we predicted that exon 6 harbors a much stronger 5′ splice-site than exon 7. We wondered whether a stronger 5′ splice-site on exon 7 could avoid the inhibitory effects of U2AF65 on exon 7 inclusion. To test this hypothesis, we generated a mutant in which 5′ splice-site of exon 7 (GGA) is replaced by 5′ splice-site sequence of exon 6 (AUG) (5′E7-E6) (Fig. 2A). It is worth noting that a single G nucleotide substitution at the last position of exon 7 has been previously shown to be able to strongly improve exon 7 inclusion (35). As predicted, Fig. 2B shows that the mutations on both SMN1 and SMN2 pre-mRNA induced an exclusive exon 7 inclusion (lanes 1 and 4), and that the mutations completely escaped the U2AF65 effects on exon 7 splicing (lanes 3 and 6). Thus, a stronger 5′ splice-site on exon 7 prevents U2AF65 effects on exon 7 splicing. The results were confirmed in another 5′ splice-site mutant, in which the 5′ splice-site of exon 7 is substituted by a conserved sequence (AAG) (5′E7-con) (Fig. 2A). This mutant also has G nucleotide at the 3′ end of exon 7. Similar to with the 5′E7-E6 mutant, 5′E7-con mutant minigenes in SMN1 and SMN2 pre-mRNA also produced exon 7 included isoform predominantly (Fig. 2C, lanes 1 and 4), preventing U2AF65 effects. Furthermore, U2AF65 did not promote exon 7 skipping in these mutants (lanes 3 and 6). Therefore, we conclude that a stronger 5′ splice-site sequence abolishes the activity of U2AF65 in the exon 7 skipping.
Fig. 2.
A stronger 5′ splice-site on exon 7 prevents U2AF65 effects on exon 7 splicing. (A) The splice-site sequences of wild type, 5′E7-E6, and 5′E7-con are shown. (B) RT-PCR analysis from SMN1-5′E7-E6 and SMN2-5′E7-E6 minigenes harboring cells that were expressed with Flag-U2AF65. (C) RT-PCR analysis with RNAs extracted from SMN1-5′E7-con and SMN2-5′E7-con minigenes, along with the Flag-U2AF65 minigene.
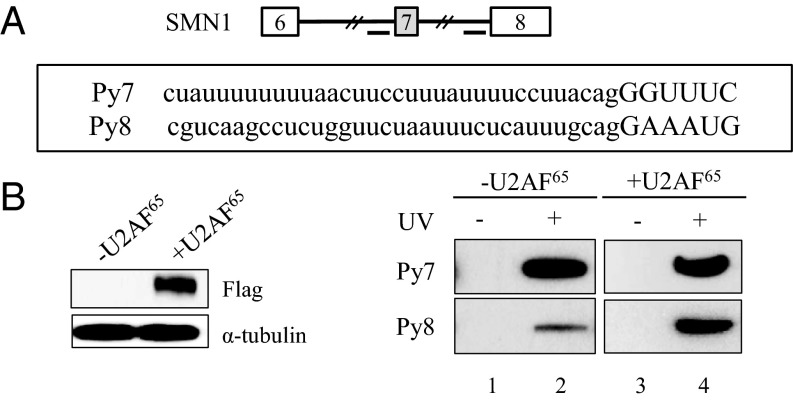
U2AF65 Promotes Its Own Binding Only on the Weaker Py Tract in SMN Pre-mRNA.
U2AF65 was previously shown to interact with the Py tract to promote prespliceosome assembly (36). It was also demonstrated that stronger binding of U2AF65 enhances pre-mRNA splicing. We have noticed that the Py tract of exon 7 (Py7) harbors more frequent U residues than that of exon 8 (Py8) (Fig. 3A). We wondered whether and how overexpression of U2AF65 affects its own binding on the Py tract of both exon 7 and exon 8. To answer the question, we incubated the Py tract RNA of exon 7 and exon 8 (Py7 and Py8) with either U2AF65 overexpressed cell lysates or untreated cell lysate as a control. After incubation, we performed UV-crosslinking analysis and then immunoprecipitation, using anti-U2AF65 antibody (MC3). Fig. 3B shows that, on the Py8 RNA, there is a significantly increased U2AF65 binding in the U2AF65 overexpressed cell lysates. However, on the Py7 RNA, the binding of U2AF65 was not altered. The results demonstrate that increased expression of U2AF65 promotes its own binding only on the weaker Py tract, but not on the stronger Py tract. One possibility is that U2AF65 binding to the stronger Py tract is already saturated, and therefore there is no binding increase even with increased U2AF65 expression.
Fig. 3.
