Significance
Biosensing technologies have significant impact on medical diagnostics but difficulties in the handling of complex biospecimens, portability, and nonlinearity in dynamic detection range present considerable technical bottlenecks in their translation into clinical settings. Here, we present the nanoplasmonic electrical field-enhanced resonating device (NE2RD) that detects and quantifies multiple biotargets from distinct clinical specimens (i.e., saliva, serum, and whole blood) with a broad linear dynamic range. Unlike conventional platforms, the NE2RD does not require lengthy sample-preparation steps, skilled personnel, or expensive infrastructure. Further, as a model clinical validation study, we monitored chemotherapy effects on viral load for coinfected patients on a single platform. Therefore, the portable NE2RD can be broadly applied to primary care and point-of-care settings with multiple clinical applications.
Keywords: nanoparticle, biodetection, point-of-need, label-free, multiple biotargets
Abstract
Recent advances in biosensing technologies present great potential for medical diagnostics, thus improving clinical decisions. However, creating a label-free general sensing platform capable of detecting multiple biotargets in various clinical specimens over a wide dynamic range, without lengthy sample-processing steps, remains a considerable challenge. In practice, these barriers prevent broad applications in clinics and at patients’ homes. Here, we demonstrate the nanoplasmonic electrical field-enhanced resonating device (NE2RD), which addresses all these impediments on a single platform. The NE2RD employs an immunodetection assay to capture biotargets, and precisely measures spectral color changes by their wavelength and extinction intensity shifts in nanoparticles without prior sample labeling or preprocessing. We present through multiple examples, a label-free, quantitative, portable, multitarget platform by rapidly detecting various protein biomarkers, drugs, protein allergens, bacteria, eukaryotic cells, and distinct viruses. The linear dynamic range of NE2RD is five orders of magnitude broader than ELISA, with a sensitivity down to 400 fg/mL This range and sensitivity are achieved by self-assembling gold nanoparticles to generate hot spots on a 3D-oriented substrate for ultrasensitive measurements. We demonstrate that this precise platform handles multiple clinical samples such as whole blood, serum, and saliva without sample preprocessing under diverse conditions of temperature, pH, and ionic strength. The NE2RD’s broad dynamic range, detection limit, and portability integrated with a disposable fluidic chip have broad applications, potentially enabling the transition toward precision medicine at the point-of-care or primary care settings and at patients’ homes.
Biosensing platforms have enabled various applications in different fields of clinical medicine such as biomarker/drug discovery and initiation and monitoring of therapy (1–3). However, material cost, accessibility, ease of operation, lack of portability, and complexity in readout remain major challenges for developing robust diagnostic assays (SI Appendix, Table S1). Recent advances in nanotechnology and biosensing have created new avenues to address these issues (4–9). Technically, they have provided integration of high-throughput sampling with readout systems for quantitative detection of disease-specific biotargets. Therefore, they have demonstrated great potential to revolutionize medical diagnostics. However, from a clinical and technological perspective, existing platforms still face several challenges. First, lengthy assay time hinders physicians from making early clinical decisions. Second, examining clinical samples with diverse pH range, ionic content, and ionic strength requires extensive sample preprocessing steps to avoid signal fluctuations and inaccuracies, and this requirement increases the complexity of current systems. Third, temperature is an extrinsic factor that needs to be tightly controlled for reliable measurements, thus resulting in additional cost for developing diagnostic technologies. Fourth, a wide dynamic linear range is needed to distinguish concentrations of biotargets reliably. Fifth, sensitivity is hindered by the complex composition of biological specimens, thus requiring labeling (10, 11), which further increases assay cost significantly. Sixth, since a panel of parameters is monitored for complex diseases, detection of multiple biotargets is critically needed. Seventh, portability is another important parameter to deploy a biosensing platform at point-of-care (POC) settings. These practical barriers seriously reduce the broad clinical applicability of current platforms for diagnostic implementations.
The development of various sensitive detection modalities has addressed some of these requirements. The mass-sensitive piezoelectric and microcantilever-based systems that use surface oscillations or changes in surface stress for molecular interactions and mass input (12, 13) have simplified sample-preparation steps. Likewise, electrical detection systems such as electrochemical sensors and interdigitated electrodes sense changes in molecular charges (14–17) and can provide affordable, simple, and high-throughput measurements. Further, plasmonic-based platforms that use an electrical field around metal surfaces/nanoparticles coupled with light (5, 18–22) have minimized readout complexity and demonstrated quantitative and sensitive measurements (23–26). In particular, surface plasmons on 3D-oriented surfaces generate highly sensitive spots that provide a sevenfold stronger electrical field than 2D-oriented plasmonic sensors (27–29). However, these advances have only reduced the effort required for sample preparation and the readout complexity, and some critical challenges remain unaddressed as summarized in Table 1.
Table 1.
Comparison of the NE2RD with the most sensitive platform technologies for biotarget detection
| Parameters | Magneto-nanosensor chip (51) | SMC (Singulex) (52) | Simoa (Quanterix) (52) | Bio-barcode assay (53) | ChIP-NMR biosensor (54) | Advanced SPR biosensor (55) | Plasmonic ELISA (56) | Plasmonic gold chip (36) | NE2RD |
| Target | Lactoferrin, survivin, CEA, VEGF, EpCAM, G-CSF, TNF-α, and eotaxin | cTnI, cytokines, amyloid-β, IL-22 | PSA, TNF-α, Tau | PSA | S. aureus, mouse macrophages, breast cancer cells, and multiple protein biomarkers | E. coli O157:H7 | PSA and HIV-1 capsid antigen p24 | Insulin Ab, GAD65 Ab, and IA2 Ab | IFN-γ, carbamazepine, casein, E. coli, human lung adenocarcinoma epithelial cells, HBV, DENV-1, DENV-2, TVV-1, KSHV and HIV-1 |
| Assay time | >2 h 20 min | Four 96-well plates per day (for single assay, not multiplex) | Five 96-well plates per day (for single assay, not multiplex) | >3 h 30 min | 30 min for incubation and 10 min for analysis | 30 min | >5 h 30 min | <2 h | 1 h for incubation and 10 min for analysis |
| Multiple biotarget detection | Only protein biomarkers | Not available | 10-plex | Not available | Bacteria, mammalian cells, and protein biomarkers | Not available | Only two protein biomarkers | Only autoimmune antibodies | Protein biomarkers, protein allergens, drugs, bacteria, eukaryotic cells, and viruses |
| Readout | Magnetic field-based detection | Fluorescent-based system (digital and analog at high concentrations) | Fluorescent-based system (digital and analog at high concentrations) | Scanometric detection for DNA | NMR signal | Long-range surface plasmons enhanced by magnetic nanoparticles | Optical-based detection | NIR-FE | Plasmonic signal on a 3D-oriented substrate |
| Limit of detection | 1 pg/mL or 5 fM (with mono labeling) | fg–pg/mL | fg–pg/mL | 330 fg/mL | 5 pg/mL (∼1 ng in 5 µL) | 50 cfu/mL | 1 ag/mL | 0.1 kU/mL | 400 fg/mL and ∼100 copies/mL |
| 10 fg/mL or 50 aM (with dual labeling) | |||||||||
| Linear dynamic range | Six orders of magnitude | Four orders of magnitude | Four orders of magnitude | Two to three orders of magnitude | Four orders of magnitude | Three to four orders of magnitude | Three to four orders of magnitude | Four to five orders of magnitude | Eight orders of magnitude |
| Sample type | PBS, serum, urine, and saliva | Plasma, serum, CSF, cell lysate, urine, human brain tissue homogenate | Serum | PBS and serum | PBS and serum | PBS | PBS | Whole serum or blood | PBS, whole saliva, serum, and whole blood |
CEA, carcinoembryonic antigen; CFS, cerebrospinal fluid; EpCAM, epithelial cell adhesion molecule; GAD65, glutamic acid decarboxylase-65; G-CSF, granulocyte-colony stimulating factor; IA2, islet antigen-2; NIR-FE, near-infrared fluorescence-enhanced spectroscopy; PSA, prostate-specific antigen; TNF-α, tumor necrosis factor-alpha; VEGF, vascular endothelial growth factor.
