Significance
Tripartite motif (TRIM) proteins are a large family of E3 ubiquitin (Ub) ligases, with many members having important roles in innate immunity. TRIM21 is a cytosolic antibody receptor that recognizes the Fc portion of antibodies bound to incoming virions. On binding to these immune complexes, TRIM21 triggers the catastrophic disassembly of viral capsids at the proteasome, terminating viral infection. Simultaneously, TRIM21 “senses” the presence of the virus and provokes signaling cascades that activate the transcription factor NF-κB, ultimately alerting surrounding cells to the infection. Here, we uncover the stepwise ubiquitination mechanism catalyzed by TRIM21, as well as the various cofactors required, that allows these two antiviral activities to occur synchronously at the proteasome.
Keywords: TRIM21, NF-kB, Ube2W, Ube2N/Ube2V2, Poh1
Abstract
Tripartite motif (TRIM) 21 is a cytosolic antibody receptor that neutralizes antibody-coated viruses that penetrate the cell and simultaneously activates innate immunity. Here we show that the conjugation of TRIM21 with K63-linked ubiquitin (Ub-63Ub) catalyzed by the sequential activity of nonredundant E2 Ub enzymes is required for its dual antiviral functions. TRIM21 is first labeled with monoubiquitin (monoUb) by the E2 Ube2W. The monoUb is a substrate for the heterodimeric E2 Ube2N/Ube2V2, resulting in TRIM21-anchored Ub-63Ub. Depletion of either E2 abolishes Ub-63Ub and Ub-48Ub conjugation of TRIM21, NF-κB signaling, and virus neutralization. The formation of TRIM21-Ub-63Ub precedes proteasome recruitment, and we identify an essential role for the 19S-resident and degradation-coupled deubiquitinase Poh1 in TRIM21 neutralization, signaling, and cytokine induction. This study elucidates a complex mechanism of step-wise ubiquitination and deubiquitination activities that allows contemporaneous innate immune signaling and neutralization by TRIM21.
TRIM21 is a cytoplasmic Ig receptor belonging to the tripartite motif family of E3 ubiquitin (Ub) ligases. Many TRIM proteins (there are ∼100 members in humans) have reported roles in innate immunity (1). TRIM21 intercepts incoming, cytoplasmic antibody-coated pathogens and neutralizes them by mediating their VCP- and proteasome-dependent degradation (2, 3). In addition, TRIM21 initiates a signaling cascade, resulting in the transcriptional up-regulation of inflammatory cytokines and a potent antiviral state in surrounding cells (4). Such antiviral duality has also been reported for a paralogue of TRIM21, TRIM5α (5), and more recently, TRIM19/PML (6), suggesting that an ability to “sense” and neutralize viral infection is a conserved TRIM function. The mechanism by which a TRIM protein coordinates sensor and effector functions remains unknown.
For TRIM21, these dual functions are dependent upon its activity as an E3 Ub ligase. In previous work, we have shown that the TRIM21 really interesting new gene (RING) domain is required for neutralization and catalyzes autoubiquitination with K48-linked polyubiquitin (polyUb) (Ub-48Ub, following the nomenclature suggested by ref. 7) in vitro (3), suggesting TRIM21 might synthesize Ub-48Ub to recruit the proteasome. We also demonstrated that TRIM21, with the heterodimeric E2 enzyme Ube2N/Ube2V1, catalyses the formation of unanchored K63-linked polyUb (Ub-63Ub) in vitro (4). Ube2N and Ub-63Ub were essential for TRIM21 to mount an innate signaling response, consistent with a role for unanchored Ub-63Ub in NF-κB signaling (8). Thus, the ability of TRIM21 to mediate multiple functions seems associated with its ability to synthesize multiple Ub chain types. Moreover, because TRIM21 only initiates innate signaling upon recognition of antibody-bound virus (4), this suggests that the effector and sensor functions of TRIM21 are mechanistically connected.
Four enzymes are involved in the ubiquitination cycle: the Ub activating (E1), the Ub conjugating (E2), the substrate-specifying Ub ligase (E3), and the Ub-erasing deubiquitinase (DUB). Of these, the E2 plays a central role in selecting the Ub chain linkage topology (9–11). For example, the heterodimeric RING-domain E3 ligase BRCA1-BARD1 auto-monoubiquitinates using one of the E2s UbcH6, Ube2E2, UbcM2, or Ube2W, and the monoUb can be extended into Ub-63Ub or Ub-48Ub by Ube2N/UbeV2 or Ube2K, respectively (12). PolyUb is typically attached to a substrate by the E3; however, with some proclivity, E3–E2 pairs can also generate unanchored polyUb chains in vitro (13). Unanchored polyUb has been shown to activate innate immune signaling (4, 5, 8, 14), but how it is generated and regulated in cells remains poorly understood.
Once a Ub-modified substrate arrives at the proteasome, three DUBs in the 19S regulatory particle (RP), Usp14/Ubp6, Uch37/UCHL5, and Rpn11/Poh1, trim or remove Ub modifications before substrate degradation (15). Although Ub-48Ub is a canonical degradation signal, both Ub-48Ub and Ub-63Ub are hydrolyzed by purified proteasomes in vitro (16, 17), and some studies have observed Ub-63Ub specificity in the polyUb amputation activity of Poh1 (18, 19). Whether Ub-63Ub has a role at the proteasome remains unclear; its presence at this location would seem incongruous with a model in which only Ub-48Ub-labeled proteins are genuine substrates (20, 21).
In this report, we show that the effector and sensor functions of TRIM21 are dependent on its sequential recruitment of the E2s Ube2W and Ube2N/Ube2V2, and the DUB Poh1.
Results
Ube2W and Ube2N Are Required for TRIM21 Dual Effector and Sensor Functions.
Previously, we have shown that infection of nonprofessional cells with antibody-coated adenovirus (AdV:Ab) results in TRIM21-dependent NF-κB activation and proteasome-mediated virus neutralization (4). To investigate the functional relationship between these two antiviral outcomes, we first asked whether the generation of an innate immune response via NF-κB activation precedes virus destruction by the proteasome. We treated human TE671 cells with chemical inhibitors of the proteasome (MG132, bortezomib) or NF-κB signaling (5Z-7-Oxozeaenol, IKK Inhibitor VII). As shown in Fig. 1A, 7-h treatment of human TE671 cells with MG132, bortezomib, IKK VII, or 5Z-7-Oxozeaenol inhibited the stimulation of a model NF-κB promoter that drives luciferase expression (NF-κB-Luc) after infection with AdV:Ab. However, although both MG132 and bortezomib inhibited virus neutralization by the monoclonal antihexon antibody 9C12 (Fig. 1B), IKK inhibitor VII or 5Z-7-Oxozeaenol did not. Thus, TRIM21-dependent NF-κB activation is not a requisite for virus neutralization, suggesting signaling and neutralization might be synchronous, rather than sequential.