U2AF65 promotes its own binding only on the weaker polypyrimidine tract in SMN pre-mRNA. (A) Py tract sequences of exon 7 (Py7) and exon 8 (Py8) are shown. (B) UV-crosslinking and immunoprecipitation with anti-U2AF65 antibody were performed using Py7 and Py8 RNA after incubation with cell lysate that is expressed with U2AF65. Immunoblotting analysis using an anti-flag antibody is shown.
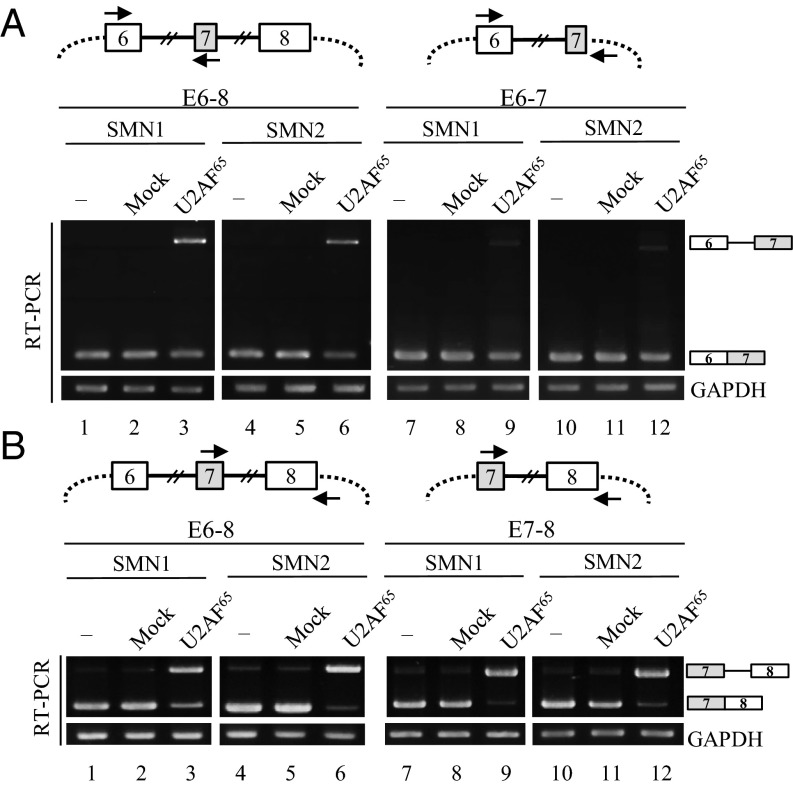
U2AF65 Inhibits Flanking Intron Splicing of Alternative Exon.
To further characterize the mechanisms of U2AF65 function, we examined its effects on intron splicing of SMN pre-mRNA. The first approach was to detect intron 6 splicing with one primer that base pairs with plasmid sequence upstream of exon 6, and the other primer that base pairs with exon 7 (Fig. 4A, Upper). In the untreated cells, we found that only intron 6 spliced isoform was detected in both SMN1 and SMN2 pre-mRNA (lanes 1 and 4). However, U2AF65 expression induced an appearance of unspliced isoform and a decrease of spliced isoform in both SMN1 and SMN2 pre-mRNA (Fig. 4A, lanes 3 and 6). Thus, U2AF65 inhibits intron 6 splicing of both SMN1 and SMN2 pre-mRNA. To test whether the 5′ splice-site of exon 7 interferes with U2AF65 effects on intron 6 splicing, we produced a minigene for SMN1 and SMN2 pre-mRNA, in which the 3′ end of exon 7, intron 7, and exon 8 was deleted (Fig. 4A, Upper). To differentiate this minigene from the other minigenes that harbor exon 6 through exon 8 (E6–E8), we named it the E6-7 minigene. The results in Fig. 4A show that E6-7 also produced unspliced intron 6 under U2AF65 treatment to a much smaller extent than did E6-8 minigenes (lanes 9 and 12). Thus, we conclude that U2AF65 inhibits intron 6 splicing and that exon 7 definition alleviates the inhibitory effects of U2AF65.
Fig. 4.
U2AF65 inhibits splicing of both intron 6 and intron 7 in SMN1 and SMN2 pre-mRNA. (A, Upper) The schemes of E6-8 and E6-7 minigenes are shown. (Lower) Intron 6 splicing of E6-8 and E6-7 minigenes is shown. U2AF65 plasmid was transfected into cells along with the minigenes. (B, Upper) The schemes of E6-8 and E7-8 minigenes are shown. (Lower) U2AF65 effects on intron 7 splicing were shown.
