Summary
Chronic infection with Plasmodium falciparum was epidemiologically associated with endemic Burkitt’s lymphoma, a mature B cell cancer characterized by chromosome translocation between the c-myc oncogene and Igh, over 50 years ago. Whether infection promotes B cell lymphoma, and if so by what mechanism remains unknown. To investigate the relationship between parasitic disease and lymphomagenesis we used Plasmodium chabaudi (Pc) to produce chronic malaria infection in mice. Pc induces prolonged expansion of germinal centers (GCs), unique compartments where B cells undergo rapid clonal expansion and express activation-induced cytidine deaminase (AID), a DNA mutator. GC B cells elicited during Pc infection suffer widespread DNA damage leading to chromosome translocations. Although infection does not change the overall rate, it modifies lymphomagenesis to favor mature B cell lymphomas that are AID dependent and show chromosome translocations. Thus, malaria infection favors mature B cell cancers by eliciting protracted AID expression in GC B cells.
Introduction
Many pathogens have been implicated as etiologic agents in the development of human cancer. For example, individuals infected with Epstein Barr Virus (EBV), Hepatitis C Virus, Human Immunodeficiency Virus, Helicobacter pylori, or Plasmodium falciparum are all at higher than average risk of developing B cell lymphoma (de Martel et al., 2012; Epeldegui et al., 2010; Kutok and Wang, 2006; Marcucci and Mele, 2011; Molyneux et al., 2012; Zucca et al., 2000; Zur Hausen, 2009). Although viruses can promote neoplasia directly, by delivering virally encoded cancer genes to target cells, the link between most pathogens and tumor development remains obscure (Karin et al., 2006; Mesri et al., 2014). For instance, endemic Burkitt’s lymphoma (eBL), a lymphoma of GC origin, is among the most common childhood cancers in Africa, and it occurs at higher incidence in areas where Plasmodium falciparum infection is endemic. This epidemiologic association is poorly understood in part because eBL cells are infected with EBV, which can induce B cell malignancy, and there is little insight into how malaria might play an additional role (Burkitt, 1961; Kutok and Wang, 2006; Magrath, 2012; Molyneux et al., 2012).
Based on histologic, molecular, and gene expression analysis it has been proposed that BL cells represent transformed GC B cells that carry t(8;14) chromosome translocations (Klein and Dalla-Favera, 2008; Kuppers et al., 1999; Magrath, 1990; Shaffer et al., 2002; Victora et al., 2012). This translocation joins c-myc to immunoglobulin (Ig) gene loci, and thereby deregulates c-myc expression. However, deregulated c-myc alone is not sufficient to produce lymphoma (Adams et al., 1985; Janz et al., 2003; Leder et al., 1986) and transformation requires additional lesions in genes encoding proteins such as p53 that regulate cell cycle checkpoints and apoptosis (Gaidano et al., 1991; Love et al., 2012). Neither p53 mutation nor EBV infection are eBL specific, as they occur broadly in lymphoid malignancies (Cesarman, 2014; Forbes et al., 2015; Koduru et al., 1997; Kutok and Wang, 2006; Saha and Robertson, 2011). In contrast, malaria is exquisitely associated with eBL (Magrath, 2012; Molyneux et al., 2012).
GC B cells are rapidly dividing cells that are unique in expressing high levels of AID, a mutator enzyme that deaminates cytidines and produces U:G mismatches in Ig gene loci to initiate somatic hypermutation (SHM) and class switch recombination (CSR) of antibodies (Muramatsu et al., 2000; Petersen-Mahrt et al., 2002; Revy et al., 2000). Although AID has a strong preference for Ig gene loci it is not entirely Ig specific, and it produces off-target mutations or DNA breaks in oncogenes including c-myc that lead to translocations in activated B cells in vitro and in IL-6 transgenic mice that develop polyclonal plasmacytosis (Klein et al., 2011; Pasqualucci et al., 2001; Ramiro et al., 2004; Robbiani et al., 2008).
