Significance
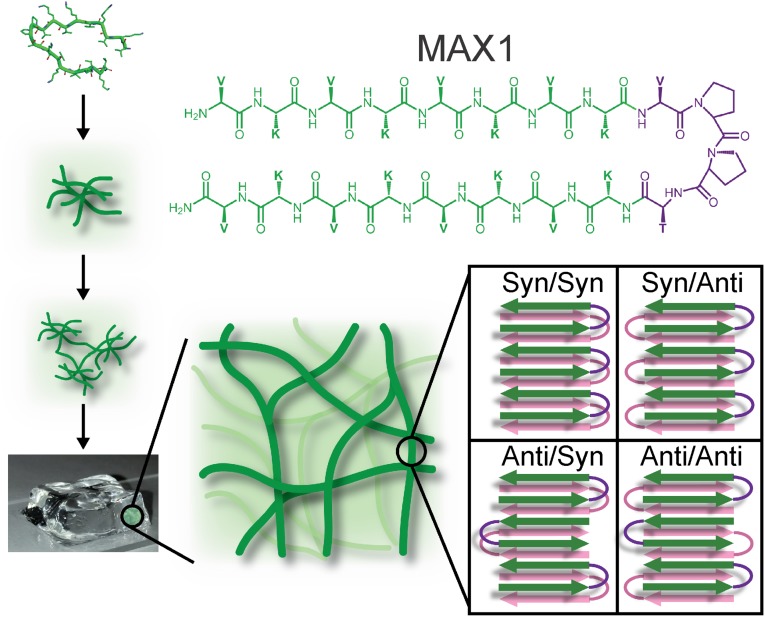
Based on the propensity of naturally occurring peptides to assemble into polymorphic fibrils, one might assume that polymorphism and peptide self-assembly go hand-in-hand. Contrary to this expectation, we find that the designed peptide MAX1 assembles with high fidelity into monomorphic fibrils that comprise a kinetically trapped hydrogel network. Our solid-state NMR data lead to a complete molecular structural model, in which MAX1 β-hairpins form a β-sheet bilayer with specific molecular conformations and intermolecular alignments within and between layers.
Keywords: polymorphism, amyloid, β-hairpin, cross-β structure, peptide design
Abstract
Most, if not all, peptide- and protein-based hydrogels formed by self-assembly can be characterized as kinetically trapped 3D networks of fibrils. The propensity of disease-associated amyloid-forming peptides and proteins to assemble into polymorphic fibrils suggests that cross-β fibrils comprising hydrogels may also be polymorphic. We use solid-state NMR to determine the molecular and supramolecular structure of MAX1, a de novo designed gel-forming peptide, in its fibrillar state. We find that MAX1 adopts a β-hairpin conformation and self-assembles with high fidelity into a double-layered cross-β structure. Hairpins assemble with an in-register Syn orientation within each β-sheet layer and with an Anti orientation between layers. Surprisingly, although the MAX1 fibril network is kinetically trapped, solid-state NMR data show that fibrils within this network are monomorphic and most likely represent the thermodynamic ground state. Intermolecular interactions not available in alternative structural arrangements apparently dictate this monomorphic behavior.
MAX1 is a 20-residue peptide designed de novo to fold into an amphiphilic β-hairpin that self-assembles to form a fibrillar network within a self-supporting hydrogel (1). The MAX1 gel exhibits shear thin-recovery rheological behavior (2), is cytocompatible toward mammalian cells, yet is inherently antimicrobial (3) and thus has applications in tissue engineering and drug delivery. In addition to exploring the utility of the gel, we seek to understand the mechanism of gelation, the macroscale morphology of its fibrillar network, and the underlying molecular structure of its fibrils.
MAX1 contains two segments of alternating lysine and valine residues, connected by a four-residue turn-forming segment. When initially dissolved in water, electrostatic repulsions among protonated lysine sidechains lead to an ensemble of monomeric random coil conformations (1). Peptide folding and self-assembly, leading to gelation (Fig. 1), can be triggered by attenuating electrostatic repulsions, by adjusting the solution pH and/or ionic strength. Increasing the solution temperature also drives hydrophobic collapse of valine sidechains, further favoring MAX1 assembly. According to circular dichroism (1), cryo-transmission electron microscopy (TEM) (4), small-angle neutron scattering (5), and dynamic light scattering coupled with rheological measurements (4), soon after the triggering event, peptides assemble into branched clusters of β-sheet–rich, semiflexible nanofibrils throughout the solution. Individual clusters contain dangling fibril ends that grow and interpenetrate neighboring clusters as the network evolves. Multiple particle tracking microrheology shows that the time at which the fibril network percolates the entire sample volume, defining the gel point, is less than 1 min at 1% (wt/vol) peptide (6). In this mechanism of gelation, the growing fibrils become kinetically trapped in the evolving network as they percolate the sample volume. Fibrils do not precipitate, but rather form a 3D random network that defines the gel state.
Fig. 1.
Self-assembly of MAX1 monomers leads to a hydrogel made from a kinetically trapped network of fibrils, each of which contains a putative double-layered β-sheet structure comprised of β-hairpins. Four possible supramolecular structures are shown, differing in the nature of intermolecular alignments within and between the β-sheets.