Here, we report the development of an innovative device, the nanoplasmonic electrical field-enhanced resonating device (NE2RD), which addresses all these challenges on a single platform. A comparison of the NE2RD capability with that of existing technologies is presented in Table 1 and in SI Appendix, Tables S1 and S2. The NE2RD employs a label-free optical readout that precisely measures collective oscillations of nanoparticles leading to an unprecedented platform sensitivity that is enabled by hot-spot regions on the surface, where the internanoparticle distance is reduced by self-assembly (Fig. 1). This strategy uniquely enables highly sensitive detection to monitor binding and capture events of biotargets. Thus, each well or chip surface of the current prototypes individually measures a change in spectral color that is translated into a quantitative biotarget concentration in a multitarget manner. Further, the NE2RD can be implemented into a disposable fluidic chip format, and the measurements are collected using a customized portable measurement tool. The NE2RD uses only fingerprick volumes of clinical samples without the need for sample preprocessing steps, thus providing a rapid assay and potentially enabling direct translation to the clinic.
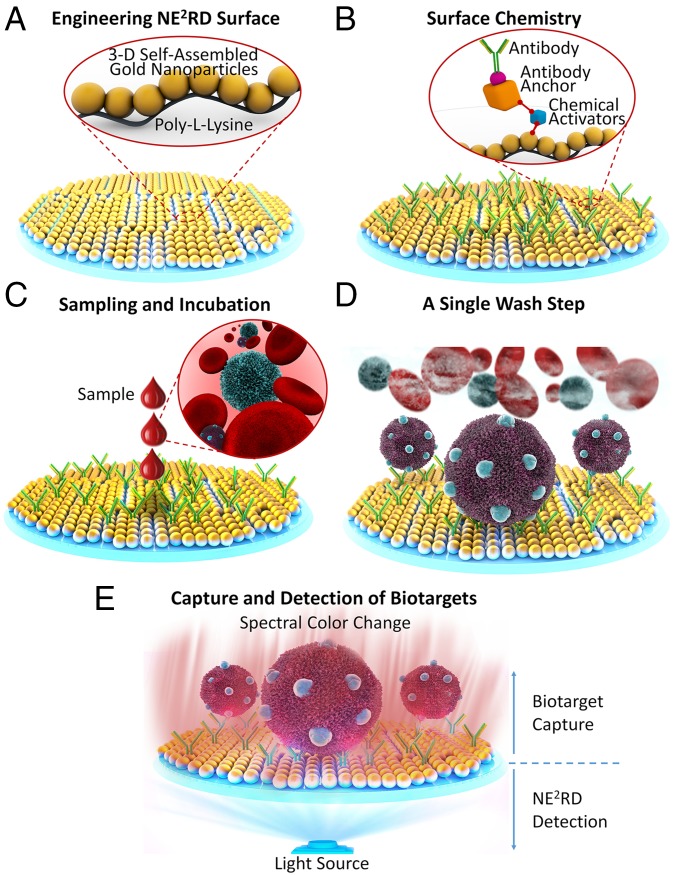
Fig. 1.
Representation of the NE2RD platform. (A) Gold nanoparticles are immobilized on polystyrene surfaces using poly-l-lysine for fabrication of the NE2RD. (B) In surface chemistry steps, several activators and antibody anchors are used to immobilize antibodies onto the surface. (C) Biological samples such as whole blood are applied to the surfaces without sample preprocessing. (D) Unbound cells and cellular components are washed out with physiological buffer. (E) Multiple biotargets of interest are captured specifically, depending on the antibodies immobilized on the surface, and spectral measurements are performed to monitor capture events, which change the spectral color of the surfaces in terms of both wavelength and extinction intensity parameters. Yusuf Yesil drew all schematics in Fig. 1.
Results
Device Design and Characteristics.
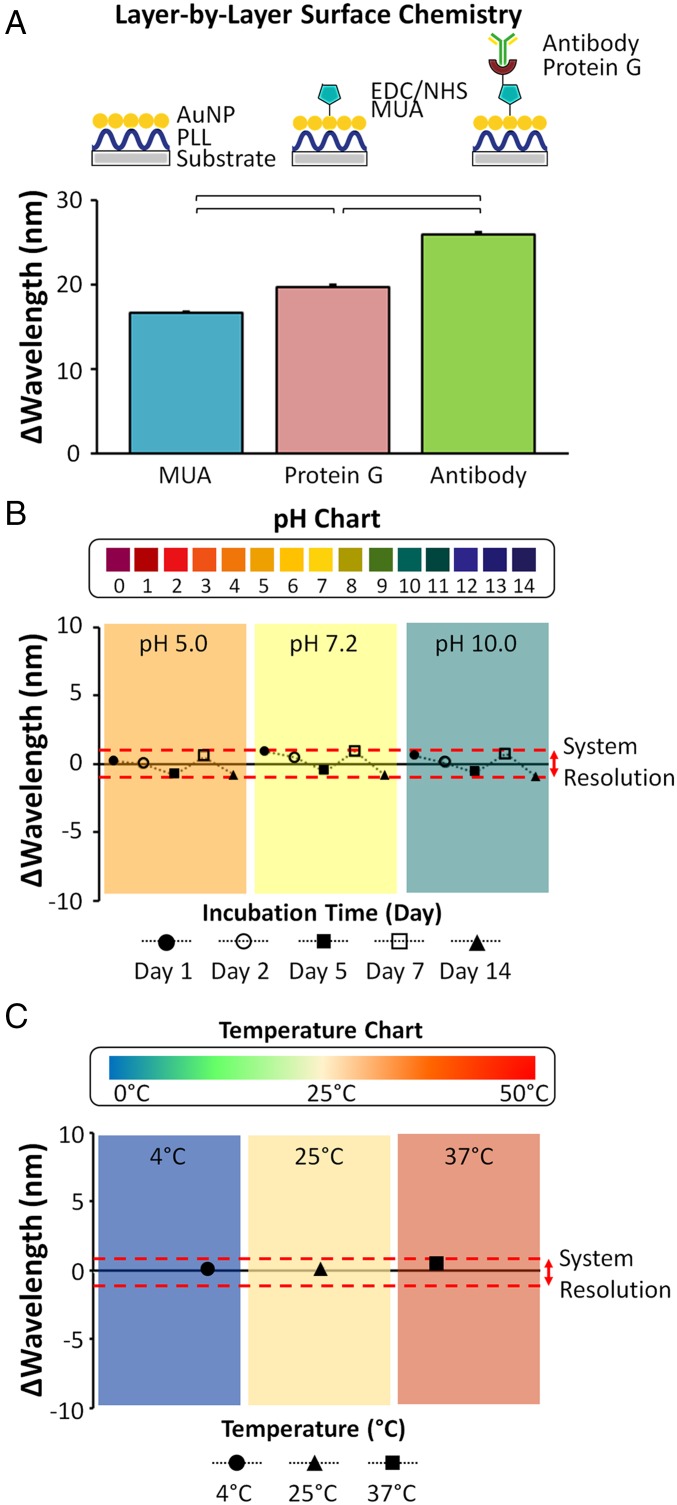
We engineered two prototypes of the NE2RD: (i) a plate (96-well) reader format and (ii) a portable reader format with a disposable fluidic chip. Conceptually, we fabricate the NE2RD surfaces on these prototypes in three steps: (i) modification of polystyrene substrates with gold nanoparticles (10-nm diameter), (ii) activation of the gold nanoparticle-decorated device surface, and (iii) immobilization of recognition elements, i.e., antibodies, to the surface. Only a fingerprick sample volume (≤100 µL) per well or fluidic chip is used to detect the biotargets. To monitor layer-by-layer modifications on the surface and the capture of the biotargets, we measured nanoplasmonic responses in terms of changes in spectral color (red- and blue-shifts in wavelength and extinction intensity parameters) facilitated by a Fourier-type expansion analysis (SI Appendix, Materials and Methods and Fig. S1). In our experiments, gold nanoparticles were first self-assembled on poly-l-lysine (PLL)-modified substrates. For instance, on the well-plate prototype, the chemical activator [i.e., 11-Mercaptoundecanoic acid (MUA)] resulted in a 16.8 ± 0.1-nm change in spectral color compared with the gold nanoparticle-coated surface (Fig. 2A). After the immobilization of protein G and antibodies (i.e., anti-casein antibody), we observed 19.8 ± 0.3-nm and 26.1 ± 0.3-nm changes, respectively. Further, these layer-by-layer constructions of surface modifications and antibody immobilizations resulted in a significant red-shift at each step (n = 96, P < 0.05) (Fig. 2A). We also analyzed the repeatability of the NE2RD results using the wavelength shifts and observed that >98% of the measurements were repeatable in surface modification experiments (SI Appendix, Fig. S2).