We further analyzed the U2AF65 effects on intron 7 splicing, using one primer that base pairs with exon 7, and the other primer that base pairs with the plasmid sequence downstream of exon 8 (Fig. 4B, Upper). The results in Fig. 4B show that U2AF65 remarkably inhibited intron 7 splicing, as shown with the significant amount of unspliced product in the E6-8 minigenes of SMN1 and SMN2 pre-mRNA (Fig. 4B, lanes 3 and 6). Furthermore, U2AF65 was able to inhibit intron 7 splicing in E7-8 minigenes on a similar level as in E6-8 minigenes (Fig. 4B, lanes 9 and 12). Thus, we conclude that U2AF65 inhibits intron 7 splicing and that the 3′ splice-site of exon 7 does not affect the inhibitory effects of U2AF65. By combining the results in Fig. 4 together, we conclude that U2AF65 inhibits splicing of both intron 6 and intron 7 and that, whereas 5′ splice-site of exon 7 reduced the inhibitory effects of U2AF65, 3′ splice-site of exon 7 did not affect its inhibition.
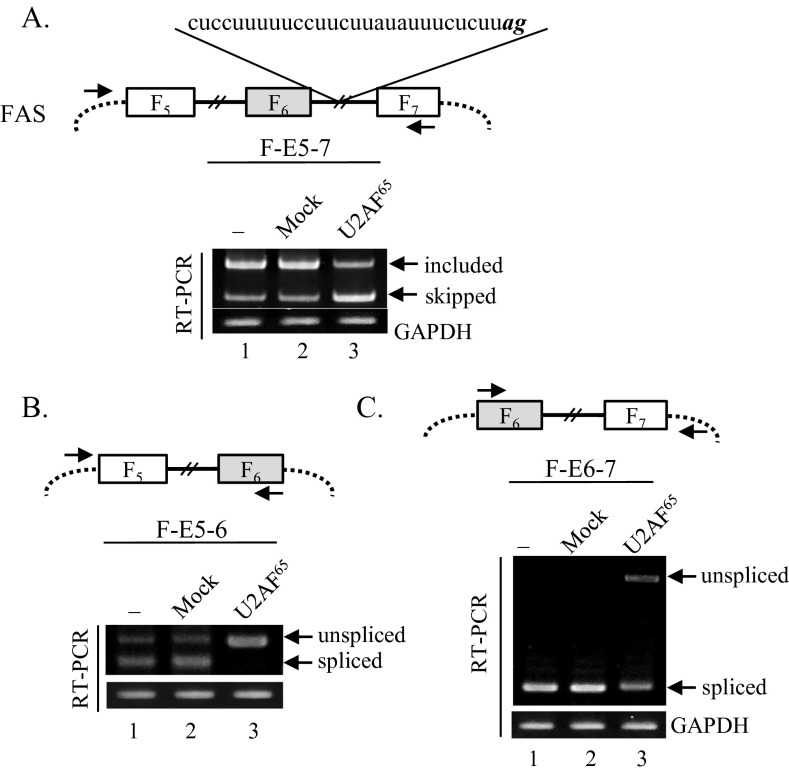
A recent report has demonstrated that upstream intronic binding of U2AF65 interferes with the immediate downstream 3′ splice-site of alternative exon or constitutive exon, causing exon skipping or inclusion (32). We found that intron 6 of SMN1/2 that includes uucuuuuuuuuuuuuuuuuuuuuuugag sequence contains pseudoexon with a potential U2AF65 binding sequence; thus, according to the report, this sequence interferes with the 3′ splice-site of exon 7, thereby promoting an exon 7 skipping event (Fig. 1B). To test whether this interfering event is the major cause of exon skipping, we analyzed U2AF65 effects on a well-established Fas minigene, which includes exon 5 through exon 7 (Upper) (37). As shown in Fig. S3A, Upper, intron 6 includes a cuccuuuuuccuucuuauauuucucuuag sequence with a pseudoexon and a strong U2AF65 binding site. According to 3′ splice-site interfering theory, U2AF65 should be able to promote inclusion of alternative exon. However, inconsistent with this prediction, Fas exon 6 skipping, but not inclusion, was strongly stimulated by U2AF65 (Fig. S3A, lane 3). Furthermore, we examined U2AF65 effects in two other minigenes in two exon contexts, one of which includes exons 5 through 6 (F-E5-6), the other of which includes exons 6 through 7 (F-E6-7) (Fig. S3 B and C, Upper). As shown in Fig. S3 B and C, U2AF65 overexpression induced the production of a high amount of unspliced isoforms of intron 5 and intron 6 (lane 3). Thus, we conclude that U2AF65 inhibits splicing of intron 5 and intron 6. Furthermore, our results indicate that U2AF65 inhibits Fas exon 6 inclusion through inhibiting splicing of intron 5 and 6, but not through interfering with the downstream 3′ splice-site.
Fig. S3.
