Significance
Histone chaperones are key regulators of transcriptional activity in damaged chromatin regions in the DNA damage response. Here we show that cytochrome c (Cc) targets the histone chaperone SET/template-activating factor (TAF)-Iβ (SET/TAF-Iβ) in the cell nucleus upon DNA damage, resulting in the blocking of the SET/TAF-Iβ function. Cc is actually translocated into the nuclei of cells treated with specific DNA damage inducers and not upon death-receptor pathway or stress-induced stimuli. Cc locks the domains engaged in histone binding of SET/TAF-Iβ, inhibiting its nucleosome assembly activity. Structural characterization of the complex between Cc and SET/TAF-Iβ provides a valuable template for designing drugs aimed at silencing the oncogenic effect of SET/TAF-Iβ.
Keywords: cytochrome c, histone chaperone, ITC, NMR, SET-TAF-Iβ
Abstract
Chromatin is pivotal for regulation of the DNA damage process insofar as it influences access to DNA and serves as a DNA repair docking site. Recent works identify histone chaperones as key regulators of damaged chromatin’s transcriptional activity. However, understanding how chaperones are modulated during DNA damage response is still challenging. This study reveals that the histone chaperone SET/TAF-Iβ interacts with cytochrome c following DNA damage. Specifically, cytochrome c is shown to be translocated into cell nuclei upon induction of DNA damage, but not upon stimulation of the death receptor or stress-induced pathways. Cytochrome c was found to competitively hinder binding of SET/TAF-Iβ to core histones, thereby locking its histone-binding domains and inhibiting its nucleosome assembly activity. In addition, we have used NMR spectroscopy, calorimetry, mutagenesis, and molecular docking to provide an insight into the structural features of the formation of the complex between cytochrome c and SET/TAF-Iβ. Overall, these findings establish a framework for understanding the molecular basis of cytochrome c-mediated blocking of SET/TAF-Iβ, which subsequently may facilitate the development of new drugs to silence the oncogenic effect of SET/TAF-Iβ’s histone chaperone activity.
The oncoprotein SET/template-activating factor (TAF)-Iβ (SET/TAF-Iβ)—also known as inhibitor-2 of protein phosphatase-2A (I2PP2A) and inhibitor of histone acetyltransferase (INHAT)—belongs to the nucleosome assembly protein (NAP) family of histone chaperones. SET/TAF-Iβ participates in numerous cellular processes, including cell cycle control (1), apoptosis (2), transcription regulation (3), and chromatin remodelling (4). Many recent studies highlight the chromatin-related properties of SET/TAF-Iβ. As a histone chaperone, SET/TAF-Iβ associates to core histones to shield their positive charge, preventing improper contact with DNA and facilitating the correct deposition of free histones onto DNA for nucleosome formation (5). Recent work points to the crucial role of chromatin dynamics in DNA damage response (6–8). In fact, histone chaperones have emerged as key players in the transient disorganization of chromatin required in the DNA repair process. Thus, the histone chaperones aprataxin-PNK–like factor (APLF) (6), antisilencing function 1 (Asf1) (9), chromatin assembly factor 1 (CAF-1) (10), death domain-associated protein 6 (DAXX) (11), facilitating chromatin transcription (FACT) (12), histone regulator A (HIRA) (13), nucleolin (14), p400 (15), and nucleosome assembly protein 1-like 1 (NAP1L1) and 1-like 4 (NAP1L4) (7) are recruited to damaged chromatin, thereby promoting histone dynamics in response to DNA damage. Recently, SET/TAF-1β was found to be recruited to DNA breaks to modulate the DNA damage response through the retention of certain chromatin proteins, resulting in chromatin compaction (16). We have identified SET/TAF-Iβ as a target protein for cytochrome c (Cc) following the release of the latter from mitochondria in human cells treated with apoptotic agents (17, 18). Although the hemeprotein Cc serves as an electron carrier between complexes III and IV in the mitochondrial respiratory chain, its role in nuclei has yet to be convincingly elucidated. It has been proposed, however, that Cc accumulation in the nucleus under apoptotic stimuli relates to nuclear pyknosis, DNA fragmentation (19), and chromatin remodelling (20). Here, we show that the SET/TAF-Iβ oncoprotein interacts with Cc in the cell nucleus in response to treatment of the cell with different well-known inducers of DNA damage and apoptosis, but not to treatment with other apoptosis-inducing agents. We also demonstrate that Cc impairs the histone chaperone activity of SET/TAF-Iβ through competitive binding, thereby preventing the formation of core histone–SET/TAF-Iβ complexes.
Results
Cc Interacts with SET/TAF-Iβ in the Nucleus Following DNA Damage.