Fig. 1.
Ube2W and Ube2N are required for TRIM21-mediated signaling and neutralization. (A) Proteasome or NF-κB signaling inhibitors prevent NF-κB activation in response to AdV/IgG. (B and C) TE671 cells, infected with AdV (MOI ∼0.1), preincubated with 9C12 or PBS, treated with the inhibitors indicated or DMSO (B), or expressing scrambled shRNA (shScr) (C–G), Ube2N-specific shRNA (shUbe2N) (C and F) or Ube2W-specific shRNA (shUbe2W) (C–E and G), percentage infection quantified by flow cytometry for GFP expression 24 h postinfection, values normalized to PBS-treated AdV control. (D and E) TE671-NF-κB-Luc cells expressing shScr or shUbe2W challenged with PBS (D and E), human IgG, AdV, AdV/IgG (D), or human TNFα (E) for 7 h. (F) Immunoblot of TE671-shScr or TE671-shUbe2N, detecting Ube2N or β-actin (loading control). (G) Quantitative RT-PCR detecting Ube2W mRNA in TE671-shScr or TE671-shUbe2W. Values calculated using the ∆∆Ct method and normalized to β-Actin mRNA. All error bars are SEM of duplicates or triplicates.
To test this idea, we considered whether TRIM21 ubiquitination, as required for innate signaling, was also required for virus neutralization. In vitro, TRIM21 synthesizes unanchored Ub-63Ub in the presence of Ube2N/Ube2V1, and both Ub-63Ub and Ube2N are necessary for TRIM21 to provoke an innate immune signal (4). In Ube2N-depleted cells, 9C12-mediated neutralization of AdV-GFP was largely diminished (Fig. 1C), suggesting the formation of Ub-63Ub precedes viral neutralization. However, proteins are typically targeted with anchored Ub chains to recruit the proteasome. Because Ube2W is described to facilitate the Ube2N/Ube2V2-mediated polyubiquitination of substrates with anchored Ub-63Ub chains (12, 22), we depleted Ube2W, using shRNA, to ask whether it too has a role in virus neutralization. Interestingly, Ube2W depletion also reversed neutralization of adenovirus by 9C12 (Fig. 1C), suggesting both Ube2W and Ube2N/Ube2V2 are cofactors for TRIM21-mediated virus neutralization. Furthermore, we observed that Ube2W depletion abolished potent activation of NF-κB-Luc on challenge with Adv:Ab (Fig. 1D). In contrast, we measured no decrease in NF-κB-Luc activation in response to TNFα in Ube2W-depleted cells (Fig. 1E). Western blots confirmed depletions of Ube2N (Fig. 1F), and RT-qPCR confirmed depletions of Ube2W (Fig. 1G). Together, these experiments suggest Ube2N and Ube2W are nonredundant cofactors for both sensing and effector responses of TRIM21, an unexpected observation because Ube2W is reported to anchor polyUb to substrates (22, 23) and Ub-63Ub chains are not thought to recruit proteins to the proteasome (17).
Ube2W Monoubiquitinates TRIM21.
We next investigated whether TRIM21 can act as an E3 enzyme for Ube2W. We expressed full-length TRIM21 bearing an N-terminal MBP tag that improved protein solubility. We incubated purified MBP-TRIM21 with E1, Ube2W, and Ub. As increasing amounts of Ube2W were titrated into the reactions, we observed an increase in modified MBP-TRIM21 (Fig. 2A, Top). Probing with an anti-Ub antibody revealed a band of the same size as modified MBP-TRIM21, suggesting the adduct was a monoUb (Fig. 2A, Bottom). There were additional bands in the Ub blot with sizes coincident with mono- and diubiquitinated Ube2W (Fig. 2A, Bottom) (22).
Fig. 2.
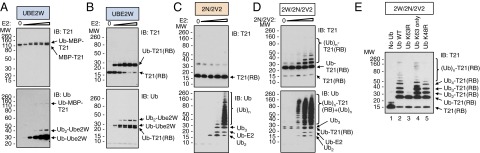
TRIM21 autoubiquitinates with Ube2W and Ube2N/Ube2V2 in vitro. (A and B) Ube2W monoubiquitinates TRIM21 (T21). Purified MBP-T21 (A) or T21RING-Box (RB) (B), E1, Ub, and ATP were incubated with increasing concentrations of Ube2W. Reaction products separated by LDS-PAGE and visualized by IB detecting T21 (Top) or Ub (Bottom). (C and D) Ube2W anchors polyUb to T21. Ubiquitination reactions as in B, but with increasing concentrations of Ube2N/Ube2V2 (2N/2V2) in the absence (C) or presence (D) of Ube2W (2W). (E) T21(RB) autoubiquitinates with Ub-63Ub. T21(RB) incubated with E1, ATP, 2W, 2N/2V2, and WT Ub or Ub bearing lysine-to-arginine substitutions at residue 63 (K63R), residue 48 (K48R), or at all Ub lysine residues except residue 63 (K63 only).
To support our interpretation that TRIM21 undergoes auto-monoubiquitination using Ube2W, we purified untagged TRIM21 corresponding to the RING and B-Box2 domains (residues 1–129) only, and incubated purified TRIM21RING-Box with E1, Ube2W, and Ub. As Ube2W was titrated into these reactions, we observed the pronounced appearance of a band corresponding to modified TRIM21RING-Box (Fig. 2B, Top). These reactions were driven to near-completion, with the appearance of modified TRIM21RING-Box correlating with a decrease in the levels of unmodified TRIM21RING-Box (Fig. 2B, Top). Probing the same blots with an anti-Ub antibody confirmed the modified TRIM21RING-Box band to be monoubiquitinated TRIM21RING-Box (Fig. 2B, Bottom). As in the previous reactions, we found that Ube2W was also mono- and diubiquitinated (Fig. 2B, Bottom). These experiments demonstrate that TRIM21 is an E3 ligase capable of auto-monoubiquitination with the E2 Ube2W in vitro.