U2AF65 stimulates exon 6 skipping of Fas pre-mRNA through inhibiting splicing of intron 5 and intron 6. (A, Upper) Scheme of Fas minigene. The primers used to detect alternative splicing of exon 6 are shown with arrows. Fas exons are indicated as F5, F6, and F7. (Lower) RT-PCR results of exon 6 splicing with U2AF65 treatment are shown. GAPDH was used as a control. (B, Upper) Scheme of a Fas minigene (F-E5-6) that includes exon 5 through exon 6. The primers used to detect intron 5 splicing are shown with arrows. (Lower) RT-PCR analysis for intron 5 splicing with U2AF65 expression is shown. (C, Upper) Scheme of a Fas minigene (F-E6-7) that contains exon 6 through exon 7. (Lower) RT-PCR analysis for intron 6 splicing under U2AF65 treatment is shown.
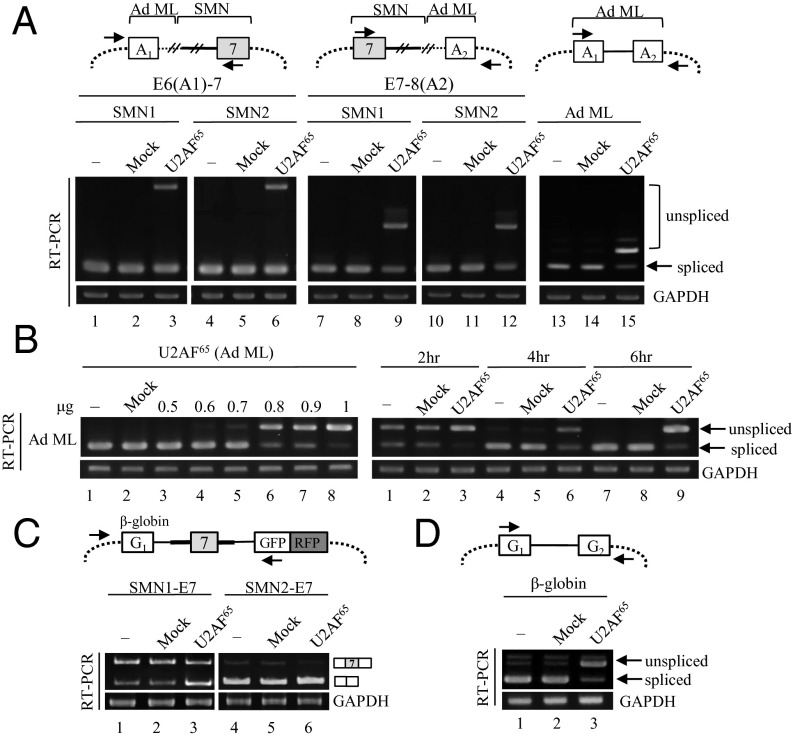
Overexpression of U2AF65 Inhibits Splicing of Ad ML and β-globin Pre-mRNA.
It is well known that U2AF65 is required for pre-mRNA splicing, as demonstrated by in vitro depletion-adding back experiments. However, inhibitory effects of U2AF65 on splicing have not yet been documented. Thus, we first hypothesized that the inhibitory effects of U2AF65 are specific to SMN intron 6 and intron 7 and that there must be essential RNA sequences that are required for intron 6 and intron 7 splicing of SMN pre-mRNA. To test this hypothesis, we generated two chimeric minigenes. The first minigene was produced from the E6-7 minigene by replacing exon 6 and part of intron 6 with Ad ML exon 1 and part of intron 1 [E6 (A1)-7], and the second minigene was generated from the E7-8 minigene by replacing exon 8 and the upstream part of intron 7 with Ad ML exon 2 and the downstream part of intron 1 [E7-8(A2)] (Fig. 5A, Upper). The results in Fig. 5A show that U2AF65 was able to promote an unspliced isoform to a significant level in these two minigenes (lanes 3, 6, 9, and 12). We therefore initially concluded that the deleted parts in E6-7 and E7-8 are not needed for the inhibitory activity of U2AF65. However, when we express U2AF65 with the Ad ML minigene, we observe that U2AF65 is also able to inhibit splicing of Ad ML pre-mRNA (Fig. 5A, lane 15), suggesting U2AF65 even inhibits general splicing. To confirm the results, we expressed Ad ML pre-mRNA with U2AF65 plasmid in different concentrations. Fig. 5B shows that U2AF65 did not inhibit splicing with a lower amount of plasmid (lane 3); however, unspliced product appeared at higher amounts of plasmid (lane 4). Furthermore, we found that the production of unspliced RNA is dosage-dependent on the U2AF65 plasmid (Fig. 5B, Left, lanes 4–9). To further confirm the results, we asked whether inhibition of U2AF65 on splicing is time-dependent. We are concerned that as Ad ML pre-mRNA is almost completely spliced 48 h after transfection, we were not able to observe the stimulatory effects of U2AF65. As shown in Fig. 5B (Right), whereas a high percentage of unspliced isoform was detected at 2 h transfection (lane 1), spliced isoform was primarily observed at 4 h, as well as 6 h transfection (lane 4 and 7). However, we found that U2AF65 can strongly inhibit splicing even at 4 h and 6 h transfection (lanes 6 and 9). Strikingly, U2AF65 was even able to repress splicing at a 2-h point, when splicing was not yet complete. Therefore, we conclude that U2AF65 repress pre-mRNA splicing of Ad ML pre-mRNA in cells in a dosage- and time-dependent manner.