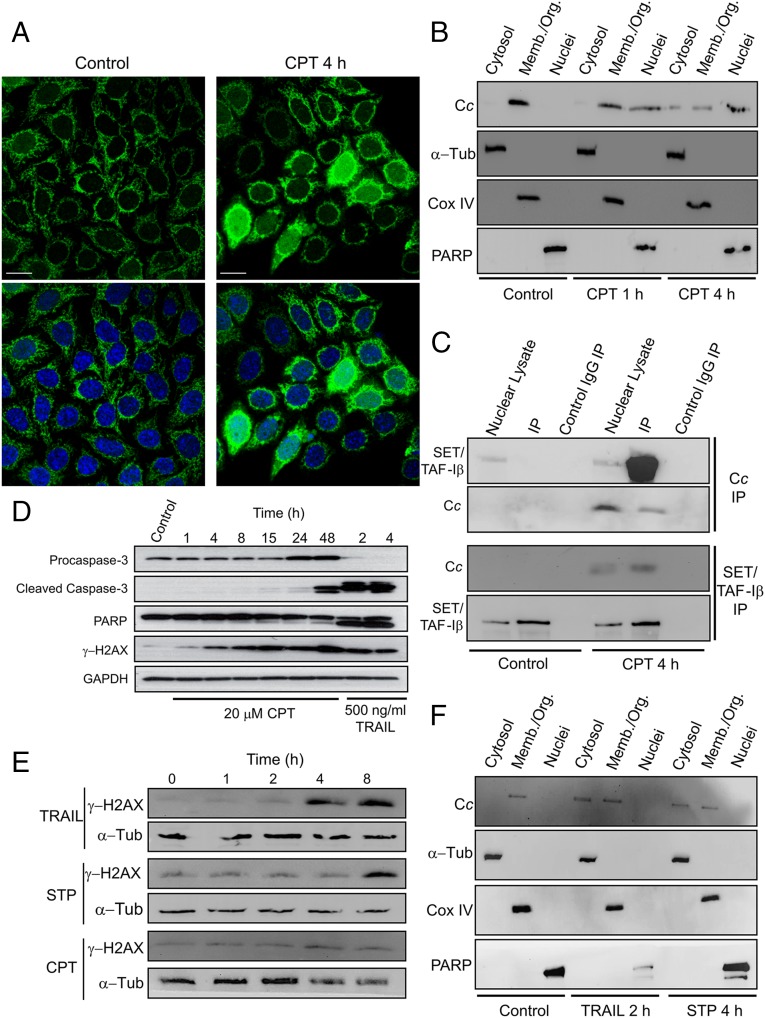
DNA damage can be induced by ionizing radiation or topoisomerase inhibitors (e.g., CPT). Subcellular localization of Cc in Heltog cells—a HeLa cell line constitutively expressing green fluorescent protein (GFP)-tagged Cc—showed Cc-GFP predominantly in mitochondria of untreated cells (Fig. 1A, Left). Confocal analysis of cells treated with 20 μM CPT for 4 h revealed a translocation of Cc-GFP from mitochondria to the cytoplasm and the nucleus. The nuclear localization of Cc-GFP has been confirmed by colocalization with the Hoechst staining (Fig. 1A, Right). Subcellular fractionation of Heltog cells treated with 20 μM CPT showed endogenous Cc accumulation in the cell nucleus after 4 h (Fig. 1B). In fact, Cc appeared in the nucleus after 1 h treatment, as previously observed in HeLa cells treated with either UV irradiation or CPT (20). Codetection in the nuclear cell fraction with nuclear-specific poly (ADP ribose) polymerase (PARP) confirmed the Cc translocation into the nucleus (Fig. 1B). Notably, following treatment with CPT, Cc translocation into the nucleus occurs before caspase-3 activation (Fig. 1D). To further demonstrate the relationship between endogenous SET/TAF-Iβ and Cc in response to DNA damage, in-cell interaction between them was examined using immunoprecipitation (IP). An antibody against Cc was used to extract associated proteins in nuclear lysates of Heltog cells treated with 20 μM CPT for 4 h. As shown in Fig. 1C, Upper, SET/TAF-Iβ coimmunoprecipitated with endogenous Cc after CPT treatment (lane 5), whereas untreated cells (control) did not show any band corresponding to SET/TAF-Iβ (lane 2). To confirm the IP specificity, nuclear lysates from untreated and CPT-treated cells were probed with the SET/TAF-Iβ antibody (Fig. 1C, Upper, lanes 1 and 4, respectively), whereas the negative controls (Fig. 1C, Upper, lanes 3 and 6) did not show any band when IgG was used. Cc IP was confirmed by immunoblotting with the anti-Cc antibody (Fig. 1C, Upper). To confirm our findings, a reverse IP using the SET/TAF-Iβ antibody was performed, and endogenous Cc was then pulled down (Fig. 1C, Lower). To examine whether the observed Cc:SET/TAF-Iβ interaction is restricted to cells experiencing DNA damage, the effect of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) and staurosporine (STP)—two well-known apoptosis-inducing agents for which DNA damage is not an initiating event—was studied. TRAIL is a member of the TNF superfamily that can induce cell death either by the extrinsic pathway or by the BH3 interacting-domain death agonist (Bid)-mediated mitochondrial pathway (21). STP is a broad-spectrum protein kinase inhibitor that promotes intracellular stress-induced apoptosis (22). Apoptosis induction with TRAIL or STP may result in DNA damage in later stages of treatment. Therefore, cell cultures were first tested to determine whether they exhibited DNA damage following TRAIL- or STP-induced apoptosis. To this end, Heltog cell lysates were treated with 100 ng/mL TRAIL or 1 μM STP for 1, 2, 4, and 8 h and immunoblotted against Ser139-phosphorylated histone H2AX (γ-H2AX). γ-H2AX is an extremely sensitive marker of DNA damage as it accumulates rapidly in response to double-strand breaks (DSBs). As shown, the DNA-damage–dependent accumulation of γ-H2AX occurs after 4 or 8 h of treatment with TRAIL and STP, respectively (Fig. 1E). By contrast, CPT promotes histone H2AX phosphorylation after 1 h. Because no significant DNA damage was observed following exposure to TRAIL for 2 h or STP for 4 h, these exposure times were selected to further explore Cc localization. Consequently, following the exposure of Heltog cell cultures to 100 ng/mL TRAIL for 2 h or 1 μM STP for 4 h, subcellular fractionation was applied, indicating Cc as having been translocated from mitochondria to cytosol, but not to the nucleus (Fig. 1F). This corresponds to findings in previous studies of Cc release from mitochondria in response to TRAIL and STP (23, 24). Nevertheless, our study shows endogenous Cc as having been unable to reach the cell nucleus following treatments with TRAIL or STP (Fig. 1F). This contrasts with the presence of Cc in the nucleus observed in response to CPT-induced DNA damage (Fig. 1 A and B). The translocation of Cc into the nucleus, along with the former’s interaction with SET/TAF-Iβ, has been studied following the induction of DNA damage with indotecan, a noncamptothecin inhibitor of topoisomerase I (25), and doxorubicin, a topoisomerase II inhibitor (26) (SI Appendix, Fig. S1). Altogether, these results indicate that a specific DNA-damage stimulus triggers nuclear translocation of Cc, thereby permitting binding to its nuclear target SET/TAF-Iβ.