Monoubiquitinated TRIM21 Is a Substrate for Ube2N/Ube2V2.
Previously we have shown in vitro that in the presence of E1, Ub, and Ube2N TRIM21 can catalyze the formation of unanchored Ub-63Ub (4). We next investigated whether this pattern of activity is altered in the presence of Ube2W. Similar to previous experiments with full-length protein, incubation of TRIM21RING-Box with Ube2N/Ube2V2 resulted in the production of Ub chains that were not conjugated to TRIM21 (Fig. 2C). However, in the presence of Ube2W, polyubiquitination of TRIM21RING-Box was observed (Fig. 2D). This was accompanied by an overall increase in the levels of polyubiquitin chains, as detected by an anti-Ub antibody, suggesting Ube2W also potentiates the reaction between Ube2N/Ube2V2 and TRIM21 (Fig. 2D, Bottom). Extension of monoubiquitinated TRIM21RING-Box by Ube2N/Ube2V2 was dose-dependent, as increasing Ube2N/Ube2V2 concentrations increased the length of the conjugated Ub chain (Fig. 2D, Top).
Repeating the reaction with intermediate E2 concentrations and various Ub mutants confirmed that, as expected for Ube2N/Ube2V2, polyubiquitination of TRIM21RING-Box occurred through a K63 linkage (Fig. 2E). K63R mutant Ub was not conjugated to monoubiquitinated TRIM21 (Fig. 2E, lane 3). Conversely, both a mutant Ub bearing only one lysine at residue 63 (Fig. 2E, lane 4) and mutant Ub bearing arginine in place of lysine 48 (Fig. 2E, lane 5) were as efficiently incorporated into TRIM21-linked polyubiquitin as WT Ub (Fig. 2E, lane 2), confirming K63-linkage homogeneity of these chains.
Ube2W and Ube2N/Ube2V2 Cooperatively Polyubiquitinate a Lysine-Less TRIM21RING-Box.
It has recently emerged that Ube2W does not require lysines for Ub conjugation and preferentially transfers Ub to the primary N-terminal α-amino group of several proteins in vitro, including Ube2W itself (22–24). To assess whether TRIM21 is also ubiquitinated by Ube2W independent of lysine residues in vitro, we purified TRIM21RING-Box proteins bearing lysine-to-arginine substitutions at all six lysine residues in TRIM21RING-Box: three in the RING domain (Lys45, Lys61, Lys77) and three in the B-Box2 domain (Lys105, Lys108, Lys119). TRIM21RING-Box-6KR (K45/61/77/105/108/119R) was purified and incubated with E1, Ube2W, and Ub, with or without ATP. Coomassie Blue-stained LDS-PAGE revealed that the mutant protein was fully active with Ube2W, as it was converted into a modified TRIM21RING-Box species as efficiently as WT TRIM21RING-Box (Fig. 3A, lanes 2 and 4). Mutation of the lysine residues also had no effect on Ub ligase activity, as TRIM21RING-Box-6KR was able to generate free Ub-63Ub in the presence of Ube2N/Ube2V2 (Fig. 3B, lanes 2 and 4). The addition of Ube2W to the Ube2N/Ube2V2-containing reactions resulted in a change in high molecular weight (HMW) reaction products, consistent with TRIM21RING-Box-6KR auto-polyubiquitination (Fig. 3C). To confirm this interpretation, we incubated increasing amounts of WT or 6KR mutant TRIM21RING-Box with either Ube2W or Ube2W and Ube2N/Ube2V2, and probed immunoblots with a TRIM21 antibody. Both TRIM21 variants were efficiently modified in the presence of Ube2W (Fig. 3 D and E, Left). The addition of Ube2N/Ube2V2 to these reactions led to the polyubiquitination of TRIM21 (Fig. 3 D and E, Right). Thus, lysine residues are not required for ubiquitination of TRIM21RING-Box by Ube2W and Ube2N/Ube2V2 in vitro, in agreement with the N terminus of TRIM21 being a potential target for ubiquitination by Ube2W.
Fig. 3.
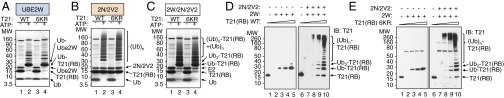
A lysine-less TRIM21RING-Box autoubiquitinates with Ube2W and Ube2N/Ube2V2 in vitro. (A–C) WT or mutant TRIM21RING-Box-6KR proteins bearing lysine-to-arginine at residues 45, 61, 77, 105, 108, and 119 were incubated with E1 and Ub in the presence or absence of ATP and Ube2W (A), Ube2N/Ube2V2 (B), or Ube2W and Ube2N/Ube2V2 (C). Reactions products resolved by LDS-PAGE and visualized by Coomassie stain. (D and E) Autoubiquitination of WT (D) or 6KR (E) TRIM21RING-Box in the presence of Ube2W (Left) or Ube2W and Ube2N/Ube2V2 (Right). Reaction products resolved by LDS-PAGE and visualized by immunoblot detecting TRIM21. Reactions containing TRIM21RING-Box in the absence of any E2 served as controls.
Ube2W and Ube2N Are Necessary for Ubiquitination of TRIM21 in Cells.
We next asked whether the requirement for Ube2W and Ube2N during TRIM21-mediated neutralization of virus infection (Fig. 1) correlates with a dependence on these enzymes for ubiquitination of TRIM21 in cells. To test this, we sought evidence for ubiquitinated TRIM21 by expressing TRIM21 with an N-terminal 6xHis-tag, together with HA-tagged Ub in control TE671 cells or cells depleted of either Ube2W or Ube2N. Cell lysate was incubated with Ni-NTA beads and captured protein blotted for total Ub. A strong, HMW polyubiquitinated smear was observed in control cells expressing 6xHis-TRIM21 and HA-Ub but was absent in cells that were depleted of Ube2N (Fig. 4A) or Ube2W (Fig. 4B), suggesting TRIM21 is ubiquitinated in cells by these two E2s. We next probed purified proteins from control or Ube2N-depleted lysates with antibodies specific for either K48 or K63 isopeptide bonds. To control for antibody linkage specificity, we loaded recombinant polyubiquitin linked by either K48 or K63 linkages. Importantly, we observed robust labeling of TRIM21 by both K48- and K63-linked polyUb in control cells, but not in cells depleted of Ube2N (Fig. 4C).