Fig. 2.
Evaluation of surface modification, durability, and pH and temperature effects on the well-plate prototype of the NE2RD. (A) Layer-by-layer surface chemistry approaches were performed to immobilize antibodies onto the NE2RD modified with surface activators [MUA, N-Ethyl-N'-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), and N-hydroxysulfosuccinimide (NHS)], antibody anchors (protein G), and antibodies (anti-casein monoclonal antibodies). The wavelength shift is the signal unit of the NE2RD, and the corresponding shifts were subtracted from the gold nanoparticle baseline value (519 nm) (n = 96, P < 0.05). H. Cumhur Tekin drew the schematic in Fig. 2A. (B) We assessed the NE2RD response to different buffers with various pH values. The pH value of each solution is indicated in the plot, and the red dashed lines in the plot represent the system resolution (1 nm). Here, we compared the results in different pH conditions with system resolution to understand any significant signal fluctuations. The changes in wavelength in different pH conditions are not significant different than the system resolution, indicating that the NE2RD surfaces are stable under the investigated conditions (SI Appendix, Fig. S3 A–C) (n = 16, P > 0.05). (C) We evaluated the NE2RD response to diverse temperature values. The temperature of each solution is shown in the plot, and the red dashed lines in the plot indicate the system resolution (1 nm). Here, we compared the results in different temperature conditions with system resolution to understand any significant signal fluctuations. The changes in wavelength in different temperature conditions are not statistically different than the system resolution, indicating that the NE2RD surfaces are stable under the investigated conditions (SI Appendix, Fig. S3D) (n = 16, P > 0.05). To perform statistical analysis, one-way ANOVA with Tukey's post hoc test was used with Bonferroni's multiple comparison test for equal variances for multiple comparisons. Horizontal brackets represent statistically significant differences between groups. Error bars represent mean ± SEM.
Device Durability and Response to pH, Ionic Strength of Samples, and Environmental Temperature Factors.
For broad applicability in clinical practice, biosensors need to operate at various ambient conditions. To evaluate the performance of our platform under various conditions, we tested the system for varying internal (e.g., pH, ionic content, and strength) and external (e.g., temperature) characteristics. The performance of the unmodified well-plate NE2RD prototype was first evaluated with diverse buffers ranging from pH 5.0 to pH 10.0, covering the ionic strength of various biological specimens including urine (30), tears (31), and sweat (32). A wide change in ionic strength and pH did not affect the performance of the device significantly, and the signal fluctuations in spectral color (wavelength) were smaller than the spectral resolution of the platform (1 nm) (n = 16, P > 0.05) (Fig. 2B and SI Appendix, Fig. S3 A–C). Thus, the NE2RD provided reliable measurements under varying ionic conditions. Overall, we focused on whole blood, serum, and whole saliva; other biospecimens in this pH range can be potentially tested using this platform technology. Further, the device performance was stable for up to 14 days of testing when the devices were incubated in buffers, which have a wide range of pH values (Fig. 2B and SI Appendix, Fig. S3 A–C). Platform responses to temperature changes ranging from 4 °C to 37 °C were also evaluated, and these changes had no significant effect on the performance (n = 16, P > 0.05) (Fig. 2C and SI Appendix, Fig. S3D).
Device Benchmarking, Limit of Detection, and Linear Dynamic Range.
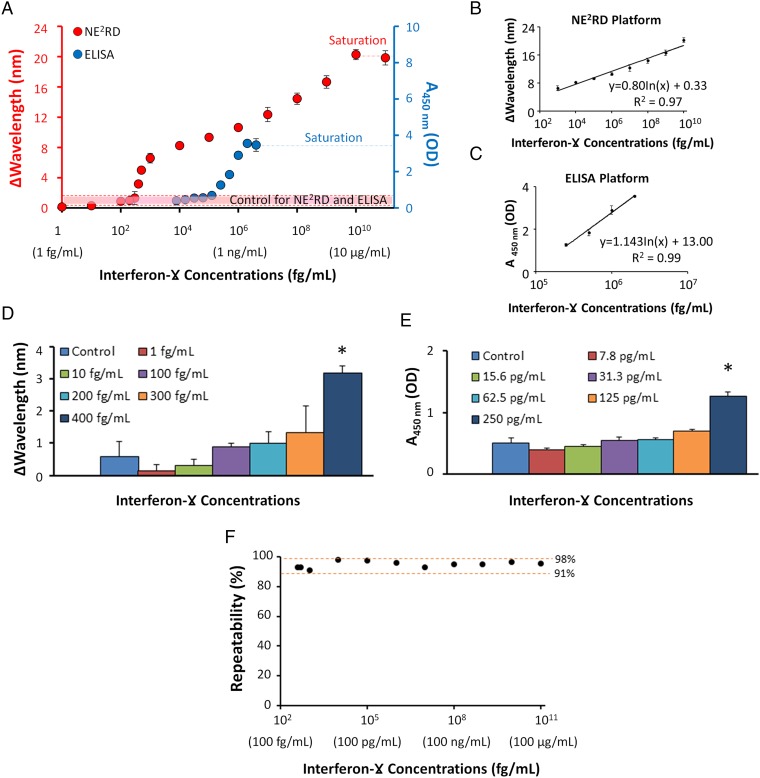
The ELISA platform is currently used as a gold-standard method (33) for biotarget detection and quantification in clinical and laboratory settings. Since quantitative readout is a key requirement for biosensing applications, we compared the performance of the well-plate NE2RD prototype with the standard ELISA (SI Appendix, Table S1). IFN-γ is a well-known biomarker for tuberculosis detection in IFN-γ–release assay (IGRA), and it was used as a model biotarget in these experiments (34, 35). We first immobilized anti–IFN-γ monoclonal capture antibodies on the substrate, and then, applied increasing concentrations of IFN-γ spiked into PBS to the surfaces (SI Appendix, Materials and Methods). To process the samples, the NE2RD requires a single loading and a single wash step by manual pipetting, whereas ELISA requires multiple steps (i.e., four loading and three wash steps) in which a biotinylated anti–IFN-γ antibody was used for signal enhancement including a chemical reaction between streptavidin-conjugated horseradish peroxidase (HRP) and 3,3′,5,5′-Tetramethylbenzidine for color development. We observed that the NE2RD quantitatively detected IFN-γ down to a sensitivity of femtograms per milliliter without any labels, tags, or signal amplification (Fig. 3A). We further observed a broad linear dynamic range with the NE2RD spanning up to eight orders of magnitude (Fig. 3B and SI Appendix, Fig. S4), whereas the ELISA platform could span a detection range of less than three orders of magnitude (Fig. 3C). Specifically, low-end concentrations were compared statistically with controls (PBS without IFN-γ) in both approaches for limit of detection (LOD). ELISA provided a detection limit down to 250 pg/mL (n = 3, P < 0.05), whereas the NE2RD detected concentrations as low as 400 fg/mL (n = 6, P < 0.05) (Fig. 3 D and E). Thus, the NE2RD provided a detection limit nearly three orders of magnitude better than that of ELISA. In the NE2RD, the wavelength changes were further analyzed to demonstrate the repeatability of the measurements across the range of detection; more than 91% of measurements were repeatable (Fig. 3F). Hence, the NE2RD has a superior detection limit, does not require labels or secondary tags to enhance the signal, and provides a broader linear dynamic range than the conventional biosensing approaches (SI Appendix, Table S1) such as sandwich-ELISA (36).
