Abstract
A precise biological mechanism by which cadmium acts as a developmental toxicant is unknown but is suggested to include an epigenetic basis. In prior work, we analyzed CpG island methylation levels within gene promoters (n = 16,421) in leukocytes collected from mothers and their infants from a pregnancy cohort in Durham County, North Carolina. The CpG methylation levels were examined in relationship to prenatal exposure to cadmium and/or cotinine to identify genes and pathways influenced by in utero exposure. In the present article, we provide an enhanced description of the data collection and processing to facilitate cross-study comparisons. Data are available within the Gene Expression Omnibus database (GSE67976).
Keywords: Cadmium, Epigenetics, Prenatal, Exposure, CpG methylation
Brief overview
1. DNA methylation assessment
1.1. Sample selection
To facilitate reproducibility and to disseminate methods to as large an audience as possible, this article provides additional details on the study design and analysis methods. The materials and methods presented here represent an expansion of those detailed in the original study [1].
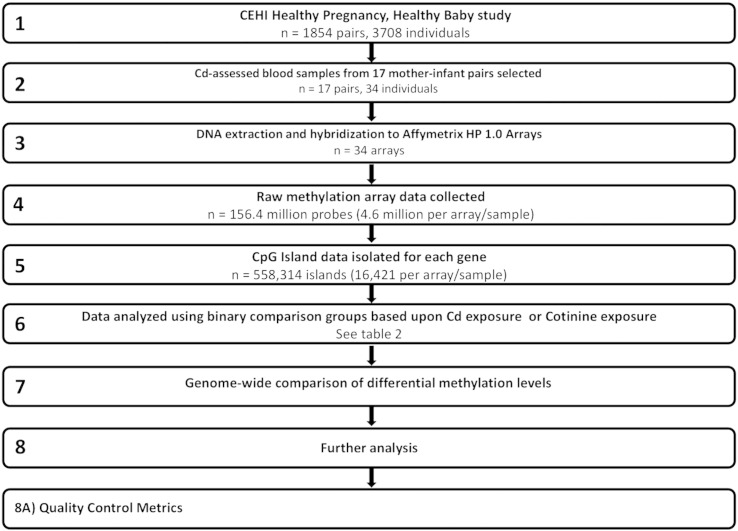
Study samples were collected from within the Children's Environmental Health Initiative's (CEHI) Healthy Pregnancy, Healthy Baby study in Durham County, North Carolina [1] (Fig. 1, Step 1). The CEHI study population includes 1854 mother–infant pairs and is a prospective cohort study of pregnant women living in Durham County, North Carolina recruited between the years of 2005 and 2011. The study was approved by institutional review boards at both Duke University and the University of North Carolina-Chapel Hill. All subjects consented to have maternal venous blood and their newborn's cord blood drawn at delivery for use for toxicant assessment and genetic analysis. Population eligibility requirements included: planned delivery at Duke University Medical Center, at least 18 years of age, English literacy, residence in Durham County, and no presence of multi-fetal gestation or known fetal genetic or congenital anomalies. A detailed description of post-blood-collection processing, including the analysis of cadmium (Cd) and cotinine levels, is reported in Edwards et al. [2].
Fig. 1.
Flowchart depicting methodology for sample collection and data processing from Sanders et al. [1].
From the total CEHI study population of 1854 mother–infant pairs, a total of 17 maternal–fetal pairs were selected for the DNA methylation analysis (Fig. 1, Step 2). The rationale for sample selection was to stratify the pairs by higher or lower Cd exposure for comparison. A median Cd blood level of 0.40 μg/l and a 75th percentile level of 0.56 μg/l were reported in the larger CEHI cohort [2]. The limits of detection for Cd reported in the study were 0.20 μg/l and 0.08 μg/l Cd depending on the laboratory of analysis. The corresponding cotinine detection limit was either 0.02 or 2.0 ng/l, depending on the laboratory of analysis. Inter-laboratory differences were accounted for using a rank permutation method [1], [3]. The Cd and cotinine levels of the mother–infant pairs selected for DNA methylation assessment are reported in the prior study [1].