Full structural models for naturally occurring amyloid (7–10) and prion (11) fibrils have been developed from solid-state NMR data, but less is known about fibril structures within designed peptide hydrogels. Disease-associated amyloid and prion fibrils are known to be polymorphic at the molecular structural level (7, 8, 12, 13), implying that structures within peptide and protein fibrils are generally not determined uniquely by amino acid sequences and do not necessarily represent thermodynamic ground states. A model for fibrils formed by the designed peptide RADA16-I has been proposed by Cormier et al. based on solid-state NMR data (14), in which RADA16-I monomers form single β-strands within a double-layered cross-β structure. Solid-state NMR spectra of this hydrogel-forming peptide also indicate coexistence of several distinct fibril structures, suggesting that polymorphism may also be a trait of designed sequences.
Here, we use solid-state NMR to develop a full structural model for MAX1 fibrils, including molecular conformation, β-sheet organization, and intersheet interactions, with experimental restraints on all levels of structure. We find that MAX1 self-assembles with high fidelity to form monomorphic fibrils with well-defined and uniform structures. This structural homogeneity suggests that although the evolution of the fibril network is governed by kinetics, the molecular structure within MAX1 fibrils most likely represents the thermodynamic ground state. Additionally, results described below represent, to our knowledge, the first complete experimentally based model for a cross-β fibril structure comprised of β-hairpins.
Results
MAX1 Fibrils Are Monomorphic.
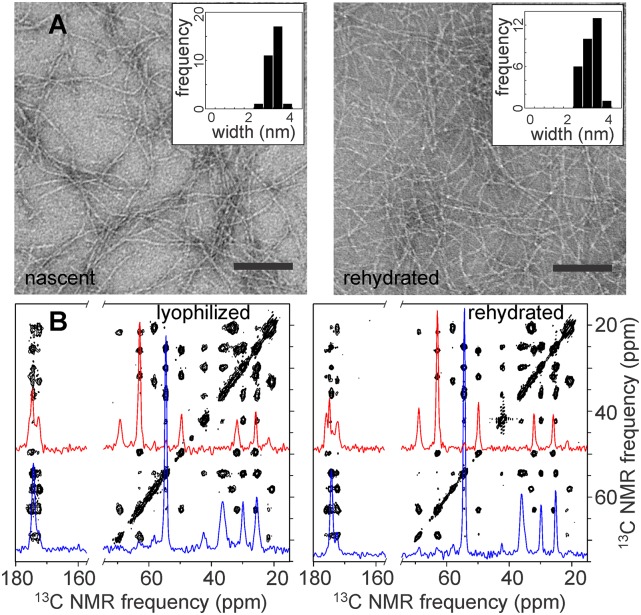
TEM images of nascent MAX1 assemblies (Fig. 2A, Left) reveal fibrils of uniform appearance, with diameters of ∼3.5 nm. Along with previous atomic force microscopy (AFM) data, which indicate a fibril height of 2.5 nm (15), these dimensions are consistent with models in which β-hairpins assemble into a double-layered cross-β structure, i.e., a pair of ribbon-like β-sheets in which β-strands run approximately perpendicular to and hydrogen bonds run approximately parallel to the fibril growth axis. Solid-state NMR experiments were performed on lyophilized and lyophilized/rehydrated MAX1 hydrogels. After rehydration of a lyophilized hydrogel, TEM images show quite similar fibril morphology and diameters (Fig. 2A, Right), indicating that MAX1 fibril structures are not perturbed during preparation of solid-state NMR samples.
Fig. 2.
Characterization of MAX1 fibrils by TEM and solid-state NMR. (A) Negatively stained TEM images of nascent MAX1 fibrils (Left) and fibrils in a rehydrated hydrogel after lyophilization (Right). (Insets) Average fibril widths for each sample were ∼3.5 nm (N = 30). (Scale bar, 100 nm.) (B) Two-dimensional solid-state 13C–13C NMR spectra of MAX1 sample IX as a lyophilized gel (Left) and a rehydrated gel (Right). One-dimensional slices at Cα chemical shifts of P11 and T12 (red) and K8 and K11 (blue) are shown. Spectra were obtained at 187.8 13C NMR frequency with MAS at 17.00 kHz and 200-ms spin diffusion mixing periods.
Fig. 2B shows 2D solid-state 13C–13C NMR spectra of MAX1 fibrils that were uniformly 15N,13C-labeled at residues 8–13 and 13C-labeled at the carbonyl site of DP10 (sample IX in Table 1). A single set of cross-peak signals is observed, corresponding to a single set of 13C chemical shifts for all uniformly labeled residues. Two-dimensional spectra of lyophilized and rehydrated gel samples are identical except that NMR linewidths are somewhat reduced in the rehydrated state (0.7–0.9 ppm versus 0.8–1.2 ppm full width at half maximum), presumably due to small-amplitude local motions that require hydration. Together with the TEM images, these data show that lyophilization does not perturb the molecular structure within MAX1 fibrils. Importantly, the 2D spectra indicate that no significant nonfibrillar material is present and that all MAX1 molecules within the fibrils have nearly identical conformations and structural environments. This rules out the possibility that MAX1 fibrils contain mixtures of the supramolecular structures shown in Fig. 1, strongly arguing against the possibility that MAX1 fibrils are inherently polymorphic, in contrast with disease-associated amyloids (7, 8, 12, 13).