Fig. 5.
Overexpression of U2AF65 inhibits pre-mRNA splicing of Ad ML and β-globin. (A, Upper) The schemes of chimeric minigenes are shown. Introns from SMN pre-mRNAs are shown with thicker lines; introns from Ad ML pre-mRNAs are shown with thinner lines. A1 and A2 indicate exons from Ad ML pre-mRNA. (Lower) Analysis of intron splicing in different minigenes are shown. (B) (Left) Intron splicing of Ad ML pre-mRNA with a different amount (µg) of U2AF65 plasmid. (Right) Intron splicing of Ad ML pre-mRNA at 2-, 4-, and 6-h points with U2AF65 expression. (C, Upper) Scheme of SMN1/2-E7 plasmids. (Lower) RT-PCR analysis of RNAs extracted from 293A cells that expressed SMN1/2 E7 and Flag-U2AF65 or control plasmid. (D, Upper) Scheme of β-globin pre-mRNA. (Lower) Intron splicing of β-globin under U2AF65 expression is shown.
As the second approach to identifying RNA sequence requirements in SMN pre-mRNA for splicing inhibition, we used a previously reported construct that includes upstream β-globin exon 1 and downstream GFP/RFP exons as a reporter for the splicing of middle test exon (38). We inserted SMN exon 7 and its upstream 573-nt intron and downstream 424-nt intron between the β-globin and GFP/RFP exons (SMN1/2-E7; Fig. 5C, Upper). As shown in Fig. 5C, U2AF65 promoted a significant increase in exon 7 skipping of SMN1-E7 minigene (Fig. 5C, lane 3). Similarly, in the SMN2-E7 minigene, although low expression of the exon 7-included isoform made it hard to detect its decrease, the decrease of the exon 7-included isoform is still detectable (Fig. 5C, lane 6). We further asked whether the exon 7 skipping in the β-globin and SMN chimeric pre-mRNA was induced by splicing inhibitory effects of U2AF65 on β-globin pre-mRNA. As shown in Fig. 5D, we found that U2AF65 treatment promoted unspliced isoform on β-globin pre-mRNA (lane 3). Thus, we conclude that U2AF65 inhibits not only SMN pre-mRNA splicing but also general splicing when expressed in the cells, and that the exon-skipping stimulatory effects of U2AF65 are induced by its intrinsic inhibitory activity.
Discussion
U2AF65 has been previously shown to play important roles in general splicing procedure through promoting ATP-dependent 3′ spliceosome assembly (14, 36). In alternative splicing, U2AF65 has been reported to be a regulatory target of multiple splicing factors (30, 31), but its role in alternative splicing has not been clear. From systematic evolution of ligands by exponential enrichment (SELEX) and RNA seq data, it was demonstrated that U2AF65 has a high specificity for CU-rich sequences and that U2AF65 specifically contacts Py tract in vivo (10, 39–41). Recent reports show that U2AF65 is able to regulate alternative splicing in the case that Py tract-linked pseudo-splice-sites exist in the flanking introns of alternative exon (32) and that upstream intronic binding events interfere with the immediate downstream 3′ splice-site selection. All of the conclusions were drawn by analyzing siRNA-knockdown effects of U2AF65 on the endogenous alternative splicing. We tested U2AF65 effects on two pre-mRNAs: SMN and Fas. Whereas SMN pre-mRNA includes an intronic U2AF65 binding event at the upstream intron of flanking exon, Fas pre-mRNA includes it at the downstream intron. In contrast to the previous model, our results show that U2AF65 overexpression in cells promotes alternative exon skipping. The conflicting results may be a result of one or more experimental variables, as two studies differed substantially in the endogenous or minigene pre-mRNA used, siRNA treatment, or overexpression of U2AF65. However, our results do not rule out the possibility that, in the endogenous pre-mRNA, the upstream intronic binding of U2AF65 interferes with immediate downstream 3′ splice-site selection.