Fig. 3.
Comparison of the NE2RD performance with ELISA in terms of sensitivity and linear dynamic range parameters. (A) IFN-γ was analyzed as a model biomarker, and standard curves for IFN-γ detection were generated using the well-plate prototype of the NE2RD and ELISA. The shift in wavelength is the signal unit of the NE2RD, and the absorbance value at 450 nm is the signal unit of ELISA. PBS without IFN-γ was used as control in both approaches and is shown as a dashed red line. The surfaces of NE2RD reached saturation at a concentration of 10 µg/mL, whereas ELISA saturated at a concentration of 4 ng/mL. (B) In the NE2RD measurements, IFN-γ concentrations ranging from 1 pg/mL to 10 µg/mL were replotted to demonstrate the linear dynamic range using wavelength and extinction intensity data (SI Appendix, Fig. S4). The NE2RD has a linear dynamic range of eight orders of magnitude. (C) In ELISA measurements, IFN-γ concentrations ranging from 250 pg/mL to 2 ng/mL were replotted to demonstrate the linear dynamic range. ELISA provides a linear dynamic range of less than three orders of magnitude. (D and E) We evaluated the biotarget detection in the low-end concentration regime. The NE2RD limit of detection (LOD) was 400 fg/mL, whereas the ELISA LOD was 250 pg/mL. The asterisk represents a statistically significant difference compared with control and the other samples using one-way ANOVA with Tukey's post hoc test followed by Bonferroni's multiple comparison test for equal variances for multiple comparisons (n = 6–12, P < 0.05). Error bars represent mean ± SEM. (F) Data of IFN-γ measurements were analyzed further to demonstrate the repeatability of the NE2RD results. Concentrations from the LOD (400 fg/mL) to the highest concentration (10 µg/mL) were evaluated using the formula defined in SI Appendix, Materials and Methods. More than 91% of the measurements observed in IFN-γ experiments were repeatable.
Performance in Detecting Multiple Biotargets from Diverse Clinical Specimen Types.
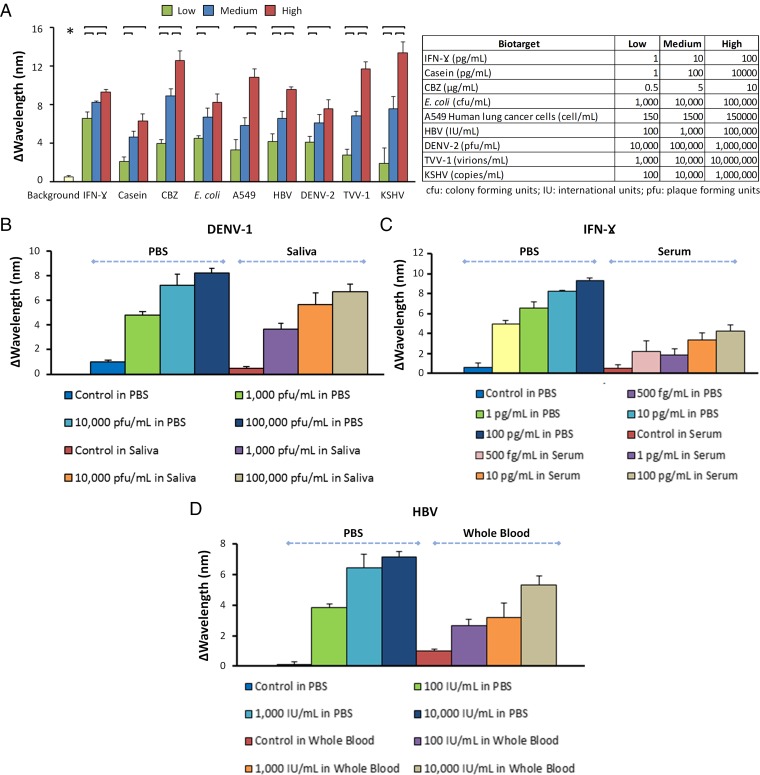
In addition to a diagnostic assay’s detection performance and dynamic range, quantitative detection of multiple biotargets is a critical parameter for an ideal biosensing platform. A multiple biotarget detection assay that senses, detects, and quantifies a wide set of biotargets such as proteins, drugs, viruses, bacteria, and eukaryotic cells will have a significant impact on clinical diagnostics and biomedical research, allowing timely monitoring of disease progression and response to therapies. Here, we tested the well-plate prototype of the NE2RD with a diverse panel of nine different biotargets spanning a wide range in size, from proteins to viruses to bacteria and cells spiked in PBS. These biotargets were a protein biomarker (IFN-γ), a drug (carbamazepine; CBZ), a protein allergen (casein), bacteria (Escherichia coli and Staphylococcus aureus), eukaryotic cells (A549 human lung adenocarcinoma epithelial cell line), and viruses (dsDNA virus, positive-stranded RNA virus, and dsRNA virus) including hepatitis B virus (HBV), dengue virus type 2 (DENV-2), Trichomonas vaginalis virus-1 (TVV-1), and Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) (Fig. 4A and SI Appendix, Materials and Methods). These biotargets were chosen to demonstrate the multiple biotarget detection and versatility of the NE2RD, and they are not specific to a single disease. To demonstrate the platform performance, surfaces were first modified with specific antibodies toward each biotarget. These biotargets were then detected on these active surfaces across a broad range of concentrations (designated as “low,” “medium,” and “high”) including six replicates for each concentration. Notably, increasing concentrations of each biotarget resulted in an incremental change in wavelength. Each wavelength shift was also analyzed using statistical assessments between these various concentrations and their background data (i.e., PBS without biotargets) (Fig. 4A). We observed that the background data were statistically lower than all the biotarget concentrations (n = 65, p < 0.05) (Fig. 4A).
Fig. 4.
Detection of multiple biotargets in diverse clinical specimen types. (A) Using a well-plate prototype of the NE2RD, we detect a panel of nine diverse biotargets: a protein biomarker (IFN-γ), a protein allergen (casein), a drug (CBZ), a bacterium (E. coli), a eukaryotic cell (A549 human lung cancer cell), and four different viruses (HBV, DENV-2, TVV-1, and KSHV). Shifts in wavelength are subtracted from the previous surface chemistry step (i.e., antibody immobilization). Biotarget concentrations (low, medium, and high) spiked into PBS are shown in the table (SI Appendix, Materials and Methods). Background data were collected from PBS without biotargets. Background data (*) are statistically lower than all sample concentrations (n = 65, P < 0.05). For statistical analysis, we used one-way ANOVA with Tukey's post hoc test followed by Bonferroni's multiple comparison test for equal variances for multiple comparisons (n = 6, P < 0.05). Horizontal brackets represent statistically significant differences between groups. (B–D) Evaluation of different clinically relevant specimen types (PBS, simulated saliva, serum, and whole blood) for the detection of DENV-1, IFN-γ, and HBV, respectively. PBS, simulated saliva, serum, and whole blood without biotargets were used as controls for each set of experiments. Error bars represent mean ± SEM.