Table 1.
Isotopic labeling patterns of MAX1 samples
| Sample number | Selectively labeled residues | Uniformly 15N,13C-labeled residues |
| I | 1-13C-V1; 15N-V20 | |
| II | 1-13C-V3; 15N-V18 | |
| III | 1-13C-V5; 15N-V16 | |
| IV | 1-13C-V7; 15N-V14 | |
| V | 1-13C-V1,V16* | |
| VI | 1-13C-V3,V16* | |
| VII | 1-13C-V5,V16* | |
| VIII | 1-13C-V7,V16* | |
| IX | 1-13C-DP10 | K8,V9,P11,T12,K13 |
| X | V1,V20,P11,T12 | |
| XI | V20,P11,T12 |
Mixture (1:1) of peptide molecules labeled at these two sites, with one 13C label in each molecule.
MAX1 Adopts a β-Hairpin Conformation.
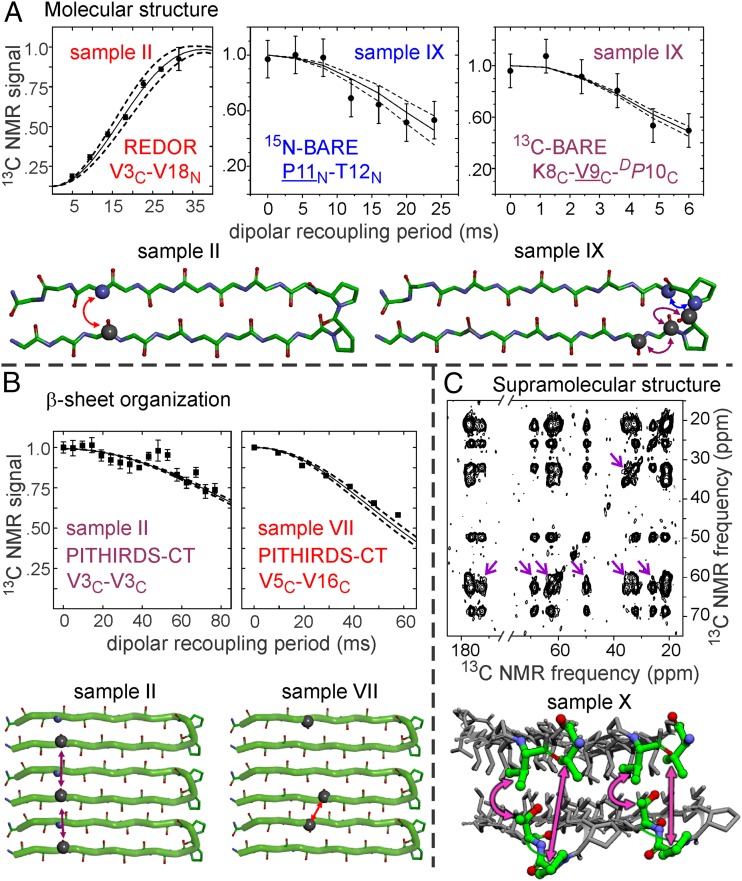
Experimental restraints at three distinct levels of structure, depicted in Fig. 3, were obtained from a series of solid-state NMR measurements on samples with the labeling patterns in Table 1. The first step was to obtain restraints on the conformation of individual MAX1 molecules. Conformational information is contained in 13C and 15N chemical shifts (SI Appendix, Table S1), which were measured in 1D and 2D solid-state NMR spectra (Fig. 2 and SI Appendix, Figs. S1, S5, and S6). Backbone torsion angles were predicted from chemical shifts for certain sites in residues 9–13, using the TALOS+ program (16). 13C chemical shifts for backbone carbonyl sites of all valine residues in samples II–IX were found to be in the 173.3–173.9-ppm range, significantly upfield from the random-coil value of 176.3 ppm (17) and hence consistent with β-strand conformations at valines in residues 3–9 and 16.
Fig. 3.
Measurements of magnetic dipole–dipole couplings that provide experimental restraints on internuclear distances in MAX1 fibrils. (A) 15N–13C REDOR data from sample II and 15N-BARE and 13C-BARE data from sample IX support a β-hairpin conformation. (B) 13C PITHIRDS-CT data from samples II and VII indicate a Syn alignment of MAX1 β-hairpins within each β-sheet. The relevant residues and labels (C for carbonyl 13C, N for amide 15N) are indicated in each plot. The detected sites in BARE data are underlined. Solid lines in all plots are averaged simulations using atomic coordinates from the final ensemble of 20 MAX1 fibril structures, with dashed lines indicating 1 SD. Error bars on experimental data points represent uncertainties due to the root-mean-squared noise in the solid-state NMR spectra. (C) Two-dimensional solid-state 13C–13C NMR spectrum of MAX1 sample X as a rehydrated gel, with a 500-ms RAD mixing period, obtained at 100.8-MHz 13C NMR frequency with 9.00-kHz MAS. Arrows indicate long-range interresidue cross-peaks that serve as restraints on supramolecular structure in MAX1 fibrils.