Here we show that in the absence of p53, Pc infection results in development of mature B cell lymphoma. Malaria infection alters lymphomagenesis to favor development of mature GC or post-GC origin B cell lymphomas and also destabilizes the genome in rapidly dividing AID expressing GC B cells.
Results
Plasmodium infection induces AID-expressing germinal centers
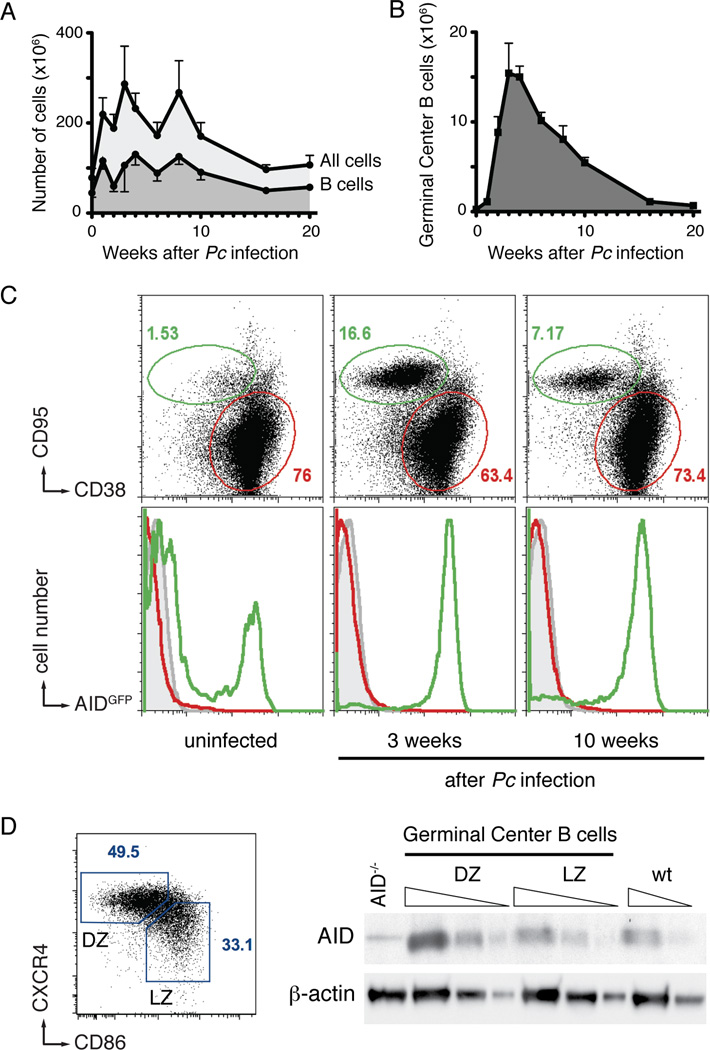
We used Plasmodium chabaudi (Pc) to establish chronic malaria infection in mice. In agreement with previous reports (Achtman et al., 2007; Horne-Debets et al., 2013), parasitemia peaked between 7 and 10 days after infection, and was followed by an increase in total spleen cellularity (mean of 286.6 mio cells/spleen, 4-fold expansion at 3 weeks), including B lymphocytes (mean of 130.4 mio B cells/spleen, 3-fold expansion at 4 weeks; Figures 1A and S1A). Strikingly, splenic GC B cells expanded by 63-fold (mean of 15.4 mio GC B cells/spleen at 3 weeks), and this response was sustained for over 10 weeks (Figure 1B). We conclude that Plasmodium infection induces a robust and long-lasting expansion of GC B cells.
Figure 1. B cell responses to Plasmodium infection.
A and B- Total spleen cellularity, total B cells (A), and germinal center B cells (B) following Pc infection. Mean with standard deviation is shown; at least five mice for each time point.