Fig. 4.
K48- and K63-linked ubiquitination of cellular TRIM21 requires Ube2W and Ube2N. (A–C) TE671 expressing a scrambled shRNA (Scr), Ube2N-specific shRNA, or Ube2W-specific shRNA (B) and the indicated combinations of 6xHis-TRIM21 and HA-Ub. Cell lysates were incubated with Ni2+ affinity resin, and input and pull-down (PD) fractions were resolved by LDS-PAGE and detected by IB with antibodies against total Ub, Ube2N, Ub-48Ub, Ub-63Ub, or β-Actin (loading control). In C, 1 μg purified Ub-48Ub or Ub-63Ub was loaded to control for antibody specificity.
Taken together, the data so far fit a model in which TRIM21 is first monoubiquitinated by Ube2W and then polyubiquitinated by Ube2N/Ube2V2. Given the requirement for each of these E2s in the dual effector–sensor functions of TRIM21 (Fig. 1) and for Ub-48Ub and Ub-63Ub labeling in cells (Fig. 4), we hypothesized that polyubiquitination of TRIM21 is a critical initial step in TRIM21 antiviral function.
The Proteasome-Associated Deubiquitinase Poh1 Is Required for TRIM21 Signaling.
We next considered how TRIM21-anchored Ub-63Ub could activate immune signaling. We hypothesized that anchored Ub-63Ub could be liberated during proteolysis at the proteasome, enabling its binding to downstream signaling effectors such as TAB2 (5, 8). Such a mechanism would allow stimulation of innate signaling to occur synchronously with neutralization of virus, rationalizing the requirement for Ube2W in both functions (Fig. 1). Of the three DUBs associated with the proteasome 19S RP, Poh1 uniquely cleaves an isopeptide bond between the proximal Ub in a polyUb adduct and its substrate, thereby releasing a polyUb chain en bloc (16, 25, 26). To test the notion that Poh1 might deubiquitinate TRIM21, we stably depleted Poh1 in TE671 cells by shRNA expression, assessing efficiency by immunoblot (Fig. 5A). Specific purification of 6xHis-TRIM21 from control or Poh1-depleted cells revealed increased Ub-63Ub conjugation to TRIM21 after Poh1 depletion (Fig. 5B). To determine whether the persistence of Ub-63Ub on TRIM21 bore a consequence for immune signal induction, we infected control or Poh1-depleted cells with AdV:Ab and measured stimulation of NF-κB-Luc. Whereas in control cells, AdV:Ab robustly stimulated luciferase expression, cells depleted of Poh1 were largely unresponsive to challenge (Fig. 5C), supporting a role for Poh1 in TRIM21 innate signal induction.
Fig. 5.

Poh1-mediated NF-κB activation and cytokine transcription in response to AdV/IgG in human and mouse cells. (A) IB detecting Poh1 or β-Actin (loading control) in TE671 expressing a scrambled shRNA (shScr) or Poh1-specific shRNA (shPoh1). (B) Pull-down of 6xHis-TRIM21 (T21) from TE671 expressing shScr or shPoh1, and precipitated proteins assessed by IB detecting Ub-63Ub. (C) TE671-NF-κB-Luc cells expressing shScr or shPoh1 incubated with PBS, AdV, IgG, or AdV/IgG. NF-κB activation determined by luminometry, values expressed relative to PBS control in each condition. (D) TE671 expressing shScr or shPoh1 and Ub-GV-GFP, treated with MG132 for 10 h, GFP-positive cells enumerated by flow cytometry. (E) TE671 expressing shScr or shPoh1 infected with AdV (MOI ∼0.2) preincubated with increasing concentrations of 9C12. Percentage infection determined by flow cytometry detecting GFP. Values normalized to infection in the presence of PBS alone. (F) WT MEFs transfected with nonspecific siRNA (siControl) or Poh1-specific siRNA (siPoh1) and infected as in E. (G) WT or TRIM21−/− (K21) MEFs incubated with PBS, IgG, AdV, or AdV/IgG. Quantitative RT-PCR (qRT-PCR) was performed, and TNFα mRNA copies relative to TBP mRNA copies were quantified 4 h postchallenge. (H) qRT-PCR measuring Poh1 mRNA copies relative to TBP mRNA copies in WT or K21 MEFs transfected with siControl or siPoh1. (I–K) WT (I, K) or K21 (J) MEFs transfected with siControl or siPoh1, incubated with PBS, IgG, AdV, AdV/IgG (I–K), or poly(I:C) (K). qRT-PCR performed and TNFα mRNA copies relative to TBP mRNA copies quantified 4 h postchallenge. (L) IB detecting Poh1 or β-Actin (loading control) in HeLa transfected with siControl or siPoh1. (M and N) HeLa transfected with siControl or siPoh1 incubated with PBS, IgG, AdV, AdV/IgG (M and N) or poly(I:C) (N). qRT-PCR performed and IL-8 (M) or IL-6 (N) mRNAs relative to β-Actin mRNA copies quantified 4 h postinfection. Values are means of triplicates ± SEM.