Additional conformational restraints were obtained from quantitative measurements of nuclear magnetic dipole–dipole couplings with the rotational echo double resonance (REDOR) and the 15N and 13C backbone recoupling (15N- and 13C-BARE) techniques (18, 19). Examples of the data are shown in Fig. 3A. The complete data sets are shown in SI Appendix, Fig. S2 A–C. Each of these techniques includes a dipolar recoupling period, during which the relevant couplings are activated by an appropriate radiofrequency (rf) pulse sequence, before the detection of solid-state NMR signals. Dipole–dipole coupling strengths are inversely proportional to the cubes of the distances between coupled nuclear spins. Shorter distances (i.e., stronger couplings) produce more rapid decays of the NMR signals with increasing recoupling period.
15N–13C REDOR measurements on samples I–IV (Fig. 3A and SI Appendix, Fig. S2A) indicate distances of 4.3–4.5 Å between carbonyl 13C labels at residues 1, 3, 5, and 7 and amide 15N labels at residues 20, 18, 16, and 14, respectively. These distances are consistent with hydrogen bonding between 13C-labeled and 15N-labeled residues, as expected in a β-hairpin conformation. The intramolecular nature of these hydrogen bonds is supported by REDOR measurements on a MAX1 fibril sample in which molecules with the labeling pattern of sample II were diluted in unlabeled molecules (SI Appendix, Fig. S3). In particular, the REDOR data show that the β-hairpin structure extends to the N- and C-termini of MAX1, although the broadened carbonyl 13C line for V1 (SI Appendix, Fig. S1B) and the REDOR data for sample I (SI Appendix, Fig. S2A) suggest somewhat greater disorder and a broader distribution of 15N–13C distances at the termini. Importantly, the intramolecular hydrogen bonds must involve pairs of valine residues, rather than pairs of lysine residues, a finding that places strong constraints on the turn conformation.
Evidence from solid-state NMR for β-hairpin conformations in fibrils formed by the related peptide MAX8 has been reported previously by Leonard et al. (20). Their data only define distances from V3 to K17 and V18.
Further restraints on the conformation in residues 8–12 of MAX1 were obtained from 15N- and 13C-BARE measurements on sample IX. As previously described (19), 15N-BARE measurements probe 15N–15N dipole-dipole couplings of the backbone amide nitrogen of residue k to amide nitrogens of residues k-1 and k+1, which depend on backbone torsion angles ψk−1, ϕk, and ψk. (When only residue k − 1 or k+1 is 15N-labeled, the data depend only on ψk−1 or ψk, or equivalently on one 15N–15N distance.) 13C-BARE measurements probe 13C–13C dipole–dipole couplings of the backbone carbonyl carbon of residue k to carbonyl carbons of residues k − 1 and k + 1, which depend on backbone torsion angles ϕk, ψk, and ϕk+1. Experimental 15N-BARE and 13C-BARE curves for sample IX (Fig. 3A and SI Appendix, Fig. S2 B and C) show unique signal decays for each labeled residue, consistent with a β-turn conformation in residues 8–12. Thus, MAX1 adopts a well-defined β-hairpin conformation in the fibrillar solid state, without competing alternative folds that could lead to polymorphism.
Evidence for Syn Alignment Within β-Sheets.
As illustrated in Fig. 1, several supramolecular structures with different β-hairpin orientations are consistent with information derived from scattering and microscopy experiments. In both Syn/Syn and Syn/Anti structures, β-hairpins within a given β-sheet have their turns on the same edge of the β-sheet. In the Syn/Syn structure, the two β-sheets have their turns on the same side of the fibril, whereas in the Syn/Anti structure, the two β-sheets have their turns on opposite sides. An ideal Syn/Syn structure has twofold axes of rotational symmetry perpendicular to the fibril growth axis and parallel to the β-sheet plane, whereas an ideal Syn/Anti structure has approximate twofold symmetry about the fibril growth axis (see below). In both Anti/Syn and Anti/Anti structures, β-hairpins within a given β-sheet have their turns on alternating edges of the β-sheet. In the Anti/Syn structure, facially associated β-hairpins from the two β-sheets have their turns on the same side of the fibril, whereas in the Anti/Anti structure, facially associated β-hairpins have their turns on opposite sides. An ideal Anti/Syn structure has twofold symmetry axes perpendicular to the fibril growth axis and parallel to the β-sheet plane, whereas an ideal Anti/Anti structure has approximate twofold symmetry about the fibril growth axis.
Experimental restraints on β-sheet organization in MAX1 fibrils were obtained from measurements of intermolecular 13C–13C dipole–dipole couplings among carbonyl 13C labels in samples I–VIII, using the constant-time, 1/3-rotor-period, π-pulse recoupling (PITHIRDS-CT) technique (21). Examples of the data are shown in Fig. 3B. The complete data sets are shown in SI Appendix, Fig. S2 D and E. In PITHIRDS-CT measurements on samples I–IV, signals decayed by 25–40% as the recoupling period increased from 0 to 76.8 ms. These signal decays indicate 13C–13C distances greater than 8 Å (21) and are similar to decays observed for natural-abundance 13C signals in unlabeled samples (22). The absence of shorter distances rules out Anti alignments of β-hairpins with all possible registries of intermolecular hydrogen bonds, because any Anti alignment would imply intermolecular 13C–13C distances of ∼6.1 Å in at least one of samples I–IV (SI Appendix, Fig. S4).