Fig. 1.
CPT-induced nuclear translocation of Cc and formation of the Cc:SET/TAF-Iβ complex. (A) Subcellular localization of Cc-GFP stably expressed (green; Upper) in Heltog cells upon treatment with 20 μM CPT for 4 h detected by confocal microscopy (60x oil objective). Nuclei were stained in blue with Hoechst. Colocalization of green Cc-GFP fluorescence and blue nuclear staining is shown in the merge images (Lower). (Scale bars, 25 μm.) (B) Subcellular fractioning showing endogenous Cc location upon treatment with 20 μM CPT for 1 or 4 h. Nontreated and CPT-treated Heltog cells were fractionated to yield cytosolic, membrane/organelle (Memb./Org.) and nuclear fractions. Purity of subcellular fractions was verified by Western blot using anti–α-Tub (50 kDa), anti-Cox IV (17 kDa), and anti-PARP (116 kDa) antibodies. (C, Upper) IP of SET/TAF-Iβ with endogenous Cc after treating Heltog cells with 20 μM CPT for 4 h. Western blot showed the detection of SET/TAF-Iβ as an ∼34-kDa band (lanes 1 and 4) in the nuclear fraction. Cc-IP of nuclear lysates from nontreated (lane 2) and CPT-treated cells (lane 5), followed by probing with the SET/TAF-Iβ antibody. Confirmation of immunoprecipitated Cc from nuclear lysates is also shown (lanes 4 and 5) under CPT treatment. (Lower) Reverse IP of endogenous Cc with SET/TAF-Iβ following CPT treatment. Detection of Cc as an ∼12-kDa band in the nuclear lysate (lane 4) and in the IP of SET/TAF-Iβ of CPT-treated cells (lane 5). Mouse IgG was used as control (lanes 3 and 6). (D) Detection of caspase-3 activation in Heltog cells treated with 20 μM CPT for 1, 4, and 24 h and 100 ng/mL TRAIL for 2 h. Cleaved caspase-3 (19 and 17 kDa) and full-length (116 kDa) or cleaved PARP (89 kDa) was detected by Western blot. Procaspase-3 (32 kDa) and α-Tub (50 kDa) antibodies were used as controls. (E) DNA-damage response upon treatment of Heltog cell cultures with 100 ng/mL TRAIL and 1 μM STP or 20 μM CPT for 0, 1, 2, 4, and 8 h. Specific antibody against phosphorylated γ-H2AX was used in Western blotting. α-Tub antibody was used as loading control. (F) Subcellular fractioning showing endogenous Cc location upon treatment with 100 ng/mL TRAIL for 2 h or 1 μM STP for 4 h.
Cc Binds to SET-TAF-Iβ and Blocks Histone Binding.
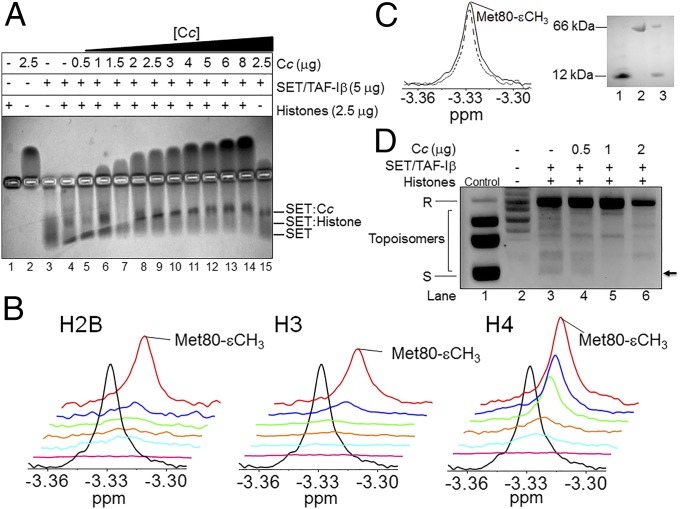
To explore the biological significance of the Cc:SET/TAF-Iβ interaction in response to a DNA-damage stimulus, we tested the ability of Cc to prevent histone binding to SET/TAF-Iβ. Hence, an electrophoretic mobility shift assay (EMSA) was performed to detect complex formation between SET/TAF-Iβ and calf-thymus histones and to further study the effect of the addition of Cc. The mobility of the histone mixture, as well as Cc and SET/TAF-Iβ, are shown in Fig. 2A (lanes 1–3). Due to their opposite charges, Cc and SET/TAF-Iβ migrated in reverse directions. Notably, the histone mixture vaguely penetrated the gel in the EMSA assay due to its propensity to form large aggregates (SI Appendix, Fig. S2A). The lower mobility of SET/TAF-Iβ following histone addition (Fig. 2A, lane 4) indicates the formation of chaperone–histone complexes. The addition of Cc at increasing concentrations to the SET/TAF-Iβ and histone mixture (Fig. 2A, lanes 5–14) made the chaperone mobility match observed for the Cc:SET/TAF-Iβ complex (lane 15), revealing that Cc competes with histones for the SET/TAF-Iβ binding site. To confirm the EMSA results, NMR measurements of reduced Cc were recorded in the presence of SET/TAF-Iβ and Xenopus laevis core histones H2B, H3, and H4. The use of isolated histones allowed for a comparison of their respective binding affinities for the chaperone in competition with Cc. However, binding to histone H2A could not be tested, as the expression of H2A was not possible. Specifically, the Met80-εCH3 NMR signal of the methionine axial ligand of Cc (Met80) was monitored. As shown, this signal broadens beyond the detection limit upon the addition of SET/TAF-Iβ (Fig. 2B). This is due to the long diffusional correlation time of Cc bound to SET/TAF-Iβ, which results in a fast signal relaxation. As expected for a binding competition, titration of the Cc:SET/TAF-Iβ solution with increasing concentrations of H2B, H3, or H4 histones led to dissociation of Cc from SET/TAF-Iβ and the recovery of the Met80-εCH3 signal. As