It has not been reported that U2AF65 interacts with enhancers or inhibitors on pre-mRNA, which is different from SR proteins and hnRNP, which interact with enhancer or inhibitor sequences on pre-mRNA. It was shown in previous reports that increased binding of U2AF65 is directly related with increased splicing activity (31, 42). However, it was also reported that enhanced splicing is not directly related with enhanced U2AF65 binding (43). Our results demonstrate that U2AF65 promotes its own binding only on a weak 3′ splice-site in mammalian cells. Surprisingly, increased U2AF65 expression did not affect its binding on a strong Py tract. In alternative splicing, the relative strength of the Py tract provides a potential that U2AF65 can regulate the alternative splicing. It has been reported that SR proteins or hnRNPs have antagonistic functions in alternative splicing (44, 45). Furthermore, the opposite effects occur based on their binding locations (45). In vitro SELEX results demonstrate that U2AF65 is able to contact various U-rich sequences on pre-mRNA; however, in vivo iClips demonstrate that U2AF65 interacts only with Py tract sequence in pre-mRNA (7, 14). Although we do agree that U2AF65 interacts with Py tract to promote prespliceosome assembly, we do not exclude the possibility that U2AF65 interacts with other sequences to inhibit splice-site selection.
U2AF65 protein is not the only protein that participates in both general and alternative splicing activity. There is a general understanding that SR proteins and hnRNP proteins regulate alternative splicing by contacting enhancers or inhibitors (46–50). However, it was also reported that hnRNP M and hnRNP L are essential in general pre-mRNA splicing. Remarkably, U1C, which is essential in the first step of spliceosome assembly as well as for stabilizing early splicing complexes, when mutated, affects a large set of alternative splicing (51). The functional relationship of SR proteins, hnRNP proteins, and general splicing factors in general and alternative splicing need to be further determined.
One of our findings is that U2AF65 inhibits flanking intron splicing of alternative exon in three- or two-exon contexts. Most strikingly, we demonstrate that U2AF65 inhibits intron splicing of even Ad ML and β-globin pre-mRNA, common pre-mRNAs that are used in general splicing mechanisms. Previous conclusion that U2AF65 is a general splicing factor came from the experiments, in which U2AF65 was depleted from nuclear extract and then purified U2AF65 protein was added back. Although our results are not contrary to the fact that purified U2AF65 protein is an essential splicing factor, the inhibitory effects of U2AF65 have not been detected yet. The difference may be a result of the difference in the assay systems, HeLa nuclear extract, and cell overexpression systems. The different results from different assay systems were also observed in other groups. Whereas SRSF1 was demonstrated to promote exon 7 splicing of SMN1 pre-mRNA using HeLa nuclear extract, its effects on exon 7 splicing were not detected in cell transfection assays (28, 52).
Materials and Methods
Plasmid Construction.
SMN1-L, SMN2-L, SMN1-S, and SMN2-S minigene constructs were produced as described previously (53, 54). We constructed 5′E7-E6 and 5′E7-con minigenes through site-directed mutagenesis, using the following primers: common primers (SMNE6.F/SMNE8.R), specific primers for 5′E7-E6 (E7E6.F/E7E6.R), and 5′con (E7CS.F/E7CS.R). E6-7 (SMN1/2) and E7-8 (SMN1/2) were generated by using the following primer sets: SMNE6.F/SMNE6-7.R (E6-7), SMN1E7-8.F, SMN2E7-8.F/SMNE8.R (E7-8), and with SMN1/2-S minigenes as templates. To produce SMN1/2-E7 constructs, we used SMN1/2-L minigenes as templates and performed PCR with SMN.F and SMN.R primers to amplify the 573-nt intron 6, exon 7, and 424-nt intron 7. Ad ML and β-globin minigenes were generated by PCR amplification using primers (Ad ML.F/Ad ML.F and globin.F/globin.R primer). pIRES2-EGFP U2AF65△RS and RS constructs were generated with following primer sets: U2AF65△RS.F/U2AF65.R and U2AF65 RS.F/U2AF65.R. All primer sequences are listed in Table S1.
Table S1.