Additionally, we tested a variety of biotargets spiked in more complex clinical matrices including unprocessed whole blood, serum, and simulated saliva (Fig. 4 B–D and SI Appendix, Materials and Methods). In DENV-1 experiments, we evaluated the detection of viruses spiked into PBS and into simulated saliva, which differs from PBS in terms of composition, ionic content, and ionic strength. We observed comparable changes in spectral color under both conditions across the same set of virus concentrations (Fig. 4B). Further, because of a variety of potential factors, such as excessive levels of serum, other proteins, and lipids, some biological fluids present a more complex microenvironment. The complexity of such biological fluids might interfere with the recognition of and interactions with biotargets. To assess the robustness and biosensing performance of the NE2RD, we also detected IFN-γ and HBV spiked into serum and whole blood, respectively (Fig. 4 C and D). In all experiments, PBS/clinical matrices without biotargets were included as controls. To reduce nonspecific binding onto the device surfaces, we used a blocking agent, i.e., 10% BSA (SI Appendix, Materials and Methods). Applying these clinically relevant samples, we again observed concentration-dependent changes in spectral color for each case. However, spiked experiments in clinical matrices showed higher standard errors than those in PBS due to the complex content of these clinical samples (Fig. 4 C and D). For instance, at the same spiking concentrations, the results for IFN-γ spiked in PBS ranged from 5.0 ± 0.4 to 9.3 ± 0.2 nm, whereas IFN-γ spiked in serum samples ranged from 2.2 ± 0.4 to 4.3 ± 0.6 nm (Fig. 4C). We observed the same trend in results for HBV spiked in PBS and whole blood (Fig. 4D). Further, the NE2RD provided quantitative measurements for whole blood- and serum-spiked samples that are statistically different from the controls (Fig. 4 C and D). In the future, the other biospecimens, including urine, which have different ionic strength and contents could be potentially tested for various biomarkers, since the NE2RD platform is stable over a broad range of pH values. Overall, these results point to the unique ability of the NE2RD to detect multiple biotargets and handle various sample types.
The applications in this study indicate that the NE2RD is a versatile platform with potential applications in multiple diseases. Proteins, allergens, drugs, bacteria, viruses, and cells are all different sizes. For many biosensing platforms, cutting across multiple size scales is much more challenging than detecting multiple types of the same biotarget. Further, similar biotargets such as virus subtypes can be captured and detected using monoclonal antibodies. In addition, by adapting specific surface-chemistry characteristics, the device can be widely tailored to capture and detect a nearly unlimited range of biotargets having reasonably well-described biomarkers and can work under essentially any bodily fluid for clinical applications.
Validation with Clinical Patient Samples and Multiplexing In Coinfections.
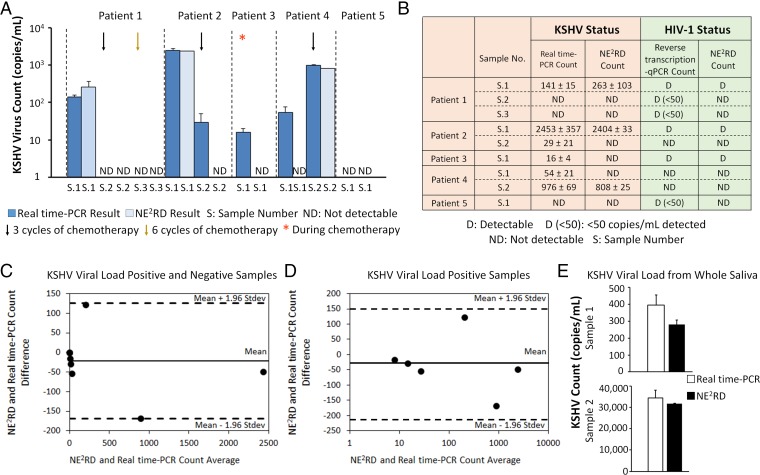
For a platform to be a viable clinical tool, it needs to translate from laboratory samples to clinical samples. Validation with clinical samples and time-dependent monitoring of biotargets are key factors for conventional clinical laboratory approaches to initiate therapy and provide information on patient status during treatment. Specifically, coinfections are critical for human health and alter the patient’s status more severely (37). For instance, Kaposi's sarcoma (KS) is the most common AIDS-related malignancy in the setting of KSHV and HIV-1 coinfection (38). Recent studies have demonstrated that KSHV coinfection significantly increases HIV replication in the acute stage and reactivates HIV in the latent stages (38, 39). Further, understanding the coinfection status of patients requires diverse and specific detection methods; multiple PCR primers, runs and reactions that increase the assay time and cost, and require large sample volumes for sample preprocessing. So far, there are no available tools that can provide a quantitative viral load rapidly and simultaneously for more than one virus type. In our experiments, we first generated standard curves including different concentrations of KSHV and HIV-1 for quantitative and qualitative measurements using the well-plate prototype of the NE2RD platform (SI Appendix, Materials and Methods and Fig. S5). For initial validation experiments, we applied a small cohort of nine serum samples from five different KS patients, who were undergoing chemotherapy, and measured changes in spectral color caused by viruses captured on the device surface (Fig. 5A). For KSHV detection, real-time PCR was used as the gold-standard comparison, and reverse transcription-PCR (RT-PCR) was used for HIV-1 detection (SI Appendix, Materials and Methods). Using the NE2RD, we observed that KSHV viral load of patient 1 decreased after both three and six cycles of chemotherapy. Additionally, we detected an increase in the viral load level of patient 4 after three cycles of chemotherapy. For patient 5, no KSHV viral load was observed. Given that the LOD of NE2RD for KSHV is 100 copies/mL, we detected the viral load trends and distinguished between positive and negative responses during chemotherapy. For instance, we observed that the KSHV viral load decreased in patient 2 after three cycles of chemotherapy and in patient 3 during the course of chemotherapy. We detected an increasing viral load in patient 4 despite three cycles of chemotherapy. During chemotherapy cycles, coinfected samples were also analyzed simultaneously to monitor the HIV status, and results were compared with the gold standard, i.e., RT-PCR (Fig. 5B). For HIV-1 viral load, the NE2RD presented a correlation with the gold-standard method above 100 copies/mL (SI Appendix, Fig. S5B).
Fig. 5.
Validation with clinical patient samples and multiplexing in coinfections. (A) Nine serum samples collected from five different individuals with KS were analyzed with real-time PCR and the NE2RD well-plate prototype. The effects of chemotherapy cycles on KSHV viral load were examined in both approaches. (B) During chemotherapy, the quantitative and qualitative coinfection status of these patients were indicated in detail. The results obtained from the NE2RD (n = 6) were compared with gold-standard methods [i.e., real-time PCR (n = 3) and quantitative reverse transcription-PCR (n = 3)] for KSHV and HIV-1, respectively. (C) All patient samples were analyzed using the Bland–Altman method for the results obtained from the NE2RD (n = 6) and real-time PCR (n = 3) for KSHV viral load. Statistical analysis showed no evidence of a systematic bias for KSHV viral load for the KS patient serum samples tested. (D) Real-time PCR reported that six of the nine samples were positive for KSHV viral load. We also reanalyzed these viral load-positive samples using the Bland–Altman method. No evidence of a systematic bias was observed between the NE2RD (n = 6) and real-time PCR (n = 3) for the six KSHV viral load-positive samples. (E) The NE2RD was used to measure the KSHV viral load in whole saliva samples collected at different times from the same KSHV-infected patient, and an increase in the patient’s viral load level was observed. No statistically significant difference was observed between the NE2RD (n = 3–10) and real-time PCR (n = 3–4) methods (P > 0.05). Statistical analysis was performed using one-way ANOVA with Tukey's post hoc test followed by Bonferroni's multiple comparison test for equal variances for multiple comparisons. Error bars represent mean ± SEM.
In the well-plate prototype of the NE2RD platform, each well requires only a minimal volume of 100 µL of serum per target to provide a dual viral load reading for the coinfections of KSHV and HIV using only adjacent wells of a single device rather than two different runs of sample-processing heavy PCR measurements. Thus, this unique advantage significantly reduces the assay time, cost, and technician effort in assays compared with the traditional PCR-based approaches (SI Appendix, Table S1). The NE2RD platform was able to detect trends in viral load for KSHV and HIV-1 during therapy, thus providing a unique, rapid dual-measurement tool for primary care type settings (SI Appendix, Fig. S5A). Further, Bland–Altman residual analysis was used to compare the results of NE2RD (n = 6) and real-time PCR (n = 3) for samples with positive and negative KSHV viral loads. The results revealed no evidence for a systematic bias between these two methods (Fig. 5C). We further analyzed samples with a positive KSHV viral load to evaluate the quantification performance of NE2RD and observed no bias compared with the results of real-time PCR (Fig. 5D). Additionally, we assessed KSHV viral load in whole saliva samples collected from a single KSHV-infected patient at different time points (Fig. 5E). As a result, an increase in the patient’s viral load level in whole saliva was monitored. The results from the NE2RD (n = 3–10) were also compared with the gold-standard (n = 3–4) method (real-time PCR), and no statistically significant difference was observed between the two methods (P > 0.05). Overall, the NE2RD provided a correlation with real-time PCR and RT- PCR methods for KSHV and HIV-1 viral load status, respectively. Hence, using the NE2RD, we obtained quantitative and qualitative clinical information before and during a course of therapy as well as detection using different clinical specimens, i.e., serum and whole saliva.