a control, BSA was added to unbound Cc at the same ratio as that used for SET/TAF-Iβ. The line width of Met80-εCH3 of the Cc remained unchanged as unbound Cc, suggesting that Cc does not interact with BSA (Fig. 2C). To avoid nonspecific interactions, 1D 1H NMR spectra of Cc, following binding to SET/TAF-Iβ and a calf-thymus histone mixture in the presence of 100 mM KCl, were recorded (SI Appendix, Fig. S2B). The same competition effect was observed as that shown in Fig. 2B: namely, titration with increasing concentrations of histones led to the dissociation of Cc from SET/TAF-Iβ and the recovery of the intensity of the Met80-εCH3 Cc signal. Circular dichroism (CD) spectra of SET/TAF-Iβ and histones H2B, H3, and H4 corroborated the proper secondary structure organization (SI Appendix, Fig. S2C). To compare the binding thermodynamics of the complexes formed by SET/TAF-Iβ with Cc and core histones, Isothermal Titration Calorimetry (ITC) experiments were performed. Measurements revealed the interaction between Cc and SET/TAF-Iβ being slightly exothermic and entropically driven (binding enthalpy, ΔH, −0.9 kcal⋅mol−1) with a dissociation constant (KD) of 3.1 μM and a Cc:SET/TAF-Iβ stoichiometry of 2:1 at 25 °C (SI Appendix, Table S1). Notably, the binding of Cc to SET/TAF-Iβ exhibited weak positive cooperativity (SI Appendix, Fig. S3), yielding a cooperativity constant (k) of 3.8 and leading to a smaller KD value and a very positive enthalpy of interaction for the second site (SI Appendix, Table S1). In other words, the binding of the first Cc molecule increases 3.8 times the affinity for the binding of a second Cc molecule to SET/TAF-Iβ. Therefore, it might be plausible that, following the binding of the first Cc molecule, SET/TAF-Iβ undergoes a conformational change that facilitates the interaction with the second Cc molecule. As before, ITC experiments were used to quantitatively assess the binding of SET/TAF-Iβ to the core histones H2B, H3, and H4 from X. laevis. As shown (SI Appendix, Fig. S3 and Table S1), H3 binds SET/TAF-Iβ with a lower affinity (KD = 25 μM) than H2B (KD = 2 μM) or H4 (KD = 6 μM). These values agree with those previously reported for the interaction of SET/TAF-Iβ with H2B (KD = 2.87 μM), but differ slightly for H3 (KD = 0.15 μM) (27). In all cases, the stoichiometry of histone:SET/TAF-Iβ interactions was 2:1 (SI Appendix, Table S1). As for NMR titrations, ITC measurements at 100 mM KCl were also recorded to suppress nonspecific interactions (SI Appendix, Table S1). At high ionic strength, the binding affinity of the two-component complexes decreases because all are electrostatically driven and their stoichiometry remains unaltered.
Fig. 2.
Competition between histones and Cc for binding to SET/TAF-Iβ. (A) EMSA showing competitive interaction of SET/TAF-Iβ with calf-thymus histones in the presence of Cc at increasing concentrations. (B) 1D 1H NMR spectra monitoring Met-80 methyl signal of reduced Cc in the presence of SET/TAF-Iβ and X. laevis core histones. Details of superimposed 1D 1H NMR spectra of 13 μM reduced Cc [either free (black) or bound to 3.5 μM SET/TAF-Iβ (pink)] in the presence of the X. laevis core histones H2B, H3, or H4 at increasing concentrations of 5 μg (cyan), 10 μg (orange), 20 μg (green), 30 μg (blue), and 40 μg (red). (C, Left) 1D 1H NMR spectra monitoring Met-80 methyl signal of 13 μM reduced Cc in the presence (dashed line) and absence (straight line) of 3.5 μM BSA. (Right) SDS/PAGE showing the Cc and BSA mixture used in the 1D 1H NMR spectrum (lane 3). Isolated Cc (lane 1) and BSA (lane 2) are shown as controls. (D) Impairment of the histone chaperone activity of SET/TAF-Iβ by Cc. Two micrograms SET/TAF-Iβ was combined with 200 ng relaxed plasmid after being treated with Topo I and incubated with 2 μg HeLa core histones. Nucleosome assembly activity was tested in the absence (lane 3) and the presence of Cc at increasing concentrations (lanes 4–6). Relaxed and supercoiled forms of circular plasmid DNA are indicated by “R” and “S,” respectively. Lane 2 corresponds to DNA plasmid relaxed after treatment with Topo I, whereas lane 1 (control) shows a supercoiled, untreated DNA plasmid. The arrow indicates the topoisomer with a high number of nucleosomes.
SET/TAF-Iβ Histone-Binding Domain Is Engaged During Binding to Cc.