1.2. Assessment of CpG methylation
After selection of the mother–infant pair samples, corresponding blood samples were retrieved for processing. DNA was extracted using Qiagen's PAXgene Blood DNA kits (Fig. 1, Step 3). The recovered DNA was stored in nuclease free water at − 80 °C between processing steps. The MethylCollector Ultra Kit from Active Motif was used to isolate only CpG methylated DNA first cut via enzymatic digestion. This kit takes advantage of the methylated-CpG island recovery assay (MIRA) based on the high affinity of the MBD2b/MBD3L1 protein complex for double-stranded CpG-methylated DNA to pull down only CpG methylated fragments [4]. These fragments were then amplified using the WGA3 kit (Sigma), to produce copies of the DNA sequences. Finally, the amplified DNA was hybridized to Affymetrix Human Promoter 1.0R arrays used to assess over 4.6 million sites (probes), with each probe comprising a 25 bp region within gene promoters (Fig. 1, Step 4). These steps were followed for all DNA samples (maternal and fetal) using the manufacturer's protocols.
The 4.6 million probe dataset per sample were then processed bioinformatically to yield summarized CpG island methylation abundances for 16,421 genes. Summarized CpG island data were generated by averaging all probe methylation abundances located within the CpG islands based upon location in the genome. CpG island locations were determined as detailed by the UCSC database [5] (Fig. 1, Step 5) and annotated according to the Human Genome 18.
1.3. Identification of differentially methylated genes
The methylation datasets (n = 34 datasets, 16,421 CpG islands/dataset) recovered from the previous step were then classified into a Cd-exposed or unexposed group (Fig. 1, Step 6). The Cd-exposed group was defined as > 0.20 μg/l (median) maternal blood Cd. Cotinine is a known indicator of nicotine exposure and was also analyzed in this study to examine the epigenetic effects of smoking. For the cotinine analysis, cotinine exposure or unexposed groups were defined as those with any detectable value versus no detectable level. The rationale for this choice was to assess all forms of smoking exposure, including second-hand smoke.
CpG island methylation was then compared within the binary comparison groups using an ANOVA. The criteria for differential methylation between CpG islands were as follows: (1) minimum absolute change of 30% DNA methylation between the exposed and unexposed groups; and (2) p < 0.05. Table 1 presents general study information. The results of the ANOVA analyses are summarized in Table 2.
Table 1.
General Study Information.
| Subject area | Epigenetics, DNA methylation |
|---|---|
| Cell type | Leukocyte |
| Population description | 17 Mother–Infant Pairs |
| Total number of samples | 34 |
| Technologies utilized | |
| DNA extraction | Qiagen PAXgene Blood DNA kit |
| Methylated DNA collection kit | Active Motif MethylCollector Ultra kit |
| DNA amplification kit | Sigma WGA3 kit |
| Microarray type | Affymetrix Human Promoter 1.0R Arrays |
| Pathway analysis | Ingenuity pathway analysis software and Database for Annotation, Visualization, and Integrated Discovery (DAVID) |
| Measurement Variable | DNA methylation levels at promoter-based CpG Islands |
| Link to Public Data | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67976 |
Table 2.
Numbers of differentially methylated genes (DMGs) identified comparing cadmium exposed versus unexposed, cotinine exposed versus unexposed, or shared cadmium/cotinine exposed versus unexposed subjects.
| Maternal dataset (n = 17 subjects) | Fetal dataset (n = 17 subjects) | Shared fetal, maternal dataset DMGs | |
|---|---|---|---|
| Cadmiuma-associated DMGs | 92 DMGs | 61 DMGs | 0 DMGs |
| Cotinineb-associated DMGs | 134 DMGs | 366 DMGs | 12 DMGs |
| Shared cadmium and cotinine-associated DMGs | 0 DMGs | 30 DMGs |
Cadmium exposure was classified as above/below 0.20 μg/l (n = 10 exposed/n = 7 unexposed).
Cotinine was classified as detectable/undetectable (n = 11 detectable subjects/n = 6 undetectable).