List of primers
| Name | Sequences |
| SMN.F | 5′-CGACGCGTCGGGTTCAAGTGATTCTCCTGCC-3′ |
| SMN.R | 5′-CGGGATCCCGCCAGAGGCTTGACGAATTCC-3′ |
| SMNE6.F | 5′-TACTCGGATCCATAATTCCCCCACCACCTCC-3′ |
| SMNE8.R | 5′-CTAACCTCGAGAACAGTACAATGAACAGCCATG-3′ |
| E7CS.F | 5′-TTTCTCATTTCCAGGAAATGCTGGCA-3′ |
| E7CS.R | 5′-TGCCAGCATTTCCTGGAAATGAGAAA-3′ |
| E7E6.F | 5′-ATTCCTTAAATTAAATGGTAAGTCTGCCAGCATTATG-3′ |
| E7E6.R | 5′-CATAATGCTGGCAGACTTACCATTTAATTTAAGGAAT-3′ |
| U2AF65.F | 5′-TTTTTGAATTCATGTCGGACTTCGACGAGTTC-3′ |
| U2AF65.R | 5′-TTTTTGGATCCCTACCAGAAGTCCCGGCGG-3′ |
| U2AF65△RS.F | 5′-TTTTTGAATTCCCACCCCCAGGCTTTGAGCACA-3′ |
| U2AF65 RS.R | 5′-TTTTTGGATCCCTACACGTCCCAGTATTTACGGAC-3′ |
| U2AF65 (RT).F | 5′-ATTTCTTCAACGCCCAGATG-3′ |
| U2AF65 (RT).R | 5′-ACAACCCCAGGCACATAGAC-3′ |
| Ex5.F | 5′-CTATCATGCTGGCTGCCT-3′ |
| Ex8.R | 5′-CTACAACACCCTTCTCACAG-3′ |
| pcI.F | 5′-GCTAACGCAGTCAGTGCTTC-3′ |
| GFP.R | 5′-CTGAAGCACTGCACGCCGTA-3′ |
| SMNE6.F | 5′-CTGAAGCACTGCACGCCGTAC-3′ |
| pcDNA.R | 5′-CTAGAAGGCACAGTCGAGGCT-3′ |
| SMNE7 F. | 5′-AAGGAAGGTGCTCACATTCC-3′ |
| SMNEx7 R. | 5′-GGAATGTGAGCACCTTCCTT-3′ |
| SMNE6-7.R | 5′-CTTAGCTCGAGAATTTAAGGAATGTGAGCACCT-3′ |
| SMN1.E7-8.F | 5′-ATCTCGGATCCTTTCAGACAAAATCAAAAAGAAGG-3′ |
| SMN2.E7-8.F | 5′-ATCTCGGATCCTTTTAGACAAAATCAAAAAGAAGG-3′ |
| Ad ML.F | 5′-TCGTAGCTAGCACTCTTGGATCGGAAACCCGT-3′ |
| AD ML.R | 5′-ATTAGCTCGAGACTGGAAAGACCGCGAAGAG -3′ |
| globin.F | 5′-TCCTAGCTAGCACATTTGCTTCTGACACAACTG-3′ |
| globin.R | 5′-ATTGTCTCGAGCCTGAAGTTCTCAGGATCCAC-3′ |
| FasE5.F | 5′-ATGTGAACATGGAATCATCAAGG-3′ |
| FasE6.R | 5′-CCCAAACAATTAGTGGAATTGG-3′ |
| FasE6.F | 5′-GATCCAGATCTAACTTGGGG-3′ |
| Tau.F | 5′-CAACGCCACCAGGATTCCAGCAAA-3′ |
| Tau.R | 5′-ATGTTGCCTAATGAGCCACACTTG-3′ |
| E1.F | 5′-CTCAGATCTACCATTGGTGCACC-3′ |
| pFlare GFP.R | 5′-GGTGCAGATGAACTTCAGG-3′ |
| GAPDH.F | 5′-ACCACAGTCCATGCCATCA-3′ |
| GAPDH.R | 5′-TCCACCACCCTGTTGCTGTA-3′ |
RT-PCR.
Forty-eight hours after transfection, cells were harvested and total RNAs were isolated using RiboEx reagent (Geneall). To generate cDNA, reverse transcription was carried out using oligo dT oligomer and ImProm-II TM reverse transcriptase (Promega). One microgram of total RNA was used per reverse transcription reaction. A primer set (Ex5.F/Ex8.R) was used to PCR-amplify endogenous SMN1 and SMN2 transcripts. Minigene-specific spliced products were amplified with Taq polymerase and the following primer combinations: SMN1/2-L (pCI.F/GFP.R), SMN1/2-S (SMNE6.F/pcDNA.R), and SMN1/2-E7 (E1.F/pFlare GFP.R). To analyze splicing of the intron 6 and 7 in SMN1/2 E6-E8, SMN1/2 E6-7, and SMN1/2 E7-8 minigenes, we used the following primer sets: SMN1/2 E6-7 (SMNE6.F/SMNE7.R) and SMN1/2 E7-E8 (SMNE7.F/pcDNA.R). To analyze pre-mRNA splicing of Ad ML, β-globin, and Fas, the following primer sets were applied: Ad ML.F/pcDNA.R, globin.F/pcDNA.R, and FasE5.F/pcDNA.R. U2AF65 mRNAs were amplified with primers U2AF65 (RT).F and U2AF65 (RT).R. All of primer sequences are listed in Table S1.
UV-Crosslinking Immunoprecipitation Assay.
A UV-crosslinking immunoprecipitation assay was performed as previously described (54). A mixture containing 100 pmol 5′-biotinylated RNA (Bioneer), cell lysates, 16 mM creatine phosphate, 0.4 mM ATP, and 2.6 mM MgCl2 was incubated for 10 min at 30 °C and then was irradiated with UV in a Stratalinker (Stratagene) at 80,000 µJ for 5 min on ice. After incubating with MC3 antibody, protein A agarose beads (Upstate) were added to immunoprecipitate U2AF65. Luminol/peroxide solution was used to detect the RNA–protein complex.