C- AID expression is confined to germinal center B cells. Representative flow cytometry plots of splenocytes from Plasmodium infected AIDGFP mice. Top row shows the relative expansion of germinal center (green) over non-germinal center (red gate) B cells over time (gated on B220+). Bottom row shows the expression of AIDGFP in non-B cells (B220−, grey), non-germinal center B cells (B220+ CD38+ CD95−, red), and germinal center B cells (B220+CD38−CD95+, green). At least three mice for each time point.
D- AID protein in malaria GC B cells. Gating strategy (left) and semiquantitative Western Blot analysis (right) identifying AID in both light zone (LZ) and dark zone (DZ) cells sorted 3 weeks post inoculation. Triangles indicate threefold serial dilution. AID−/− and wild type (wt) control lanes are from in vitro activated B cells of the respective genotypes. Representative of two independent experiments.
See also Figure S1.
Ig somatic mutation and class switch recombination are initiated by AID, a cytidine deaminase (Muramatsu et al., 2000; Petersen-Mahrt et al., 2002; Revy et al., 2000). This enzyme is typically restricted to activated B cells in GCs, but malaria infection produces widespread B cell activation (Scholzen and Sauerwein, 2013). To determine if AID is restricted to GCs during Pc infection, we examined AIDGFP reporter mice (Crouch et al., 2007). We found that AID expression was restricted to GC B cells and was sustained over a period of at least 10 weeks (Figures 1C and S1B). To confirm AID protein expression, we sorted GC B cells into dark and light zone cells and performed western blot analysis. As expected AID was found mainly in dark zone cells, which contained three times more AID than light zone cells or in vitro activated B cells (Figure 1D). We conclude that AID is primarily expressed in Plasmodium-induced GC B cells.
Widespread chromosome translocations in malaria germinal centers
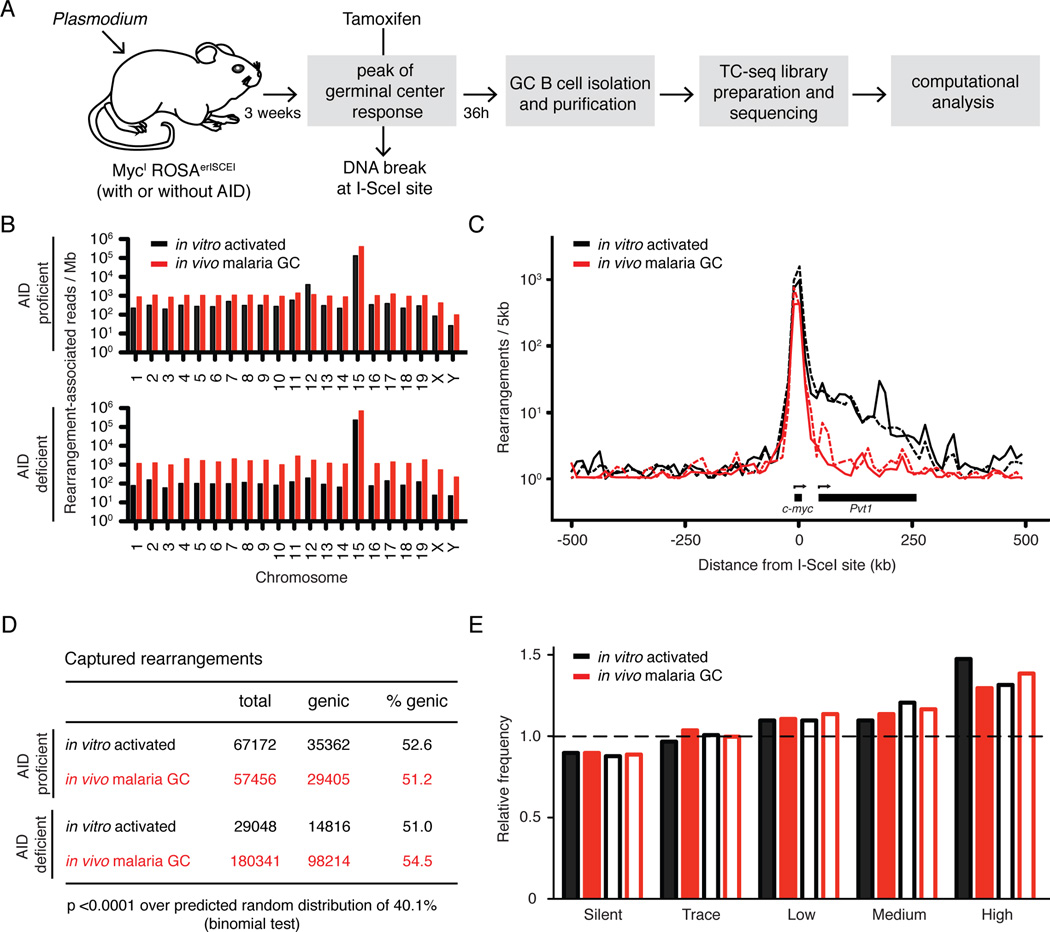
AID and replication associated stress can independently produce DNA breaks and translocations in cultured B cells (Barlow et al., 2013; Hakim et al., 2012; Klein et al., 2011). To map the DNA damage caused by Plasmodium infection in vivo, we adapted a previously described next-generation translocation capture and sequencing method (TC-Seq; (Klein et al., 2011)). Mice with an I-SceI recognition site at c-myc (MycI) were bred to a ROSAerISCEI transgene (encoding for I-SceI fused to the estrogen receptor