Benchmarking of the NE2RD Disposable Fluidic Chip-Integrated Portable Setup.
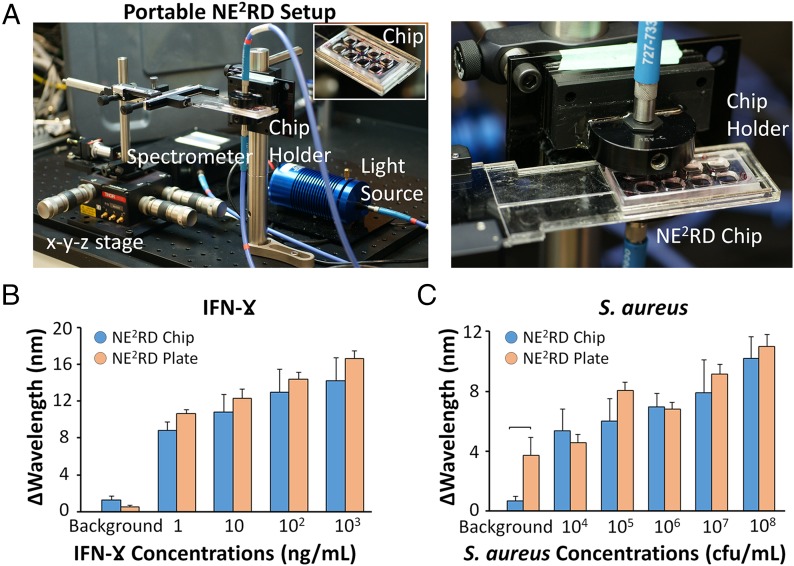
In POC settings, portability is a crucial parameter for the implementation of a biosensing platform. Thus, we fabricated a fluidic chip prototype of the NE2RD and measured the changes in spectral color using a customized portable tool, whose components weigh less than 3 lb (Fig. 6A and SI Appendix, Materials and Methods and Fig. S6). On the surface of the chip prototype, we used the surface chemistry steps described for IFN-γ and S. aureus (SI Appendix, Materials and Methods). In IFN-γ experiments, we evaluated the chips with concentrations ranging from 1 ng/mL to 1 µg/mL spiked in PBS (Fig. 6B). We also compared these results with the results obtained from the NE2RD plate prototype and observed no statistically significant difference between the results (n = 2–6, P > 0.05) (Fig. 6B). In S. aureus experiments, the disposable chips were tested with concentrations ranging from 104–108 cfu/mL spiked in PBS (Fig. 6C). Comparing these results with those of the well-plate–based NE2RD, we observed no statistically significant difference (n = 2–6, P > 0.05) (Fig. 6C). However, the results of chip prototype presented higher standard errors than those in the plate format (Fig. 6 B and C). For instance, in IFN-γ experiments, the results of fluidic chip format ranged from 14.2 ± 2.5 to 8.8 ± 1.0 nm, whereas the results of the well-plate prototype ranged from 16.7 ± 0.8 to 10.6 ± 0.4 nm (Fig. 6B). The same trend was observed in the S. aureus experiments (Fig. 6C). To determine the LOD, each wavelength shift was analyzed using statistical assessment between these various concentrations and their background (i.e., PBS without biotargets) (Fig. 6 B and C). In S. aureus experiments, we observed higher background signal in the well-plate prototype. The background signals can vary within an SEM because all the plates/chips are prepared in the laboratory using manual chemical modification steps. In addition, we also analyzed the repeatability of the chip prototype for IFN-γ and S. aureus experiments and observed that the repeatability factor for the measurements was >78% (SI Appendix, Figs. S7 and S8). Overall, these observations indicate that the NE2RD chip can be employed as a portable platform for multiple biotarget detection.
Fig. 6.
Evaluation of the NE2RD disposable fluidic chip-integrated portable set-up. (A) Images of the portable set-up and the NE2RD chip. The chip is placed between the light source and spectrometer, which are coupled with fiberoptic cables. The chip holder is manually controlled by x-y-z stage to measure changes in spectral color. Details are given in SI Appendix, Materials and Methods and Fig. S6. IFN-γ and S. aureus samples are evaluated using the fluidic chip and well-plate NE2RD prototypes. Background data were collected from PBS without biotargets (IFN-γ or S. aureus). (B) In IFN-γ experiments, concentrations ranging from 1 ng/mL to 1 µg/mL spiked in PBS were tested. In both prototypes, no statistical difference between the groups was observed (n = 2–6, P > 0.05). (C) In S. aureus experiments, concentrations ranging from 104–108 cfu/mL spiked in PBS were evaluated in both prototypes. The background signal obtained from the well-plate prototype was higher than that from the chip format (n = 3–6, P < 0.05). No statistical difference was observed in the sample concentrations from the two prototypes (n = 2–6, P > 0.05). For statistical analysis, we used one-way ANOVA with Tukey's post hoc test followed by Bonferroni's multiple comparison test for equal variances for multiple comparisons (P < 0.05). Horizontal brackets indicate statistically significant differences between groups. Error bars represent mean ± SEM.
Discussion
Biosensing assays have considerable impact on medical diagnostics to control diseases and to initiate and monitor the course of therapy. Since diseases have complex structures and networks that are affected by many biofunctional units, including various biomarkers, the ability to detect multiple biotargets has great utility in developing clinically useful diagnostic approaches for medical practice. Recently, nanoparticle-based detection strategies have been used to develop biosensing technologies by using their plasmonic features (26) and their aggregation strategies for biotarget detection (40). Although a well-plate–based nanoplasmonic platform was used to detect HIV (41), here we show, for the first time to our knowledge, that the NE2RD provides (i) an unprecedented multitarget capability whereby we detect multiple biotargets (i.e., protein biomarkers, drugs, protein allergens, bacteria, eukaryotic cells, and distinct viruses); (ii) versatility in detecting targets from distinct clinical matrices such as saliva, serum, and whole blood; and (iii) portability potentially allowing applications at the POC. The NE2RD platform is implemented in a disposable fluidic chip format and is integrated with a portable measurement tool for diagnostic applications in the POC settings. Further, the platform has a linear dynamic range up to eight orders of magnitude compared with ELISA's dynamic range of less than three orders of magnitude. Overall, we have created a platform that combines an inexpensive assay with rapid, quantitative multiple-biotarget detection. This device has the potential to become a POC diagnostic platform and represents a significant advance over the diagnostic tools and assays currently in clinical use.
Cost, Antibodies, and Translation.
The NE2RD uses affordable materials by integrating polymeric substrate and gold nanoparticles with a polystyrene surface to capture biotargets. Polystyrene surfaces are inexpensive substrates that are compatible with optical measurements. The costs of goods used for the fabrication and surface modification are provided in SI Appendix, Table S3; these costs potentially can be also reduced further with mass production. Further, automated preparation of surfaces by a robotic arm could decrease variations in the standard errors of the background signal and could eliminate batch-to-batch variations. Additionally, antibodies used in this work provide a specific, sensitive, and inexpensive method to detect biotargets. Based on our results using commercial and customized antibodies, we found these antibodies to be sensitive and specific to our biotargets. Some surface chemistry approaches such as Protein G-based strategies are reported to have long-term shelf life exceeding 8 months (42). The shelf life of immobilized antibodies potentially can be prolonged using various biological materials including trehalose, glycerol (43), and silk (44, 45) as well as by biopreservation (46) and dry storage approaches (47).