2. Quality control and confounder adjustment
2.1. Controlling for confounding demographic variables
The effect of covariates on differential DNA methylation was assessed in both maternal and newborn DNA based on the known association of maternal age, race, and infant sex with DNA methylation patterns. Covariates were dichotomized where maternal age was defined as ≥ 30 versus < 30 (referent), maternal race was defined as non-Hispanic Black versus non-Hispanic White (referent) and one Hispanic mother was excluded from these analyses due to insufficient sample size and infant sex (male = referent) (Fig. 1, Step 8A). CpG island methylation was compared within the binary comparison groups using an ANOVA with criteria identical to those above. Differentially methylated CpG islands were identified for each demographic (maternal age, race, or infant sex) and excluded from the sets of identified Cd- and cotinine-associated differentially methylated genes.
2.2. White blood cell gene comparison
White blood cells comprise different types of cells with differential DNA methylation patterns that influence cell differentiation, including differentiation into different blood cell types from progenitor cells [6]. The DMGs identified by Sanders et al. were compared to the list of 500 DMGs known to predict cell type in an adult population reported by Houseman et al. [6]. The maternal dataset DMGs showed no overlap while the fetal dataset DMGs had one overlapping DMG associated with Cd, and four with cotinine suggesting a minimal influence of white blood cell composition on the DNA methylation patterns [1].
3. Validation of DNA methylation results
Validation of the DNA methylation results obtained using the MIRA assay was carried out at both the gene-specific and genome-wide levels. These methods are described in detail by Sanders et al. [1] and are summarized here. Bisulfite conversion, conversion of unmethylated cytosine residues to uracil, with the Zymo EZ DNA Methylation-Lightning kit was used for gene-specific analysis for 15 infant samples. These changes were then quantified into appreciable methylation values using the qPCR-based EpiTect MethyLight Assay for the PRR13 gene and compared to the MIRA results using spearman rank correlation, which yielded significant results. The PRR13 gene was chosen specifically because it was one of the top five most significant cell-death-associated genes for infants with a fold-change in DNA methylation > 1.3 and p < 0.03.
Genome-wide analysis was done by comparing the MIRA results from two maternal samples with results from a separate array, the Illumina Infinium HumanMethylation450 BeadChip array. The two maternal samples were selected based on stratified Cd levels and matched with respect to maternal age, race (non-Hispanic Black), and Medicaid coverage. For quality control, DNA methylation proportion values with p-values < 0.0001 located within CpG islands were required. The two samples were compared based on Cd exposed versus unexposed status, yielding DNA methylation proportion values for each gene. Each gene was then compared to its MIRA fold-change counterpart using the spearman rank correlation. Among Cd-associated genes, there was a significant association between the Illumina Infinium HumanMethylation450 BeadChip array data and the MIRA data.
4. Conclusions
The dataset described here reflects DNA methylation abundances located within CpG islands of gene promoters. There are 34 datasets reflecting 17 mother–infant pairs selected for analysis based on their levels of blood cadmium at delivery. The present article provides further details on the experimental design, data generation, and quality control from the Sanders et al. study [1]. The prior study demonstrated associations between CpG island methylation and Cd levels during pregnancy and provided evidence for the transcription factor occupancy theory of DNA methylation patterning related to prenatal exposure to environmental contaminants. Taken together, these data suggest that prenatal exposure to toxic metals is associated with altered DNA methylation patterning in fetal cord DNA.
Acknowledgments
This research was funded by grants from the NIEHS (T32-ES007018, P42-ES005948, and R01-ES019315), the USEPA (RD83329301), and in part by the North Carolina Translation and Clinical Sciences Institute (NC TRaCS) grants UL1RR025747, KL2RR025746, and TLRR025745 from the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health.
References
- 1.Sanders A.P. Cadmium exposure and the epigenome: exposure-associated patterns of DNA methylation in leukocytes from mother–baby pairs. Epigenetics. 2014;9(2):212–221. doi: 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards S.E. Cadmium levels in a North Carolina cohort: identifying risk factors for elevated levels during pregnancy. J. Expo. Sci. Environ. Epidemiol. 2014 doi: 10.1038/jes.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgette L.F., Reiter J.P. Nonparametric Bayesian multiple imputation for missing data due to mid-study switching of measurement methods. J. Am. Stat. Assoc. 2012;107(498):439–449. [Google Scholar]
- 4.Rauch T.A., Pfeifer G.P. The MIRA method for DNA methylation analysis. Methods Mol. Biol. 2009;507:65–75. doi: 10.1007/978-1-59745-522-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karolchik D. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houseman E.A. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]