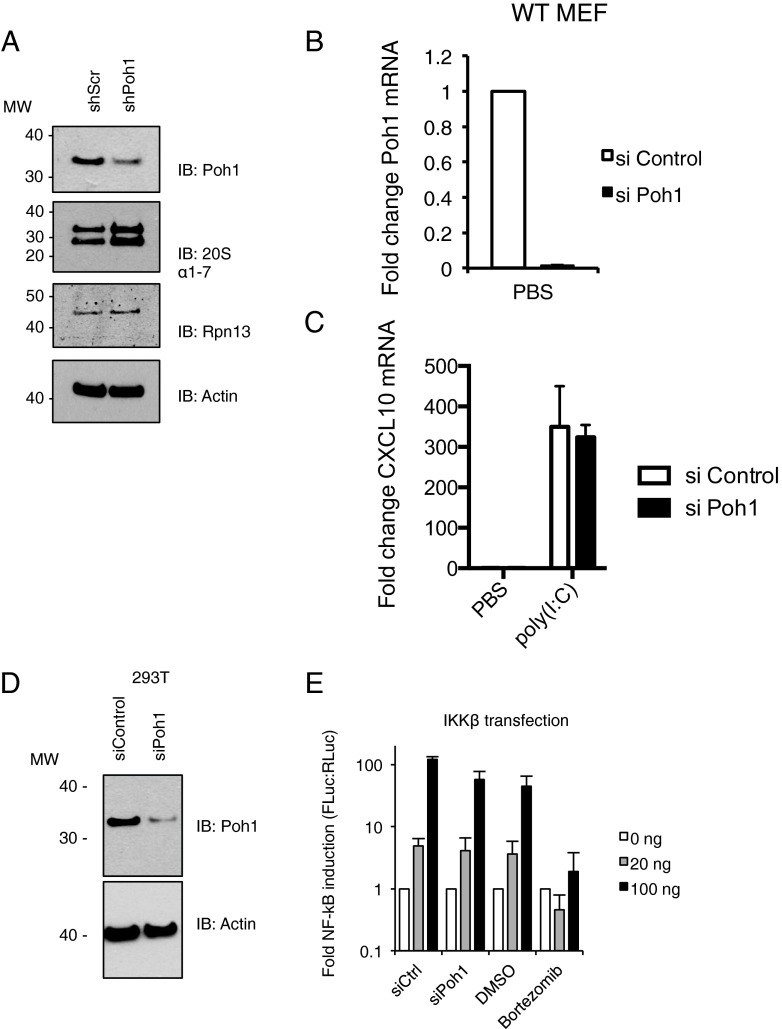
Protein levels of the 20S core α1–7 subunits (Fig. S1A) or the 19S RP-associated Ub acceptor Rpn13/Adrm1 (Fig. S1A) were not diminished in Poh1-depleted cells. Moreover, Poh1 depletion did not alter the degradation of the model proteasome degradation substrate Ub-G76V-GFP (27); Poh1 depletion neither caused accumulation of Ub-G76V-GFP in DMSO-treated cells nor affected the magnitude of stabilization by MG132 (Fig. 5D). Together, this suggests stable Poh1 depletion does not nonspecifically disrupt proteasomes. However, the ability of TRIM21 to facilitate 9C12-mediated virus neutralization was diminished in cells depleted of Poh1 (Fig. 5E), suggesting Poh1 is a necessary cofactor for both of TRIM21’s antiviral functions. We repeated this experiment in mouse embryonic fibroblasts (MEFs), using the mouse monoclonal 9C12, and observed a similar substantial reduction in Adv neutralization, suggesting Poh1 requirement is conserved across mammalian species (Fig. 5F).
Fig. S1.
(A) IB detecting proteasome components Poh1, 20S alpha subunits 1–7, Rpn13, or β-Actin (loading control) in TE671 expressing shScr or Poh1-specific shRNA (shPoh1). (B and C) MEFs transfected with nonspecific siRNA (siControl) or Poh1-specific siRNA (siPoh1). Quantitative RT-PCR performed and Poh1 (B) or CXCL10 (C) mRNA relative to TBP mRNA copies quantified by ∆∆Ct method (D) IB detecting Poh1 or β-Actin (loading control) in 293T transfection with siControl or siPoh1. (E) 293T transfected with siControl or siPoh1 were subsequently transfected with NF-κB-luciferase and TK-renilla luciferase and increasing amount of IKKβ and luciferase values read 24 h posttransfection. In parallel, 293T were treated with 50 nM bortezomib or DMSO vehicle for the duration of the IKKβ transfection. Firefly luciferase values normalized relative to renilla luciferase values and expressed relative to empty vector control.
To further substantiate the requirement for Poh1, we assessed its importance during inflammatory cytokine induction by TRIM21. We infected WT and TRIM21-knockout (K21) MEFs with AdV:Ab and determined immune activation by measuring TNFα transcription. As previously observed (4), potent up-regulation of TNFα transcription by AdV:Ab in WT MEFs was largely abolished in K21 MEFs (Fig. 5G), confirming the transcriptional response to be TRIM21-dependent. We transiently depleted Poh1 from these cells by siRNA, measuring Poh1 transcript expression by qPCR (Fig. 5H). In WT MEFs, the up-regulation of TNFα transcription by AdV:Ab was significantly reduced by Poh1 depletion (Fig. 5I). Conversely, the minimal TRIM21-independent TNFα transcription observed in K21 cells was Poh1-independent (Fig. 5J), suggesting NF-κB activation per se was not inhibited by Poh1 depletion. In agreement with this observation, the ability of the RIG-I agonist poly(I:C) to trigger expression of NF-κB-dependent genes was unaffected by Poh1 depletion (Fig. 5K and Fig. S1 B and C), supporting the notion that Poh1 depletion does not inhibit signaling nonspecifically. Poh1 was also required for cytokine transcription in human cells infected with AdV:Ab. Depletion of Poh1 in HeLa cells (Fig. 5L) significantly repressed the up-regulation of IL-8 (Fig. 5M) and IL-6 (Fig. 5N) transcription on AdV:Ab challenge. As in MEFs, Poh1 depletion had no effect on NF-κB stimulation by the RIG-I agonist poly(I:C) (Fig. 5N). Together, these data suggest Poh1 has a specific role in TRIM21-mediated signaling during infection by antibody-coated virus.
In performing these experiments, we were cautious of the fact that the polyubiquitination and proteasome-mediated degradation of IκBα after its phosphorylation by IKKβ is required for NF-κB nuclear translocation (28). To further control for nonspecific effects of Poh1 depletion on NF-κB signaling, we cotransfected IKKβ and NF-κB-Luc into control 293T cells or cells transiently depleted of Poh1, assessing depletions by immunoblot (Fig. S1D). In parallel, we treated 293T cells with vehicle (DMSO) or bortezomib for the duration of the experiment. Transfection of IKKβ potently stimulated NF-κB-Luc activation in both control and Poh1-depleted cells (Fig. S1E). However, proteasome inhibition by bortezomib abolished all signaling activity (Fig. S1E), suggesting Poh1 depletion does not inhibit NF-κB signaling through proteasome inhibition (Fig. S2).