Future commercial development of the NE2RD will benefit from highly specialized quality-controlled antibodies, increasing the repeatability and success of this technology for clinical translation. The antibody cost per well is only 80 cents per assay using currently commercially available antibodies. The amount of antibody used on each test can be further reduced by bioprinting technologies (48) along with reduced cost for larger purchasing volume. This large-scale use of antibodies potentially will decrease even further the cost of antibodies used.
Ease of Operation.
The NE2RD requires only a single sample loading and a single wash step after incubation, rather than the multiple wash steps in standard ELISA.
Minimum Requirements for Process.
Only basic biology laboratory equipment, such as a pipette and spectrometer, are required for the whole measurement process, allowing the broad use of this technology globally. The next generation of the system would benefit from the elimination of the two manual steps by integration with an automation stage before measurement for sampling.
Detection of Multiple Biotargets and Sample-Type Independence.
The current prototypes of the NE2RD employs 96-well or fluidic chip formats to detect multiple biotargets simultaneously on a single device. Further, the device is compatible and stable with distinct sample conditions and provides measurements in multiple clinical samples. The next-generation device can be designed as a microarray-type platform for massively parallel detection.
Sensitivity and Linear Detection Range.
The NE2RD is able to detect concentrations as low as at a 400 fg/mL level for IFN-γ, nearly three orders of magnitude lower than the standard ELISA methods. The linear detection range of the NE2RD is five orders of magnitude broader than the ELISA. In addition to these advantages, the NE2RD has provided results comparable with those of the PCR-based assays for clinical studies in the range of 100 copies/mL or more.
Validation.
The current study demonstrates the rapid and simultaneous monitoring on a single device of patients’ coinfection status and their response to therapy using patient serum samples, and the elimination of multiple sample preprocessing steps further facilitates the translation of the technology to clinical and primary care settings. Other disease models also could be potentially validated on the same platform. In the future, clinical trials using this technology may further accelerate its clinical translation.
Easy Readout, Portability, and Quantification.
The NE2RD employs changes in spectral color on nanoparticles to provide quantitative data. To translate the readout signal into quantitative data, the device facilitates a custom software code that is run on a computer. The NE2RD chip prototype and portable setup can be used in multiple settings including primary care and POC. In the future, the current system could be integrated seamlessly with mobile platforms to make it more accessible to physicians and field healthcare operators, thus enabling measurements without the need for skilled personnel.
Materials and Methods
Engineering the NE2RD and Sampling of Biotargets.
In the well-plate prototype, we used polystyrene wells as plastic substrates for the construction of NE2RD surfaces. A poly(methyl methacrylate) (PMMA) layer, double-sided adhesive film, and a polystyrene surface were used to produce the surfaces of NE2RD chip prototype (Fig. 6A and SI Appendix, Materials and Methods). Briefly, to self-assemble gold nanoparticles (10-nm diameter), we incubated the nanoparticle solution on PLL-modified plastic substrates overnight at 4 °C. After construction of the nanoplasmonic layer, the surfaces were modified with several chemical activators to tether antibody anchors (49, 50), and then, specific antibodies for each biotarget were immobilized onto the surfaces. To minimize nonspecific binding on the NE2RD surface, 10% BSA, a blocking agent, was incubated for 1 hour before biotargets were sampled. The surfaces were washed out three times with 1× PBS using a micropipette, and biotargets were then introduced to the device surfaces, followed by washing three times with 1× PBS to remove all unbound biotargets. To measure the changes in spectral color, the surfaces were filled with 100 µL of PBS. The detailed protocols for the construction and modification of NE2RD surfaces and preparation of samples are available in SI Appendix, Materials and Methods.
Spectral Measurements and Data Analysis.
In plate and chip prototypes, detectable shifts in spectral color of the maximum extinction point of gold nanoparticles were collected for data analysis. To analyze spectral color data, we developed a MATLAB code, generating a curve fitting raw data using a Fourier-type expansion with eight harmonics. All shift data for spectral color measurements are indicated as the mean of wavelength measurements ± SEM. The details in spectral measurements, the curve-fitting method, and data analysis are available in SI Appendix, Materials and Methods.
ELISA Platform.
We performed a conventional ELISA for the detection of IFN-γ spiked in PBS samples. The detailed protocol in ELISA experiments is available in SI Appendix, Materials and Methods.
Statistical Analysis.
To assess each surface modification and biotarget binding events, we used statistical analyses using Minitab software (Release 14; Minitab Inc.). The details are available in SI Appendix, Materials and Methods.
Human Subject Consent.
The study to collect serum and whole saliva samples from KS patients (KSHV/HIV coinfected patients) was reviewed and approved by the Dana–Farber Cancer Institute/Harvard Cancer Center Institutional Review Board. Informed consent was obtained from all subjects.
Supplementary Material
Acknowledgments
We thank Mehlika Toy for collaboration and discussion in HBV studies; Mark Anthony Lifson and Jaeyong Yang for critical discussions and reviews; and Yusuf Yesil for his help in Fig. 1 and H. Cumhur Tekin for his help in Fig. 2A, schematic. This work was supported by NIH Grants R01AI093282, R01AI081534, U54EB15408, R21AI087107, R01AI076442, and U54AI057159; by NIH/National Institute of Allergy and Infectious Disease Grants R56AI091889, R01AI079085, and R01AI076442; NIH/National Cancer Institute Grant R01CA082036; by the Brigham and Women’s Hospital Bright Futures Prize; and by the Epilepsy Foundation Shark Tank Prize. M.L.V. was supported by NIH Grant 5T32AI007061.
Footnotes
Conflict of interest statement: U.D. is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions, and (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions. U.D.'s interests were viewed and managed in accordance with the conflict of interest policies.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510824112/-/DCSupplemental.