To test for a Syn alignment within individual β-sheets, samples V–VIII were prepared from 1:1 mixtures of molecules with a carbonyl 13C label at V16 and molecules with carbonyl 13C labels at V1, V3, V5, or V7. Use of 1:1 mixtures eliminated effects of intramolecular couplings in each sample. PITHIRDS-CT measurements revealed more rapid signal decay for sample VII than for samples V, VI, or VIII (SI Appendix, Fig. S2E). For sample VII, the decay by 42% at a recoupling period of 57.6 ms indicates an intermolecular distance of ∼6.4 Å, after taking into account the random distribution of V5-labeled and V16-labeled molecules within sample VII. To produce an intermolecular carbonyl–carbonyl distance that is ∼6.4 Å in sample VII, with longer distances observed in samples V, VI, and VIII, intermolecular hydrogen bonds must connect lysine residues 2 and 19, 4 and 17, 6 and 15, and 8 and 13 of neighboring peptides. This result supports an in-register Syn alignment of β-hairpins within each β-sheet (SI Appendix, Fig. S4D).
Evidence for Anti Alignment Between Two β-Sheet Layers.
Prior work suggests that individual MAX1 fibrils contain two β-sheet layers that interact through hydrophobic amino acid sidechains (15, 23–25). The distances between backbone atoms of the two layers must exceed 8 Å to accommodate amino acid sidechains in the hydrophobic interior of the fibril. Such long distances cannot be measured reliably in selectively 15N- or 13C-labeled samples with REDOR, PITHIRDS-CT, or similar dipolar recoupling techniques because the corresponding dipole–dipole coupling strengths are less than 15 Hz. To distinguish between Syn/Syn and Syn/Anti structures, we performed 2D 13C–13C rf-assisted diffusion (RAD) measurements (26, 27). When acquired with long mixing periods, 2D RAD spectra of samples that contain uniformly labeled residues typically show interresidue cross-peaks between 13C NMR signals of residues whose sidechains are within 6–8 Å of one another (7, 8, 28). Hence, sample X was designed with uniformly labeled residues at opposite ends of the MAX1 β-hairpin, with V1 and V20 being more than 20 Å away from P11 and T12. Given the Syn alignment within a single β-sheet, any 2D RAD cross-peaks between V1/V20 and P11/T12 must arise from interlayer proximities of these labeled residues, not from intralayer proximities.
Experimental 2D RAD spectra of sample X are shown in Fig. 3C and SI Appendix, Fig. S5. With a 500-ms mixing period, cross-peaks are clearly observed between the 13Cα chemical shift of V20 and the T12 13Cβ chemical shift as well as the P11 13Cγ and 13Cδ chemical shifts. These cross-peaks are absent from the 2D RAD spectrum with a 25-ms mixing period, as expected for interlayer 13C–13C distances (SI Appendix, Fig. S5A). Assignment of the 60.3-ppm chemical shift to 13Cα of V20 (rather than V1) is confirmed by data for a sample in which V1 was not labeled (SI Appendix, Fig. S6).
With a 500-ms mixing period, strong 2D RAD cross-peaks also occur between chemical shifts of P11 and T12, as expected for sequentially labeled residues. For example, all P11 chemical shifts have cross-peaks with 13Cβ of T12. Cross-peaks between Cβ chemical shifts of V1 and V20 are also observed (SI Appendix, Fig. S5B), consistent with the expected intramolecular and intermolecular 13C–13C distances of ∼5 Å between V1 and V20 sidechains within each β-sheet layer. Taken together, the data support an Anti alignment of β-hairpins between the two β-sheet layers.
Structural Model for MAX1 Fibrils.
Structural restraints from solid-state NMR, listed in SI Appendix, Table S2, were included as potential energy functions in two stages of structure calculations, performed with Xplor-NIH (stage 1) and NAMD (stage 2). Structure calculations included eight copies of MAX1 to form a bilayer of β-sheets composed of four monomers each. Restraints were included in Xplor-NIH calculations by standard methods (29) and in NAMD calculations by a Monte Carlo algorithm (Materials and Methods and SI Appendix). NAMD calculations allowed inclusion of explicit hydration and full electrostatics (30). Conformational and translational symmetry restraints were imposed during Xplor-NIH calculations, but not during NAMD calculations.