Considering that Cc binds to SET/TAF-Iβ and affects its interaction with core histones, we aimed at determining whether Cc recognizes the SET/TAF-Iβ histone-binding domain. SET/TAF-Iβ forms a headphone-shaped homodimer, each monomer consisting of an N terminus, a backbone helix, an “earmuff” domain, and an acidic disordered stretch (5) (SI Appendix, Fig. S4). The region of SET/TAF-Iβ responsible for histone binding, and chaperone activity comprises the lower area of the so-called “earmuff” domain (5). To examine the SET/TAF-Iβ region involved in binding to Cc, two different SET/TAF-Iβ deletion mutants were designed. The first included the N terminus along with the backbone helix [hereafter referred to as SET/TAF-Iβ(1–80) (amino acids 1–80)], whereas the second [SET/TAF-Iβ(81–277) (amino acids 81–277)] encoded the “earmuff” domain and the acidic stretch of the C terminus. Worth particular mention is that CD spectra of both mutants indicated a secondary structure similar to that of the wild-type protein (SI Appendix, Fig. S2C). ITC of Cc interactions with SET/TAF-Iβ(1–80) and SET/TAF-Iβ(81–277) showed the binding reaction in both cases (SI Appendix, Fig. S5, Upper). However, weak calorimetric signals in the thermogram of SET/TAF-Iβ(1-80) throughout the titration with Cc (SI Appendix, Fig. S5, Upper) suggest a weak interaction with an estimated KD of 44 μM (SI Appendix, Table S1). On the contrary, the affinity between Cc and SET/TAF-Iβ(81–277) was higher and yielded a KD of 9.6 μM. The results indicate a binding preference of Cc for the C terminus of SET/TAF-Iβ and explain the way in which Cc hampers the chaperone’s ability to bind to histones. To further confirm the C terminus of SET/TAF-Iβ as the region interacting with Cc, two SET/TAF-Iβ mutants were obtained by replacing three adjacent amino acid residues from the “earmuff” domain by alanine residues (SI Appendix, Fig. S4). Mutants S162A/K164A/D165A and T191A/T194A/D195A (SI Appendix, Fig. S4, green and red, respectively) failed to bind to histones and demonstrated significantly reduced histone chaperone activity (35% of wild-type activity) (5). According to the calorimetric data resulting from the titrations of the two SET/TAF-Iβ triple mutants with Cc (SI Appendix, Fig. S5, Lower), the S162A/K164A/D165A mutant shows a higher affinity toward Cc (KD = 6.3 μM) than T191A/T194A/D195A (KD = 22 μM). These findings suggest that threonine residues at positions 191 and 194 and aspartate at 195, located in the lower region of the “earmuff” domain, are involved in the interaction with Cc, as they are in the histone remodelling activity of SET/TAF-Iβ.
SET/TAF-Iβ Histone Chaperone Activity Is Impaired by Cc.
As binding to core histones is essential for the histone chaperone activity of SET/TAF-Iβ, the ability (or inability) of Cc to affect such activity was tested using a supercoiling assay (SI Appendix, Fig. S6A). The histone chaperone activity of SET/TAF-Iβ has been previously demonstrated by the same supercoiling assay used here (5). SET/TAF-Iβ assembled nucleosomes onto the plasmid, resulting in enhanced supercoiling and subsequently increased electrophoretic migration compared with the relaxed DNA (compare lanes 2 and 3 of Fig. 2D). Although highly supercoiled DNA migrated in a single band, many different topoisomers might be present (Fig. 2D). The slight lower band that runs faster in the plasmid incubated with SET/TAF-Iβ and core histones (lane 3, Fig. 2D) corresponded to a topoisomer with a high number of assembled nucleosomes due to an increase in supercoiling. Although the relaxed band did not disappear following the addition of 0.5 or 1 μg Cc (lanes 4 and 5 of Fig. 2D), the topoisomers (the bands that run faster due to supercoiling) decreased in intensity, indicating a decrease in the amount of plasmid supercoiling. These facts reveal an inhibition of the chaperone activity of SET/TAF-Iβ when mediated by Cc. Controls corresponding to the supercoiling assay are also shown (SI Appendix, Fig. S6B).
Cc Heme Crevice Faces SET/TAF-Iβ Upon Binding.
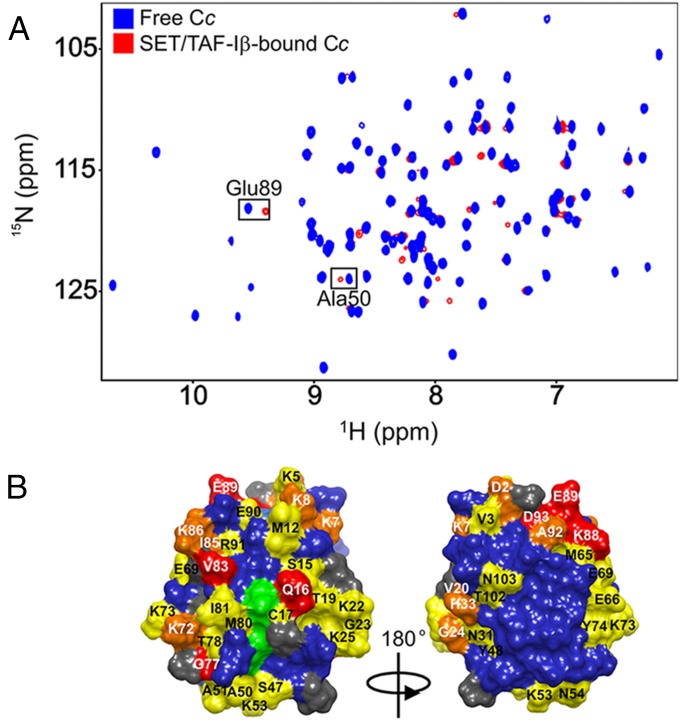
To dig into the structural features of the interaction between Cc and SET/TAF-Iβ, the Cc:SET/TAF-Iβ interaction was monitored by recording [1H, 15N] Heteronuclear Single-Quantum Correlation (HSQC) spectra of fully reduced 15N-labeled Cc, which was either free or bound to unlabeled SET/TAF-Iβ. Titration of SET/TAF-Iβ onto 15N Cc resulted in a significant broadening of the Cc resonances, thereby suggesting the formation of the Cc:SET/TAF-Iβ complex (Fig. 3A). Additionally, specific resonances from residues at the complex interface may show enhanced broadening. Consequently, the line widths (ΔΔν1/2Binding) of 15N-labeled Cc resonances were analyzed in a pure Cc sample and a 1:0.25 Cc:SET/TAF-Iβ mixture . Signals showing this effect corresponded to Thr19, Lys39, Gly41, Gln42, Tyr48, Lys79, Ile81, Ala92, and Leu94 in the 15N dimension (SI Appendix, Fig. S7A). Hence, these residues can be expected to be at or near the region of Cc interacting with SET/TAF-Iβ. In addition to specific line broadening, several amide resonances in the [1H, 15N] HSQC spectra of Cc exhibit chemical-shift perturbations (CSPs) in the presence of SET/TAF-Iβ (Fig. 3A). For example, the details of the superimposed spectra (SI Appendix, Fig. S7B) show that Ala50 and Glu89 resonances shift gradually at increasing Cc:SET/TAF-Iβ ratios (1:0.06, 1:0.12, and 1:0.25). These CSPs indicate that these residues experienced a change in their chemical environment in the presence of SET/TAF-Iβ. Therefore, they may belong to the complex interface. To identify the residues involved in the complex interface, an average CSP analysis (ΔδAvg) of the Cc amide signals was obtained. 