References
- 1.Peeling RW, Smith PG, Bossuyt PMM. A guide for diagnostic evaluations. Nat Rev Microbiol. 2008;6(11) Suppl:S2–S6. [PubMed] [Google Scholar]
- 2.Banoo S, et al. TDR Diagnostics Evaluation Expert Panel Evaluation of diagnostic tests for infectious diseases: General principles. Nat Rev Microbiol. 2008;6(11) Suppl:S16–S26. [PubMed] [Google Scholar]
- 3.Fichorova RN. Guiding the vaginal microbicide trials with biomarkers of inflammation. J Acquir Immune Defic Syndr. 2004;37(Suppl 3):S184–S193. [PMC free article] [PubMed] [Google Scholar]
- 4.Anker JN, et al. Biosensing with plasmonic nanosensors. Nat Mater. 2008;7(6):442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 5.Cooper MA. Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002;1(7):515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 6.Yildiz UH, et al. Recent advances in micro/nanotechnologies for global control of hepatitis B infection. Biotechnol Adv. 2015;33(1):178–190. doi: 10.1016/j.biotechadv.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shafiee H, et al. Emerging technologies for point-of-care management of HIV infection. Annu Rev Med. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. [DOI] [PubMed] [Google Scholar]
- 8.Tasoglu S, et al. Advances in Nanotechnology and Microfluidics for Human Papillomavirus Diagnostics. Proc IEEE. 2015;103(2):161–178. [Google Scholar]
- 9.Shafiee H, et al. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci Rep. 2014;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T, et al. An aggregated perylene-based broad-spectrum, efficient and label-free quencher for multiplexed fluorescent bioassays. Biosens Bioelectron. 2014;58:320–325. doi: 10.1016/j.bios.2014.02.083. [DOI] [PubMed] [Google Scholar]
- 11.Hu R, et al. Multicolor fluorescent biosensor for multiplexed detection of DNA. Anal Chem. 2014;86(10):5009–5016. doi: 10.1021/ac500618v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arlett JL, Myers EB, Roukes ML. Comparative advantages of mechanical biosensors. Nat Nanotechnol. 2011;6(4):203–215. doi: 10.1038/nnano.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lissandrello C, et al. Nanomechanical motion of Escherichia coli adhered to a surface. Appl Phys Lett. 2014;105(11):113701. doi: 10.1063/1.4895132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das J, et al. An ultrasensitive universal detector based on neutralizer displacement. Nat Chem. 2012;4(8):642–648. doi: 10.1038/nchem.1367. [DOI] [PubMed] [Google Scholar]
- 15.Shafiee H, et al. Acute on-chip HIV detection through label-free electrical sensing of viral nano-lysate. Small. 2013;9(15):2553–2563, 2478. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21(10):1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 17.Viswanathan S, et al. Graphene–protein field effect biosensors: Glucose sensing. Mater Today 2015 , 10.1016/j.mattod.2015.04.003. [Google Scholar]
- 18.Brolo AG. Plasmonics for future biosensors. Nat Photonics. 2012;6(11):709–713. [Google Scholar]
- 19.Sönnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat Biotechnol. 2005;23(6):741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 20.Tokel O, et al. 2015 Portable microfluidic integrated plasmonic platform for pathogen detection. Sci Rep 5:9152. [Google Scholar]
- 21.Sevimli S, Inci F, Zareie HM, Bulmus V. Well-defined cholesterol polymers with pH-controlled membrane switching activity. Biomacromolecules. 2012;13(10):3064–3075. doi: 10.1021/bm300846e. [DOI] [PubMed] [Google Scholar]
- 22.Inci F, Celik U, Turken B, Özer HÖ, Kok FN. Construction of P-glycoprotein incorporated tethered lipid bilayer membranes. Biochemistry and Biophysics Reports. 2015;2:115–122. doi: 10.1016/j.bbrep.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haes AJ, Zou SL, Schatz GC, Van Duyne RP. Nanoscale optical biosensor: Short range distance dependence of the localized surface plasmon resonance of noble metal nanoparticles. J Phys Chem B. 2004;108(22):6961–6968. [Google Scholar]
- 24.Beeram SR, Zamborini FP. Selective attachment of antibodies to the edges of gold nanostructures for enhanced localized surface plasmon resonance biosensing. J Am Chem Soc. 2009;131(33):11689–11691. doi: 10.1021/ja904387j. [DOI] [PubMed] [Google Scholar]
- 25.Feuz L, Jönsson P, Jonsson MP, Höök F. Improving the limit of detection of nanoscale sensors by directed binding to high-sensitivity areas. ACS Nano. 2010;4(4):2167–2177. doi: 10.1021/nn901457f. [DOI] [PubMed] [Google Scholar]
- 26.Tokel O, Inci F, Demirci U. Advances in plasmonic technologies for point of care applications. Chem Rev. 2014;114(11):5728–5752. doi: 10.1021/cr4000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Güngör K, Ünal E, Demir HV. Nanoplasmonic surfaces enabling strong surface-normal electric field enhancement. Opt Express. 2013;21(20):23097–23106. doi: 10.1364/OE.21.023097. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, et al. A 3D localized surface plasmon resonance biosensor for the study of trivalent arsenic binding to the ArsA ATPase. Biosens Bioelectron. 2012;38(1):19–26. doi: 10.1016/j.bios.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicoletti O, et al. Three-dimensional imaging of localized surface plasmon resonances of metal nanoparticles. Nature. 2013;502(7469):80–84. doi: 10.1038/nature12469. [DOI] [PubMed] [Google Scholar]
- 30.MedlinePlus Medical Encyclopedia 2013 Urine pH test. Available at www.nlm.nih.gov/medlineplus/ency/article/003583.htm Accessed May 8, 2015.
- 31.Rahman MQ, Chuah KS, Macdonald ECA, Trusler JPM, Ramaesh K. The effect of pH, dilution, and temperature on the viscosity of ocular lubricants—shift in rheological parameters and potential clinical significance. Eye (Lond) 2012;26(12):1579–1584. doi: 10.1038/eye.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann F, Mandol L. Studies of pH of sweat produced by different forms of stimulation. J Invest Dermatol. 1955;24(3):225–246. doi: 10.1038/jid.1955.36. [DOI] [PubMed] [Google Scholar]
- 33.Song Y, et al. Multiplexed volumetric bar-chart chip for point-of-care diagnostics. Nat Commun. 2012;3:1283. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Inci F, De Libero G, Singhal A, Demirci U. Point-of-care assays for tuberculosis: Role of nanotechnology/microfluidics. Biotechnol Adv. 2013;31(4):438–449. doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mani V, et al. Emerging technologies for monitoring drug-resistant tuberculosis at the point-of-care. Adv Drug Deliv Rev. 2014;78:105–117. doi: 10.1016/j.addr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Kumar RB, Dai H, Feldman BJ. A plasmonic chip for biomarker discovery and diagnosis of type 1 diabetes. Nat Med. 2014;20(8):948–953. doi: 10.1038/nm.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antman K, Chang Y. Kaposi’s sarcoma. N Engl J Med. 2000;342(14):1027–1038. doi: 10.1056/NEJM200004063421407. [DOI] [PubMed] [Google Scholar]
- 38.Caselli E, et al. Human herpesvirus 8 enhances human immunodeficiency virus replication in acutely infected cells and induces reactivation in latently infected cells. Blood. 2005;106(8):2790–2797. doi: 10.1182/blood-2005-04-1390. [DOI] [PubMed] [Google Scholar]
- 39.Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res. 2008;6(5):388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shafiee H, et al. 2015. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci Rep 5:8719.
- 41.Inci F, et al. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano. 2013;7(6):4733–4745. doi: 10.1021/nn3036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yakovleva J, Davidsson R, Bengtsson M, Laurell T, Emnéus J. Microfluidic enzyme immunosensors with immobilised protein A and G using chemiluminescence detection. Biosens Bioelectron. 2003;19(1):21–34. doi: 10.1016/s0956-5663(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 43.Johnson M. Antibody shelf life/how to store antibodies. Mater Methods. 2012;2:120. [Google Scholar]
- 44.Guziewicz N, Best A, Perez-Ramirez B, Kaplan DL. Lyophilized silk fibroin hydrogels for the sustained local delivery of therapeutic monoclonal antibodies. Biomaterials. 2011;32(10):2642–2650. doi: 10.1016/j.biomaterials.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, et al. Stabilization of vaccines and antibiotics in silk and eliminating the cold chain. Proc Natl Acad Sci USA. 2012;109(30):11981–11986. doi: 10.1073/pnas.1206210109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Song YS, et al. Vitrification and levitation of a liquid droplet on liquid nitrogen. Proc Natl Acad Sci USA. 2010;107(10):4596–4600. doi: 10.1073/pnas.0914059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramachandran S, Fu E, Lutz B, Yager P. Long-term dry storage of an enzyme-based reagent system for ELISA in point-of-care devices. Analyst (Lond) 2014;139(6):1456–1462. doi: 10.1039/c3an02296j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durmus NG, Tasoglu S, Demirci U. Bioprinting: Functional droplet networks. Nat Mater. 2013;12(6):478–479. doi: 10.1038/nmat3665. [DOI] [PubMed] [Google Scholar]
- 49.Wang S, et al. Portable microfluidic chip for detection of Escherichia coli in produce and blood. Int J Nanomedicine. 2012;7:2591–2600. doi: 10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, et al. Efficient on-chip isolation of HIV subtypes. Lab Chip. 2012;12(8):1508–1515. doi: 10.1039/c2lc20706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaster RS, et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15(11):1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer SK, et al. Emerging technologies to increase ligand binding assay sensitivity. AAPS J. 2015;17(1):93–101. doi: 10.1208/s12248-014-9682-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thaxton CS, et al. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci USA. 2009;106(44):18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14(8):869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Knoll W, Dostalek J. Bacterial pathogen surface plasmon resonance biosensor advanced by long range surface plasmons and magnetic nanoparticle assays. Anal Chem. 2012;84(19):8345–8350. doi: 10.1021/ac301904x. [DOI] [PubMed] [Google Scholar]
- 56.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7(12):821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.