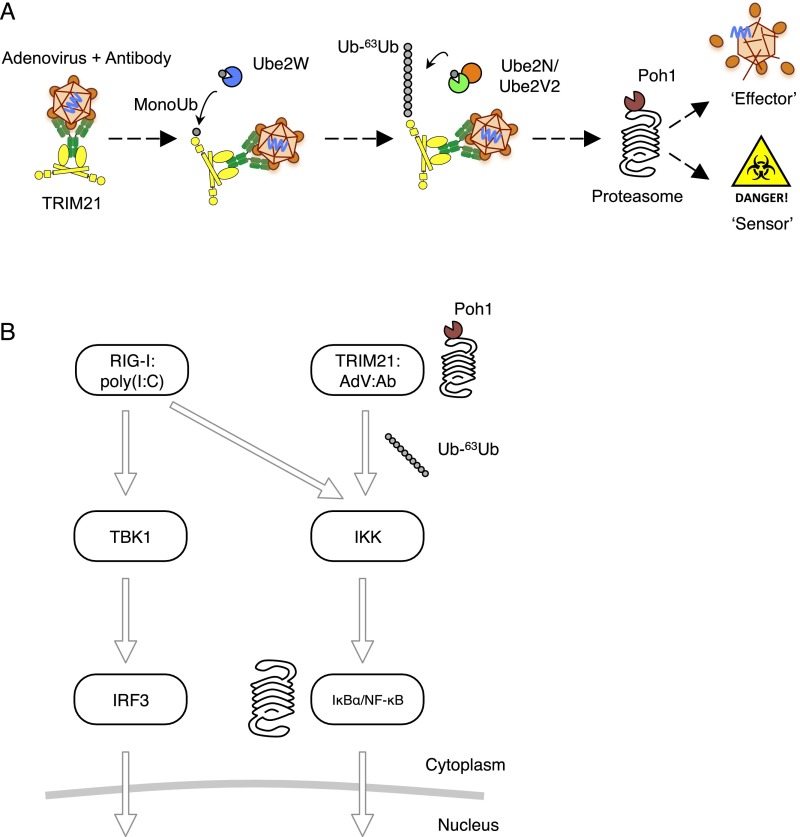
Fig. S2.
(A) Schematic of TRIM21 antiviral mechanism. TRIM21 and the E2 Ub conjugation enzyme Ube2W mediate monoubiquitination of TRIM21. This provides a substrate for Ub-63Ub chain elongation by the E2 heterodimer Ube2N/Ube2V2, resulting in TRIM21-anchored Ub-63Ub chains. The proteasome-associated deubiquitinase Poh1 liberates polyubiquitin from TRIM21, thus synchronizing both “effector” (virus degradation) and “sensor” (immune signal induction) functions of TRIM21. (B) RIG-I stimulates the NF-κB pathway via IKK and is unaffected by depletion of Poh1. IκBα is phosphorylated and degraded by the proteasome but is unaffected by depletion of Poh1. TRIM21 stimulates the NF-κB pathway via IKK and is impaired by depletion of Poh1. Thus, Poh1 is required at the indicated point of the signaling pathway.
Discussion
In this study, we find that TRIM21 sensor and effector functions both derive from a TRIM21-anchored Ub-63Ub chain, synthesized in conjunction with the E2 Ub conjugating enzymes Ube2W and Ube2N/Ube2V2. These results are in good agreement with previous reports of cooperation between Ube2W and Ube2N/Ube2V2 (12, 22) and the ability of Ube2W to mediate specific N-terminal ubiquitination of substrates (22–24). In the absence of Ube2W, Ube2N/Ube2V2 and TRIM21 synthesize unanchored Ub-63Ub in vitro. This reaction looks sufficient to generate the polyUb stimulus of NF-κB signaling because unanchored Ub-63Ub is alone able to activate TAK1 (8). Indeed, it is possible that unanchored polyUb is generated in cells via the same mechanism as it is generated in vitro. However, our finding that Ube2W is a critical component of TRIM21-mediated immune signaling, when considered alongside its Ub chain-anchoring activity (Figs. 1 and 2) (12, 22–24), suggests immunostimulatory Ub-63Ub chains are first synthesized as adducts of TRIM21. Under this model, TRIM21-anchored Ub-63Ub chains could either bind downstream receptors while anchored to TRIM21 or represent precursors of unanchored Ub-63Ub chains (8).
Our data support a requirement for TRIM21-anchored Ub-63Ub to be liberated before immune activation. Depletion of the 19S DUB Poh1 prevents neutralization and signaling and leads to increased TRIM21 polyubiquitination. Although we have not directly demonstrated that it is Ub-63Ub released by Poh1 that activates NF-κB, such a model is mechanistically attractive. The requirement for Poh1 in both neutralization and signaling could be explained if the release of Ub-63Ub by Poh1 is coupled to translocation into the proteasome, allowing degradation. Ub must be removed from a substrate before translocation into the proteolytic proteasome core particle (15). Poh1 is a degradation-coupled DUB (25, 29), with Ub-63Ub specificity in vitro (18, 19) and in cells (30). Electron microscopy experiments position Poh1’s active site directly above the pore formed by the heterohexameric ATPase ring of the 19S base, through which a substrate must pass before entering the core particle (31–33). Although the removal en bloc of a Ub chain by Poh1 is a noted feature of its mechanism (26, 34), and one that distinguishes it from other 19S DUBs (15), the reason for this singular activity is not well understood. Our finding that Poh1 is required for signaling provides a plausible rationale for its liberation of Ub-63Ub.
Our data do not address how the proteasome is recruited by TRIM21. Although we observe that Ube2W and Ube2N synthesize anchored Ub-63Ub in vitro, depletion of either E2 results in a loss of TRIM21 conjugation with both Ub-63Ub and Ub-48Ub in cells. This may indicate that Ub-63Ub formation precedes Ub-48Ub conjugation and that it is the loss of Ub-48Ub that prevents proteasomal recruitment. However, we have no direct evidence that K48 polyubiquitination is required for virus degradation by TRIM21. Several E2s have been shown to synthesize Ub-48Ub with TRIM21 in vitro (3, 35), suggesting Ub-48Ub-conjugating activity in cells might be redundant. Unraveling the role of Ub-48Ub in TRIM21 function may also be complicated by the fact that downstream pathways induced by TRIM21 may require Ub-48Ub conjugation. For instance, activation of NF-κB requires Ub-48Ub-dependent IκBα degradation (28).