15N Cc resonances from Gln16, Gly77, Val83, Lys88, Glu89, and Asp93 experience significant chemical shifts (ΔδAvg ≥ 0.075) following binding to SET/TAF-Iβ (SI Appendix, Fig. S7C). Strikingly, the majority of perturbed Cc residues are located on the side of the exposed heme periphery (Fig. 3B). Notably, a similar surface patch of Cc is involved in the interactions with cytochrome c oxidase (28) and cytochrome bc1 (29). Interestingly, the interaction between Cc and the SET/TAF-Iβ chaperone implicated 10 lysine residues—namely 5, 7, 8, 22, 25, 53, 72, 73, 86, and 88 (Fig. 3B)—thereby evidencing the key role of electrostatic forces in the formation of Cc:SET/TAF-Iβ complexes. Similarly, lysine residues are also involved in the interaction with cytochrome bc1 (29, 30). Given the well-known “lysine masking activity” of SET/TAF-Iβ that prevents the acetylation of histone lysines inside the INHAT complex (3), it is plausible that SET/TAF-Iβ also recognizes the lysine residues from Cc. Intriguingly, the Cc residues thought to play an important role in the interaction between the hemeprotein and apoptosis protease-activating factor-1 (Apaf-1)—namely, Lys7, Lys25, Lys39, and Lys72 (31)—perfectly match those identified in this study as interacting with SET/TAF-Iβ. Furthermore, whereas studies of the complex formed between Cc and prosurvival protein Bcl-xL identified both His26 and Gly41 as the most important Cc residues affected upon binding (32), the present study shows a change only in Gly41 following the addition of SET/TAF-Iβ to the Cc sample. To clarify the role of the redox state of Cc, this interaction was also analyzed using the oxidized form of Cc by combining NMR and ITC techniques. Most of the residues of oxidized Cc involved in the binding to SET/TAF-Iβ were the same as those found for its reduced form (SI Appendix, Fig. S8A). Notably, specific Cc resonances were not detectable upon complex formation as they broadened beyond the detection limit (SI Appendix, Fig. S8A). Similarly, the ITC titration using oxidized Cc resulted in similar thermogram profiles and binding affinities as those of the reduced Cc (SI Appendix, Fig. S8B). Altogether, this indicates that the redox state of Cc does not influence its binding to its nuclear target SET/TAF-Iβ.
Fig. 3.
NMR titrations of 15N-labeled reduced Cc with SET/TAF-Iβ. (A) Superimposed [1H-15N] 2D HSQC spectra of 15N-labeled Cc, which is either free or bound to dimeric SET/TAF-Iβ at Cc:SET/TAF-Iβ molar ratio of 1:0.25. (B) Mapping of Cc residues perturbed upon binding to SET/TAF-Iβ. Cc surfaces are rotated 180° around vertical axes in each view. Residues are colored according to their ΔδAvg (ppm): ≥0.075 (red), between 0.075 and 0.050 (orange), between 0.050 and 0.025 (yellow), and < 0.025 (blue). Unassigned and Pro residues are in gray, whereas the heme group is in green.
Structural Look into the Cc:SET/TAF-Iβ Complex and NMR-Based Molecular Docking Models.
With the aim of defining Cc:SET/TAF-Iβ complex interface regions, NMR restraint-driven docking was performed. CSPs obtained from NMR analysis of the Cc:SET/TAF-Iβ complex at a 1:0.25 ratio were used as input data. From the 500 solutions obtained from the NMR-based docking (SI Appendix, Fig. S9A), Cc geometric centers are represented around Robertson (ribbon) diagrams of SET/TAF-Iβ. In agreement with the two binding sites observed by ITC titrations, the models predicted by the NMR restraint-driven docking yielded two differentiated clusters. One of these clusters included the vast majority of highest scoring models (SI Appendix, Fig. S9A, cluster 1), whereas the second cluster encompassed fewer than 50 solutions (SI Appendix, Fig. S9A, cluster 2). Interestingly, nearly all of the probe solutions are located in the same region of SET/TAF-Iβ—namely, between its “earmuff” domains. This finding supports the data reported in earlier sections of this study suggesting a Cc-binding preference for the SET/TAF-Iβ histone-binding domain. Indeed, the highest-scoring solution from the first cluster revealed how Cc leans its heme group to approach the lower region of a SET/TAF-Iβ “earmuff” domain (SI Appendix, Fig. S9B). The surface representation of both proteins shows them to be in close contact. A secondary set of Cc structures with higher energies (cluster 2) was found near the backbone helices (SI Appendix, Fig. S9C).