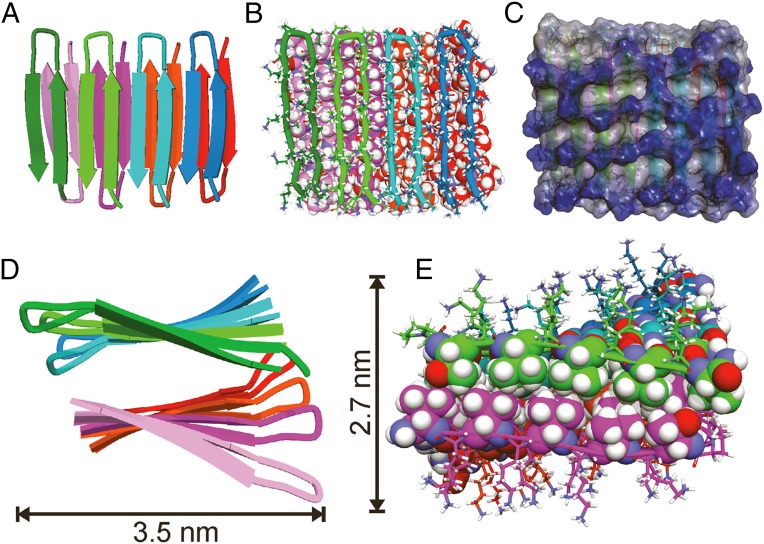
Fig. 4 shows several representations of the final structural model of a MAX1 octamer (specifically, the final frame from NAMD calculations). Overall dimensions are consistent with the apparent fibril diameters in Fig. 2A, providing further support for a structure that consists of two β-sheet layers. A bundle of structures (SI Appendix, Fig. S7C), representing 20 frames from the latter half of the NAMD calculations, has been deposited in the Protein Data Bank, PDB ID code 2N1E. Values of backbone torsion angles in residues 1–9 and 12–19 are typical of β-strands (average ϕ angles between −144° and −120° from K2 through K9 and from K13 through K19; average ψ angles between 119° and 140° from V1 through V9 and from V12 through V18). Backbone torsion angles are ϕ = 29° ± 4°, ψ = −118° ± 9° for DP10 and ϕ = −87° ± 16°, ψ = 12° ± 35° for P11. These values are similar to the torsion angles of an ideal type II′ β-turn (31).
Fig. 4.
Structural model for MAX1 fibrils. (A) Cartoon representation, viewed perpendicular to the β-sheets and the fibril growth axis, with each monomer colored differently for clarity. MAX1 assembles in the Syn/Anti configuration. (B) Same view, with a tube representation of monomers in the upper β-sheet layer and a Corey-Pauling-Koltun (CPK) representation of monomers in the lower layer. MAX1 β-hairpins in the upper layer are slightly translated along the fibril growth axis relative to β-hairpins in the bottom layer, allowing for optimal packing of hydrophobic sidechains. (C) Same view, represented as an interpolated charge surface. Lysine sidechains on the hydrophilic outer faces of MAX1 fibrils produce a high density of positive charge, shown in blue. (D) Cartoon representation viewed along the fibril growth axis, illustrating left-handed β-sheet twist. (E) Same view after removal of the first monomers in the upper and lower β-sheets, with stick representations of lysine and proline residues and CPK representations of valine and threonine residues. Interdigitation of sidechains perpendicular to the fibril growth axis results in hydrophobic nesting of valine and threonine methyl groups.
As shown in Fig. 4 A and B, β-strands in one β-sheet layer are displaced along the fibril growth axis relative to β-strands in the other layer, i.e., β-hairpins across the bilayer do not directly oppose one another. This displacement allows rows of valine sidechains from the two layers to interdigitate. Interdigitation in the direction perpendicular to the fibril growth axis is also present, as shown in Fig. 4E, producing a tightly packed, dry, hydrophobic interface between β-sheet layers. No water molecules or ions were found in the fibril core throughout NAMD calculations, whereas lysine sidechains on the exterior were fully solvated (SI Appendix, Fig. S8). The high density of lysine-borne positive charge on solvent-exposed surfaces (Fig. 4C) presumably keeps individual fibrils soluble during gel formation, preventing higher-order fibril assembly that would lead to precipitation.
Displacement of β-strands along the fibril growth axis removes the ideal twofold symmetry of the fibril structure, because (as depicted in Fig. 4A) the N-terminal β-strands of the “top” layer lie above the β-hairpins of the “bottom” layer, whereas the C-terminal β-strands of the bottom layer lie beneath the β-hairpins of the top layer. Fig. 4D shows a left-handed β-sheet twist along the fibril axis, which developed during the NAMD calculations along with proper hydrogen bonding geometries (SI Appendix, Fig. S7). Left-handed twist is a common feature of amyloid fibrils (8, 13). Twisting of MAX1 fibrils may not be observable in TEM and AFM images due to the approximately cylindrical dimensions of the peptide fibrils.
Discussion
Significance of Structural Homogeneity.
Kinetically trapped fibril networks play important biological roles, for example in cytoskeletal networks of actin (32, 33) and functional amyloid such as curli-derived protein matrices important in bacterial colonization (34). The formation of kinetically trapped networks is often accompanied by local structural heterogeneity at the nanoscale. Amyloid fibrils formed in vitro are generally polymorphic unless special fibril growth protocols are used, such as repeated seeding (8) or long periods of incubation with agitation to promote convergence to a single structure with lowest free energy (35). Prior solid-state NMR studies of the gel-forming peptides RADA16-I and MAX8 suggested polymorphism or incomplete molecular structural order within the hydrogels (14, 20).