An important question raised by our results is why TRIM21 uses a complex mechanism of sequential ubiquitination in which anchored Ub-63Ub chains are a precursor to proteasome recruitment. One reason may be to ensure that efficient degradation of the viral particle does not antagonize the ability of TRIM21 to trigger innate signaling. Linking TRIM21 function closely with a controlled process of ubiquitination and deubiquitination allows for exquisite regulation, synchronizing signaling with virus degradation, and could limit spurious unanchored Ub-63Ub production as a result of stochastic TRIM21:Ube2N interaction in cells, in the absence of infection. Several TRIM proteins are thought to synthesize Ub-63Ub with immunomodulatory function, including TRIM5α (5) and TRIM25 (36). It will be interesting to consider whether sequential ubiquitination and deubiquitination are general features of TRIM biology, evolved to coordinate sensor and effector antiviral duality.
Materials and Methods
Cells, RNAi.
TE671, HeLa, 293T, and MEF cells were maintained in DMEM supplemented with 10% (vol/vol) FCS, penicillin at 100 U/mL, and streptomycin at 100 μg/mL. For RNAi, cells were transfected with Poh1 SMARTpool siRNA or control oligonucleotides, using Oligofectamine or RNAiMAX. Retroviral particles were generated by cotransfection of 293Ts with pCMVi GagPol, pMDG VSVgp, and shRNA-expressing vectors using Fugene-6. DNA constructs, shRNA sequences, antibodies, and inhibitors are listed in SI Materials and Methods.
Infection Assays.
AdV-GFP incubated with or without antibody for 1 h before addition to cells. Percentage GFP determined by flow cytometry 24–48 h postinfection. For luciferase assays, cells were transfected with pGL4.32 NF-κB luciferase 48 h before infection, and relative light units (RLU) measured 7 h postinfection. For cytokine mRNA measurement, cDNA was prepared at 4 h postinfection. Gene expression assays are listed in SI Materials and Methods.
Protein Purification and In Vitro Ubiquitination Reactions.
TRIM21, E1, and E2s were expressed in Escherichia coli C41 cells as GST-TEV or His-MBP fusion proteins and purified to homogeneity. In vitro ubiquitination reactions were carried out as previously described (4). For His-tag pulldowns, 106 TE671 cells, transfected with 6xHis-TRIM21 and HA-Ub, were incubated with Ni2+ agarose and purified proteins analyzed by immunoblot (IB). Full details of enzymatic reactions and purification procedures are described in SI Materials and Methods.
SI Materials and Methods
Cells, Virus, DNA, Antibodies, and Inhibitors.
TE671, HeLa, 293T, and MEF cell lines were maintained in DMEM supplemented with 10% (vol/vol) FCS, penicillin at 100 U/mL, and streptomycin at 100 μg/mL. MEFs were obtained from WT or TRIM21- deficient C57BL/6 mice (37) and have been described (4). Adenovirus-GFP had a deletion of the E1 genomic region (VQAd CMV eGFP, Viraquest). His-tagged TRIM21 was generated by PCR and ligated into pcDNA4/His (Invitrogen). Ub-GV-GFP was generated by site-directed mutagenesis of pUbGFP. HA-Ub is described (38). Pooled human serum IgG was from Sigma. Anti-adenovirus hexon monoclonal antibody 9C12 was purified as previously described (4). Antibodies used for immunoblot were anti-Ube2N (Bio-Rad, AHP974, 1:1,000), anti-β-actin-HRP (Santa Cruz, sc47778, 1:20,000), anti-Ub-HRP (Santa Cruz, sc8017-HRP P4D1, 1:1,000), anti-Ub-48Ub (Millipore, Apu2, 1:1,000), anti-Ub-63Ub (Millipore, Apu3, 1:1,000), anti-TRIM21 [raised against human TRIM21 RING-B-Box-Coiled Coil (3), 1:1,000], anti-HA (Roche, 3F10, 1:5,000), anti-Poh1 (AbCam, EPR4258, 1:1,000), anti 20Sα1–7 (AbCam, MCP231, 1:1,000), and anti-Rpn13 [AbCam, EPR11450(B), 1:1,000]. Secondary antibodies were anti-mouse-HRP (Sigma, A0168, 1:5,000), anti-rabbit-HRP (Cell Signaling, 7074, 1:5,000). MG132 (Calbiochem) was used at 2 μg/mL, bortezomib (New England Biolabs) was used at 50 nM, IKK Inhibitor VII (Calbiochem) was used at 200 nM, and 5Z-7-Oxozeaenol (Sigma) was used at 500 nM.
Luciferase Reporter Assays.
Cells were transfected with pGL4.32 NF-κB luciferase (Promega). Two days later, cells were plated at a density of 1 × 104 cells per well in 96-well plates. After 24 h, virus and antibody were incubated for 1 h at a ratio of 1:1 before addition to cells. Viruses were added to cells at the following titers: AdV, 7.5 × 105 infectious units per well. As controls, TNF was used at a concentration of 10 pg/mL. Cells were incubated for 7 h at 37 °C before the addition of 100 μL Steadylite Plus luciferase reagent (Perkin-Elmer) and analysis on a BMG Pherastar FS plate reader. For dual reporter assays, 293T cells were transfected with Poh1-specific or control siRNA oligonucleotides, and then 1 d later replated at 105 cells per well. The next day, cells were transfected with pGL4.32 NF-κB luciferase (Promega), pTK-renilla luciferase, M5P-FLAG-IKKβ, and DNA dose normalized with empty pcDNA3.1. Firefly and renilla luciferase activities were quantified using the Dual-Luciferase Reporter Assay (Promega).
Cytokine Analysis by Quantitative RT-PCR.
Cells were infected as described earlier for luciferase reporter assays. Cells were transfected with poly(I:C) (Sigma) at 200 ng per well with Lipofectamine 2000. Cells were incubated for 7 h at 37 °C, and then were washed with PBS, and cDNA was prepared with Cells-to-CT Kit (Ambion). Gene expression was monitored by TaqMan Gene Expression assays (Applied Biosystems) on a StepOnePlus Real Time PCR System (Applied Biosystems). Gene expression assays were: human β-actin (Hs00159357), IL-6 (Hs01075666), IL-8 (Hs00174103), and Ube2W (Hs00217672), and mouse TBP (Mm00446971), TNF (Mm00443258), Poh1 (Mm00451955), and CXCL10 (Mm00445235). Relative expression was quantified by the change-in-threshold (2−∆∆CT) method.
Depletion by RNAi.