Discussion
Inefficient repair of DNA lesions results in genome instability, which can, in turn, lead to premature aging and cancer (33). Chromatin dynamics regulate transcriptional activity in response to DNA damage by promoting accessibility to DNA and serving as a docking site for repair and signaling proteins, thereby increasing repair efficiency (8, 34). Recent works reveal how histone chaperones temporarily evict histones from a damaged site to facilitate access of repair factors to DNA lesions (9). However, once a DNA break is repaired, histone chaperones return histone proteins to the repaired site (35) and promote the recovery of transcriptional activity (7, 13). The data presented in this study show the histone chaperone and oncoprotein SET/TAF-Iβ to specifically interact with Cc in cell nuclei in response to CPT-induced DNA damage. Although translocation of Cc to the nucleus had previously been observed (19, 20), the specific role of Cc in the nucleus has yet to be fully elucidated. However, this work shows that Cc translocates into the cell nucleus in response to DNA damage, but not following the TRAIL-activated extrinsic pathway or STP-triggered stress-induced apoptosis. The present study demonstrates that nuclear translocation of Cc occurs early in the DNA damage response—just 1 h after treatment. The finding suggests that Cc might be imported into the nucleus by diffusion through the nuclear pore complex, although the precise molecular mechanism is as yet unknown. As neither TRAIL nor STP trigger Cc entry into the cell nucleus, the interaction between Cc and SET/TAF-Iβ seems to be specifically linked to DNA damage. CPT selectively targets the essential mammalian enzyme Topo I in nuclei (36). CPT binds covalently to Topo I and supercoiled DNA, thereby forming a ternary complex that inhibits DNA religation and generates a greater number of DSBs (37). Recently, it has been proposed that histone chaperones play a central role in chromatin disassembly connected with DNA repair and transcription recovery following DNA damage (7, 9, 13, 35). Thus, histone chaperones such as HIRA (13), FACT (12), nucleolin (14), APLF (6) Asf1 (9), CAF-1 (10), DAXX (11), p400 (15), NAP1L1, and NAP1L4 (7) actively promote transient chromatin disorganization and histone reshaping in response to DNA damage (8). Because SET/TAF-Iβ belongs to the NAP1 family, it is tempting to hypothesize a role—similar to that described for the above-mentioned histone chaperones—for SET/TAF-Iβ in chromatin reshaping in response to DNA lesions. Such a role would also explain the Cc-mediated inhibition observed here.The results of ITC and NMR titrations herein performed reveal the oncoprotein SET/TAF-Iβ as binding specifically to Cc. Interestingly, Cc uses the surface residues surrounding its heme group to interact with the histone chaperone, as it does with its respiratory partners cytochrome c oxidase (28) and cytochrome bc1 (29) and its apoptotic partner Apaf-1 (31). Mutagenesis and NMR-based docking analysis carried out here demonstrated that Cc docks between the two histone-binding domains of SET/TAF-Iβ, thereby preventing the binding of the latter to core histones. Therefore, and as inferred from nucleosome assembly activity assays, it may be concluded that Cc not only binds to SET/TAF-Iβ, but also hampers the latter’s histone chaperone activity. It may therefore be proposed that, following DNA damage, the translocation of Cc into the cell nucleus and resulting core histone displacement may hinder SET/TAF-Iβ nucleosome assembly activity (Fig. 4). Thus, the specific inhibition of SET/TAF-Iβ by Cc could contribute not only to suppressing the former’s normal chaperone activity when apoptosis is inevitable and such activity is no longer necessary, but also to obstructing the nucleosome remodelling that follows DNA damage repair. It has been proposed that mitochondria from apoptotic cells are capable of oxidizing cytosolic Cc via cytochrome c oxidase and that this oxidized form induces caspase activation (38). Later, cytosolic Cc can be reduced by several reductants, leading to diminished caspase activation. Moreover, it is known that nuclei have reduced redox potentials, but are relatively resistant to oxidation (39). Our results suggest that the redox state of Cc does not influence its binding to its nuclear target SET/TAF-Iβ. Our finding that Cc interacts with the oncoprotein SET/TAF-Iβ upon its release from mitochondria suggests that the role of Cc in the execution of apoptosis is wider than previously held, insofar as it goes beyond caspase cascade activation, by also inhibiting prosurvival Cc partners. To our knowledge, this is the first time that a specific role for nuclear Cc has been suggested. The results presented not only reveal the molecular basis for the blocking of SET/TAF-Iβ activity by Cc, but also suggest that the inhibition of this oncoprotein could be a promising objective in the development of anticancer drugs. More specifically, an understanding of the molecular interfaces in the complex formed by SET/TAF-Iβ and its inhibitor, Cc, could facilitate the development of new drugs aimed at silencing the oncogenic effect of SET/TAF-Iβ histone chaperone activity.
Fig. 4.
Proposed model of Cc-mediated nucleosome assembly disability under DNA damage. Schematic assembly of nucleosomes by SET/TAF-Iβ under homeostasis (Upper) and its impairment by Cc upon nuclear translocation under DNA damage conditions (Lower), both showing mitochondrial outer membrane (MOM), mitochondrial intermembrane space (IMS), mitochondrial inner membrane (MIM), and matrix (MM).
Materials and Methods
The expression and purification of Cc, wild-type SET/TAF-Iβ, and its mutants and histones are described in SI Appendix, as are other experimental conditions and calculations.
Supplementary Material
Acknowledgments
We thank Dr. C. Muñoz-Pinedo for providing the HeLa Cc-GFP (Heltog) cell line; Dr. M. Horikoshi for the kind gift of plasmids encoding SET/TAF-Iβ triple mutants; Dr. T. Richmond for providing plasmids encoding H2A, H2B, H3, and H4 of X. laevis; Dr. Y. Pommier for providing the indotecan compound used; and Dr. S. G. Bartual for critical reading. This work was supported by the Spanish Ministry of Economy and Competitiveness (BFU2012-31670, BFU2013-47064-P, SAF2012-32824, and RD12/0036/0026); the European Commission (Fondo Europeo de Desarrollo Regional); the Regional Government of Andalusia (BIO198); the Ramon Areces Foundation; the European Bio-NMR Research Infrastructure (FP7); the NMR Facility at Centro de Investigación, Tecnología e Innovación de la Universidad de Sevilla; and the Biointeractomics Platform at cicCartuja.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 26578).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508040112/-/DCSupplemental.