Acknowledgments
We thank Douglas L. Black (University of California, Los Angeles) for a pFlare 9A and Michael R. Green (University of Massachusetts Medical School) for U2AF65 antibody (MC3). This work was supported by Grant NRF-2013-R1A1A2062582 (to H.S.) and Grant NRF-2013-R1A1A2061321 (to X.Z.), funded by the National Research Foundation (NRF) of the Korean Ministry of Education, Science, and Technology and the integrative aging research center of Gwangju Institute of Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500639112/-/DCSupplemental.
References
- 1.Green MR. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- 2.Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Prog Mol Subcell Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Graveley BR. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17(2):100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 5.Stamm S, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kielkopf CL, Lücke S, Green MR. U2AF homology motifs: Protein recognition in the RRM world. Genes Dev. 2004;18(13):1513–1526. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273(5282):1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89(18):8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman BE, Grabowski PJ. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402(6763):832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 12.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75(6):1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 13.Corrionero A, Raker VA, Izquierdo JM, Valcárcel J. Strict 3′ splice site sequence requirements for U2 snRNP recruitment after U2AF binding underlie a genetic defect leading to autoimmune disease. RNA. 2011;17(3):401–411. doi: 10.1261/rna.2444811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R, Valcárcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268(5214):1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 15.Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci USA. 2011;108(27):11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 17.Sugarman EA, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012;20(1):27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3(2):97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 19.Briese M, Esmaeili B, Sattelle DB. Is spinal muscular atrophy the result of defects in motor neuron processes? BioEssays. 2005;27(9):946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 21.DiDonato CJ, et al. Cloning, characterization, and copy number of the murine survival motor neuron gene: Homolog of the spinal muscular atrophy-determining gene. Genome Res. 1997;7(4):339–352. doi: 10.1101/gr.7.4.339. [DOI] [PubMed] [Google Scholar]
- 22. DiDonato CJ, Brun T, Simard LR (1999) Complete nucleotide sequence, genomic organization, and promoter analysis of the murine survival motor neuron gene (Smn). Mamm Genome 10(6):638–641. [DOI] [PubMed]
- 23.Rossoll W, Bassell GJ. Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes. Results Probl Cell Differ. 2009;48:289–326. doi: 10.1007/400_2009_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glinka M, et al. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19(10):1951–1966. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- 25.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 26.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78(1):63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 29.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 30.Cao W, Razanau A, Feng D, Lobo VG, Xie J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res. 2012;40(16):8059–8071. doi: 10.1093/nar/gks504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei WJ, et al. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012;40(17):8622–8636. doi: 10.1093/nar/gks579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao C, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21(11):997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martins de Araújo M, Bonnal S, Hastings ML, Krainer AR, Valcárcel J. Differential 3′ splice site recognition of SMN1 and SMN2 transcripts by U2AF and U2 snRNP. RNA. 2009;15(4):515–523. doi: 10.1261/rna.1273209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho S, et al. Validation of trans-acting elements that promote exon 7 skipping of SMN2 in SMN2-GFP stable cell line. Biochem Biophys Res Commun. 2012;423(3):531–535. doi: 10.1016/j.bbrc.2012.05.161. [DOI] [PubMed] [Google Scholar]
- 35.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10(8):1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355(6361):609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 37.Oh H, et al. 2013. hnRNP A1 contacts exon 5 to promote exon 6 inclusion of apoptotic Fas gene. Apoptosis 18(7):825–835.
- 38.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci USA. 2008;105(32):11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selenko P, et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11(4):965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 40.Sickmier EA, et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23(1):49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcárcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol Cell. 2012;45(3):314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 42.Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7(1):56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 43.Kan JL, Green MR. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13(4):462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang E, Mueller WF, Hertel KJ, Cambi F. G Run-mediated recognition of proteolipid protein and DM20 5′ splice sites by U1 small nuclear RNA is regulated by context and proximity to the splice site. J Biol Chem. 2011;286(6):4059–4071. doi: 10.1074/jbc.M110.199927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erkelenz S, et al. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19(1):96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10(10):242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet. 2010;11(5):345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 48.Änkö ML. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3(1):1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith CW, Valcárcel J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 51.Rösel TD, et al. RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J. 2011;30(10):1965–1976. doi: 10.1038/emboj.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16(24):3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 53.Zhang ML, Lorson CL, Androphy EJ, Zhou J. An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: Potential therapy of SMA. Gene Ther. 2001;8(20):1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]
- 54.Cho S, et al. 2014. hnRNP M facilitates exon 7 inclusion of SMN2 pre-mRNA in spinal muscular atrophy by targeting an enhancer on exon 7. Biochim Biophys Acta 1839(4):306–15.