Three × 105 HeLa or MEF cells were transfected in six-well plates with 100 nM or 30 nM Poh1 SMARTpool siRNA (Dharmacon), respectively, or control oligonucleotides (Ambion), using Oligofectamine (Life Technologies) (HeLa) or RNAiMAX (Life Technologies) (MEFs). Cells were replated at 24 h posttransfection and used in assays at 48 h posttransfection. DNA oligonucleotides encoding shRNA were annealed, cloned into pSIREN-RetroQ (SRQ, Clontech), and sequence verified. Retroviral particles were generated by cotransfection of 10 cm2 dishes of 293Ts with 1 μg pCMVi GagPol, 1 μg pMDG VSVgp, and 1.5 μg SRQ-shRNA, using Fugene-6 (Promega). shRNA sequences, 5′-3′, were: Ube2N (GAGCATGGACTAGGCTATA) (39), Ube2W (GCATGATAGGGCCTATGAA), Poh1 (GCAAGACAAGGGTCCATAT), and scrambled (GTTATAGGCTCGCAAAAGG).
Protein Purification.
MBP-TRIM21 purification has been described (4). TRIM21RING-Box (residues 1–129), Ube2W, Ube2N, and Ube2V2 were expressed in E. coli C41 cells as GST–TEV, His, or His-MBP fusion proteins. Cleared cell lysates were prepared by sonication in 50 mM Tris at pH 8, 150 mM NaCl, 2 mM DTT with the addition of 20% (vol/vol) BugBuster (Novagen) and cOmplete protease inhibitors (Roche), followed by centrifugation 16,000 × g for 30 min. Lysates were loaded onto GST beads and washed with lysis buffer, then cleaved with TEV protease overnight at 4 °C. Cleaved proteins were concentrated and run over a HiLoad 26/60 Superdex 75 size exclusion column (GE Healthcare). The peak fractions were pooled, concentrated, and frozen in aliquots at −80 °C. After TEV cleavage, a GSH tripeptide remained at the N terminus of TRIM21RING-Box. Ube2N was expressed as a His fusion protein and purified as described earlier, but using a Ni-NTA column followed by 300 mM Imidazole elution before size exclusion chromatography. Ube2W was expressed as a His-MBP TEV fusion and was purified as per TRIM21RING-Box, with the exception of loading lysate onto amylose resin before cleavage with TEV protease and subsequent size exclusion chromatography. TRIM21RING-Box (residues 1–129) 6KR was produced by site-directed mutagenesis of all six lysine residues within the construct: K45, K61, K77, K105, K108, and K119. For His-tag pulldowns, 106 TE671 cells, transfected with 6His-TRIM1 and HA-Ub, were washed in 5 mL PBS, resuspended in 500 μL ice-cold PBS, centrifuged, and lysed in 500 μL 6 M GuHCl, 0.1 M Na2HPO4/NaH2PO4 (pH 8), 10 mM Imidazole (pH 8). Lysates were sonicated for 15 s and rotated for 3 h at room temperature with 30 μL equilibrated NiNTA agarose (Qiagen). The agarose matrix was washed twice with 500 μL lysis buffer, twice with 500 μL 3:1 wash buffer:lysis buffer, once with 500 μL wash buffer (25 mM Tris, 20 mM Imidazole at pH 6.8), resuspended in 2× LDS sample buffer supplemented with 300 mM Imidazole to elute bound His-tagged proteins and 10% (vol/vol) β-mercaptoethanol as a reducing agent, and heated for 10 min at 95 °C before LDS-PAGE.
In Vitro Ubiquitination Reactions.
In vitro ubiquitination reactions were carried out in 1× ubiquitination buffer (50 mM Tris⋅HCl at pH 7.4, 2.5 mM MgCl2, 0.5 mM DTT) with the addition of 2 mM ATP, 0.5 µM His-E1, 1 µM Ube2W, Ube2N/Ube2V2, 8 µg Ub and 400 ng TRIM21RING-Box or MBP-TRIM21. Reaction mixtures were incubated at 37 °C for 1–4 h, quenched by addition of LDS sample buffer and boiling at 95 °C for 5 min. Samples were resolved by LDS-PAGE and TRIM21 or Ub detected by immunoblot.
Acknowledgments
This work was funded by the Medical Research Council (U105181010) and the European Research Council (281627IAI).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L. is a Guest Editor invited by the Editorial Board.
See Commentary on page 9797.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507534112/-/DCSupplemental.
References
- 1.McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23(1):46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Hauler F, Mallery DL, McEwan WA, Bidgood SR, James LC. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci USA. 2012;109(48):19733–19738. doi: 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallery DL, et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci USA. 2010;107(46):19985–19990. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwan WA, et al. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14(4):327–336. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Asmi F, et al. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014;10(2):e1003975. doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Mixed-linkage ubiquitin chains send mixed messages. Structure. 2013;21(5):727–740. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285(12):8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96(5):645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 11.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144(5):769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14(10):941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 13.Shin DY, Lee H, Park ES, Yoo YJ. Assembly of different length of polyubiquitins on the catalytic cysteine of E2 enzymes without E3 ligase; a novel application of non-reduced/reduced 2-dimensional electrophoresis. FEBS Lett. 2011;585(24):3959–3963. doi: 10.1016/j.febslet.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Rajsbaum R, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity. 2014;40(6):880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson AD, et al. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J Biol Chem. 2009;284(51):35485–35494. doi: 10.1074/jbc.M109.052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32(4):552–565. doi: 10.1038/emboj.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper EM, et al. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28(6):621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansour W, et al. Disassembly of Lys11 and mixed linkage polyubiquitin conjugates provides insights into function of proteasomal deubiquitinases Rpn11 and Ubp6. J Biol Chem. 2015;290(8):4688–4704. doi: 10.1074/jbc.M114.568295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 21.Finley D, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14(8):5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatham MH, Plechanovová A, Jaffray EG, Salmen H, Hay RT. Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem J. 2013;453(1):137–145. doi: 10.1042/BJ20130244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scaglione KM, et al. The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J Biol Chem. 2013;288(26):18784–18788. doi: 10.1074/jbc.C113.477596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vittal V, et al. Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat Chem Biol. 2015;11(1):83–89. doi: 10.1038/nchembio.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21(3):220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 26.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 27.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18(5):538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 28.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 29.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 30.Hao R, et al. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol Cell. 2013;51(6):819–828. doi: 10.1016/j.molcel.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109(37):14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathare GR, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci USA. 2014;111(8):2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20(7):781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14(2):172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimi R, et al. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182(12):7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 39.Duncan LM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25(8):1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]