References
- 1.Estanyol JM, et al. The protein SET regulates the inhibitory effect of p21(Cip1) on cyclin E-cyclin-dependent kinase 2 activity. J Biol Chem. 1999;274(46):33161–33165. doi: 10.1074/jbc.274.46.33161. [DOI] [PubMed] [Google Scholar]
- 2.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112(5):659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 3.Seo SB, et al. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277(16):14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- 4.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J Biol Chem. 1998;273(51):34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 5.Muto S, et al. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci USA. 2007;104(11):4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehrotra PV, et al. DNA repair factor APLF is a histone chaperone. Mol Cell. 2011;41(1):46–55. doi: 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho I, Tsai P-F, Lake RJ, Basheer A, Fan H-Y. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013;9(4):e1003407. doi: 10.1371/journal.pgen.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adam S, Polo SE. Blurring the line between the DNA damage response and transcription: The importance of chromatin dynamics. Exp Cell Res. 2014;329(1):148–153. doi: 10.1016/j.yexcr.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanae K, Horiuchi T, Matsuo Y, Katayama S, Kawamukai M. Histone chaperone Asf1 plays an essential role in maintaining genomic stability in fission yeast. PLoS One. 2012;7(1):e30472. doi: 10.1371/journal.pone.0030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127(3):481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 11.Lacoste N, et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53(4):631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Dinant C, et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell. 2013;51(4):469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155(1):94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi J, et al. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS One. 2012;7(11):e49245. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48(5):723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalousi A, et al. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Reports. 2015;11(1):149–163. doi: 10.1016/j.celrep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Fábregas J, et al. Structural and functional analysis of novel human cytochrome C targets in apoptosis. Mol Cell Proteomics. 2014;13(6):1439–1456. doi: 10.1074/mcp.M113.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Fábregas J, Díaz-Moreno I, González-Arzola K, Díaz-Quintana A, De la Rosa MA. A common signalosome for programmed cell death in humans and plants. Cell Death Dis. 2014;5:e1314. doi: 10.1038/cddis.2014.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Aviles ER, Jr, Fujikawa DG. Nuclear translocation of mitochondrial cytochrome c, lysosomal cathepsins B and D, and three other death-promoting proteins within the first 60 minutes of generalized seizures. J Neurosci Res. 2010;88(8):1727–1737. doi: 10.1002/jnr.22338. [DOI] [PubMed] [Google Scholar]
- 20.Nur-E-Kamal A, et al. Nuclear translocation of cytochrome c during apoptosis. J Biol Chem. 2004;279(24):24911–24914. doi: 10.1074/jbc.C400051200. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29(34):4752–4765. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- 22.Feng G, Kaplowitz N. Mechanism of staurosporine-induced apoptosis in murine hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2002;282(5):G825–G834. doi: 10.1152/ajpgi.00467.2001. [DOI] [PubMed] [Google Scholar]
- 23.Sarker M, Ruiz-Ruiz C, López-Rivas A. Activation of protein kinase C inhibits TRAIL-induced caspases activation, mitochondrial events and apoptosis in a human leukemic T cell line. Cell Death Differ. 2001;8(2):172–181. doi: 10.1038/sj.cdd.4400791. [DOI] [PubMed] [Google Scholar]
- 24.Johansson AC, Steen H, Ollinger K, Roberg K. Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ. 2003;10(11):1253–1259. doi: 10.1038/sj.cdd.4401290. [DOI] [PubMed] [Google Scholar]
- 25.Aris SM, Pommier Y. Potentiation of the novel topoisomerase I inhibitor indenoisoquinoline LMP-400 by the cell checkpoint and Chk1-Chk2 inhibitor AZD7762. Cancer Res. 2012;72(4):979–989. doi: 10.1158/0008-5472.CAN-11-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem. 2004;279(51):53272–53281. doi: 10.1074/jbc.M406879200. [DOI] [PubMed] [Google Scholar]
- 27.Karetsou Z, et al. Identification of distinct SET/TAF-Ibeta domains required for core histone binding and quantitative characterisation of the interaction. BMC Biochem. 2009;10(1):10. doi: 10.1186/1471-2091-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto K, et al. NMR basis for interprotein electron transfer gating between cytochrome c and cytochrome c oxidase. Proc Natl Acad Sci USA. 2011;108(30):12271–12276. doi: 10.1073/pnas.1108320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno-Beltrán B, et al. Cytochrome c1 exhibits two binding sites for cytochrome c in plants. Biochim Biophys Acta. 2014;1837(10):1717–1729. doi: 10.1016/j.bbabio.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 30.König BW, et al. Mapping of the interaction domain for purified cytochrome c1 on cytochrome c. FEBS Lett. 1980;111(2):395–398. doi: 10.1016/0014-5793(80)80835-0. [DOI] [PubMed] [Google Scholar]
- 31.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J Biol Chem. 2001;276(16):13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- 32.Bertini I, Chevance S, Del Conte R, Lalli D, Turano P. The anti-apoptotic Bcl-x(L) protein, a new piece in the puzzle of cytochrome c interactome. PLoS One. 2011;6(4):e18329. doi: 10.1371/journal.pone.0018329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Insinga A, Cicalese A, Pelicci PG. DNA damage response in adult stem cells. Blood Cells Mol Dis. 2014;52(4):147–151. doi: 10.1016/j.bcmd.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Lavelle C, Foray N. Chromatin structure and radiation-induced DNA damage: From structural biology to radiobiology. Int J Biochem Cell Biol. 2014;49:84–97. doi: 10.1016/j.biocel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140(2):183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu LF, et al. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922(1):1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 37.Jacob S, Miquel C, Sarasin A, Praz F. Effects of camptothecin on double-strand break repair by non-homologous end-joining in DNA mismatch repair-deficient human colorectal cancer cell lines. Nucleic Acids Res. 2005;33(1):106–113. doi: 10.1093/nar/gki154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown GC, Borutaite V. Regulation of apoptosis by the redox state of cytochrome c. Biochim Biophys Acta. 2008;1777(7-8):877–881. doi: 10.1016/j.bbabio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 39.Go Y-M, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780(11):1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.