Surprisingly, we find that the molecular structure within kinetically trapped MAX1 fibril networks is homogeneous and most likely represents the thermodynamic ground state for local assembly. The Syn/Anti arrangement of MAX1 hairpins allows critical intermolecular interactions that may dictate the monomorphic character of MAX1 fibrils. These interactions are not accessed in other competing structures (Fig. 1). For example, the Syn alignment of hairpins within a given β-sheet permits van der Waals and hydrophobic contacts between the turn regions of neighboring hairpins, resulting in an approximate 20% decrease in solvent-exposed surface area compared with an Anti alignment within β-sheets. The in-register nature of the Syn alignment within β-sheets maximizes intermolecular hydrogen bonding. The Anti alignment of hairpins across β-sheet layers effectively packs the N- and C-terminal valine sidechains of one monomer into the puckered interface formed by the diproline turn residues of its bilayer partner. In the competing Syn alignment, opposing β-turns would project away from the hydrophobic core and become solvent-exposed. Added over the length of the fibril, the additional favorable interactions provided by the Syn/Anti arrangement represent a significant thermodynamic driving force for the formation of monomorphic fibrils.
The β-hairpin design of MAX1, which restricts the conformational freedom of the monomer, may also contribute to the monomorphic nature of MAX1 fibrils. However, even with the β-hairpin conformation, multiple supramolecular structures are conceivable, as discussed above and shown in Fig. 1.
Comparison with Amyloid Fibril Structures.
Like MAX1 fibrils, disease-associated amyloid and prion fibrils contain cross-β structures. Early models for amyloid-β (Aβ) fibrils suggested that β-hairpins might be building blocks for amyloid structures (36, 37). However, subsequent solid-state NMR studies (7–10, 35) revealed in-register, parallel cross-β structures comprising peptide conformations in which N-terminal and C-terminal β-strand segments form separate β-sheets that contain only intermolecular hydrogen bonds, with sidechain–sidechain contacts between these β-sheets.
Recent work on Aβ aggregation intermediates by Härd and coworkers suggests that β-hairpins may be present in certain intermediates (38, 39). Metastable protofibrils formed by the “Iowa mutant” of Aβ have been found to contain antiparallel β-sheets, but still without β-hairpins (28). Laganowsky et al. have reported crystal structures of peptide oligomers derived from αB crystallin, called cylindrins (40). Cylindrins comprising tandem repeats of the αB crystallin peptide contain β-hairpins, but are not cross-β structures. The same tandem repeat peptides also form fibrils, but the peptide conformation within these fibrils has not been determined. Although cross-β structures comprising β-hairpins have not yet been demonstrated conclusively in disease-associated and naturally occurring fibrils, our results for MAX1 fibrils show that such structures certainly exist for appropriate amino acid sequences.
Concluding Remarks.
We have shown that the MAX1 hairpin is capable of assembling with high structural fidelity, affording monomorphic fibrils within a kinetically trapped hydrogel network. Knowledge of the exact arrangement of peptide molecules in the fibrillar state will facilitate future structure-based design of highly functional materials.
Materials and Methods
A series of MAX1 samples with isotopic labeling patterns in Table 1 was synthesized and purified by standard methods. Purified MAX1 was dissolved in water to afford a 4 wt % peptide solution. Hydrogel formation was initiated by addition of an equal volume of 250 mM bis-Tris propane buffer, pH 9, with 20 mM NaCl, followed by incubation at room temperature for 2 h. For solid-state NMR, lyophilized hydrogels were washed twice with deionized water and lyophilized again. Solid-state NMR samples contained 4–8 mg of MAX1.
Solid-state NMR data were acquired with Varian InfinityPlus, Bruker Avance III, and Varian Infinity spectrometers, operating at 1H NMR frequencies of 399.2, 400.9, and 746.6 MHz, respectively, using a Varian 3.2-mm magic-angle spinning (MAS) probe at the lower frequencies and a 1.8-mm MAS probe from the laboratory of Ago Samoson at the highest frequency. Sample temperatures during MAS were maintained near 24 °C with cooled nitrogen gas.
The structural model for MAX1 fibrils was generated in two stages. In the first stage, starting from a set of eight well-separated copies of MAX1 with randomized conformations, Xplor-NIH was used to generate an initial structural model, using restraints summarized in SI Appendix, Table S2. In addition, symmetry restraints were used to ensure identical conformations for all MAX1 molecules, translational symmetry within each β-sheet, and overall twofold rotational symmetry. Artificial distance and torsion angle restraints were also included to enforce hydrophobic contacts between β-sheets, β-strand conformations in residues 1–8 and 14–20, and the intermolecular hydrogen bond registry indicated by experimental PITHIRDS-CT data. In the second stage, the lowest-energy structure from 40 independent Xplor-NIH runs was solvated in a 80.7-nm3 box containing 1,654 water molecules, 77 Cl− ions, and 5 Na+ ions (for roughly 150 mM NaCl and charge neutrality). Molecular dynamics simulations were then run at 37 °C with the NAMD, using a Monte Carlo algorithm to enforce experimentally based structural restraints during these simulations. Fig. 4 shows the final structure from an 8.0-ns molecular dynamics/Monte Carlo simulation. SI Appendix, Fig. S7 compares structures from the ends of stages 1 and 2, as well as a superposition of structures from 20 equally spaced time points in the final 4.0 ns of the NAMD simulations (PDB ID code 2N1E). PyMOL and Accelerys Discovery Studio were used for molecular graphics.
Further details of sample preparation, NMR measurements, and structure calculations are given in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and the National Cancer Institute of the National Institutes of Health. Structure calculations were aided by the high-performance computational capabilities of the Biowulf Linux cluster at the NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2N1E). NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 25558).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509313112/-/DCSupplemental.
References
- 1.Schneider JP, et al. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124(50):15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 2.Yan C, et al. Injectable solid hydrogel: Mechanism of shear-thinning and immediate recovery of injectable β-hairpin peptide hydrogels. Soft Matter. 2010;6(20):5143–5156. doi: 10.1039/C0SM00642D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salick DA, Kretsinger JK, Pochan DJ, Schneider JP. Inherent antibacterial activity of a peptide-based β-hairpin hydrogel. J Am Chem Soc. 2007;129(47):14793–14799. doi: 10.1021/ja076300z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yucel T, Micklitsch CM, Schneider JP, Pochan DJ. Direct observation of early-time hydrogelation in β-hairpin peptide self-assembly. Macromolecules. 2008;41(15):5763–5772. doi: 10.1021/ma702840q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branco MC, Nettesheim F, Pochan DJ, Schneider JP, Wagner NJ. Fast dynamics of semiflexible chain networks of self-assembled peptides. Biomacromolecules. 2009;10(6):1374–1380. doi: 10.1021/bm801396e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen TH, Rajagopal K, Schneider JP, Furst EM. Microrheology of responsive hydrogel networks. AIP Conf Proc. 2008;1027:1090–1092. [Google Scholar]
- 7.Lu JX, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154(6):1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105(47):18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schütz AK, et al. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 2015;54(1):331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Melckebeke H, et al. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132(39):13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 12.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46(47):13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsbury CS, et al. Studies on the in vitro assembly of a β 1-40: Implications for the search for a β fibril formation inhibitors. J Struct Biol. 2000;130(2-3):217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 14.Cormier AR, Pang X, Zimmerman MI, Zhou HX, Paravastu AK. Molecular structure of RADA16-I designer self-assembling peptide nanofibers. ACS Nano. 2013;7(9):7562–7572. doi: 10.1021/nn401562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy KJ, Giano MC, Jin A, Pochan DJ, Schneider JP. Enhanced mechanical rigidity of hydrogels formed from enantiomeric peptide assemblies. J Am Chem Soc. 2011;133(38):14975–14977. doi: 10.1021/ja206742m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5(1):67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 18.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 2011;81:196–200. doi: 10.1016/j.jmr.2011.09.003. , and comment (1989), 81:196–200. [DOI] [PubMed] [Google Scholar]
- 19.Hu KN, Qiang W, Bermejo GA, Schwieters CD, Tycko R. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N-15N and carbonyl 13C-13C dipolar recoupling data. J Magn Reson. 2012;218:115–127. doi: 10.1016/j.jmr.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard SR, et al. Solid-state NMR evidence for β-hairpin structure within MAX8 designer peptide nanofibers. Biophys J. 2013;105(1):222–230. doi: 10.1016/j.bpj.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007;126(6):064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 22.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103(52):19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP. Tuning the pH responsiveness of β-hairpin peptide folding, self-assembly, and hydrogel material formation. Biomacromolecules. 2009;10(9):2619–2625. doi: 10.1021/bm900544e. [DOI] [PubMed] [Google Scholar]
- 24.Haines-Butterick L, et al. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proc Natl Acad Sci USA. 2007;104(19):7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled beta-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26(25):5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126(23):7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 27.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 28.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci USA. 2012;109(12):4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- 30.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattos C, Petsko GA, Karplus M. Analysis of two-residue turns in proteins. J Mol Biol. 1994;238(5):733–747. doi: 10.1006/jmbi.1994.1332. [DOI] [PubMed] [Google Scholar]
- 32.Schmoller KM, Lieleg O, Bausch AR. Internal stress in kinetically trapped actin bundle networks. Soft Matter. 2008;4:2365–2367. [Google Scholar]
- 33.Lieleg O, et al. Structural polymorphism in heterogeneous cytoskeletal networks. Soft Matter. 2009;5:1796–1803. [Google Scholar]
- 34.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133(40):16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Darden TA, Bartolotti L, Kominos D, Pedersen LG. An atomic model for the pleated β-sheet structure of Abeta amyloid protofilaments. Biophys J. 1999;76(6):2871–2878. doi: 10.1016/S0006-3495(99)77442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tjernberg LO, et al. A molecular model of Alzheimer amyloid β-peptide fibril formation. J Biol Chem. 1999;274(18):12619–12625. doi: 10.1074/jbc.274.18.12619. [DOI] [PubMed] [Google Scholar]
- 38.Härd T. Protein engineering to stabilize soluble amyloid β-protein aggregates for structural and functional studies. FEBS J. 2011;278(20):3884–3892. doi: 10.1111/j.1742-4658.2011.08295.x. [DOI] [PubMed] [Google Scholar]
- 39.Lendel C, et al. A hexameric peptide barrel as building block of amyloid-β protofibrils. Angew Chem Int Ed Engl. 2014;53(47):12756–12760. doi: 10.1002/anie.201406357. [DOI] [PubMed] [Google Scholar]
- 40.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.