ligand binding domain, er) and to either AID deficient or AID overexpressing mice (ROSAAIDer, see Experimental Procedures and Figure S2). Compound MycI/IROSAerISCEI/erISCEIAID−/− and MycI/IROSAerISCEI/AIDer mice were infected with Plasmodium and treated with Tamoxifen at the peak of the GC response, to induce the nuclear shuttling of erISCEI in the absence or presence of AID, and translocations between the DNA break at MycI on chromosome 15 and lesions in the rest of the genome were captured by TC-Seq (Figure 2A). We recovered 191150 and 65905 unique rearrangements to the I-SceI site in MycI from 105 million AID deficient or proficient malaria GC B cells, respectively (see Experimental Procedures). In agreement with previous in vitro experiments (Klein et al., 2011), malaria rearrangements were preferentially intrachromosomal (95.2% vs. 94.3% of all rearrangement-associated reads, in the absence or presence of AID respectively; Figure 2B), and enriched at the I-SceI site (Figure 2C). Irrespective of AID expression, rearrangements were significantly increased in genic regions over the predicted random distribution of 40.1% (Figure 2D), and favored more actively transcribed genes (Figure 2E). Consistent with this, the asymmetric distribution of events in the direction of transcription at the I-SceI site was less pronounced in malaria GCs than in vitro stimulated B cells, which express higher levels of c-myc (Figures 2C and S2E; (Klein et al., 2011; Shaffer et al., 2001; Thomas and Rothstein, 1989)). We conclude that the overall distribution of chromosome rearrangements induced by Plasmodium in vivo is similar to the one observed in retrovirally-infected cells in vitro.
Figure 2. Translocations in malaria GC B cells.
A- Schematic diagram of the protocol to identify genomic rearrangements induced by Plasmodium in vivo.
B- Chromosome distribution of rearrangement-associated reads captured by I-SceI breaks on chromosome 15.
C- Profile of rearrangements around the I-SceI site. Dotted lines represent AID deficient samples.
D- Proportion of genic rearrangements.
E- Frequency of rearrangements at genes with various levels of transcription. Empty bars represent AID deficient samples. Dashed line represents the expected frequency based on random model.
For all, rearrangements from malaria GC B cells are compared to cultured B cells (Klein et al., 2011). See also Figure S2.
AID-independent damage involves sites of DNA replication-associated fragility
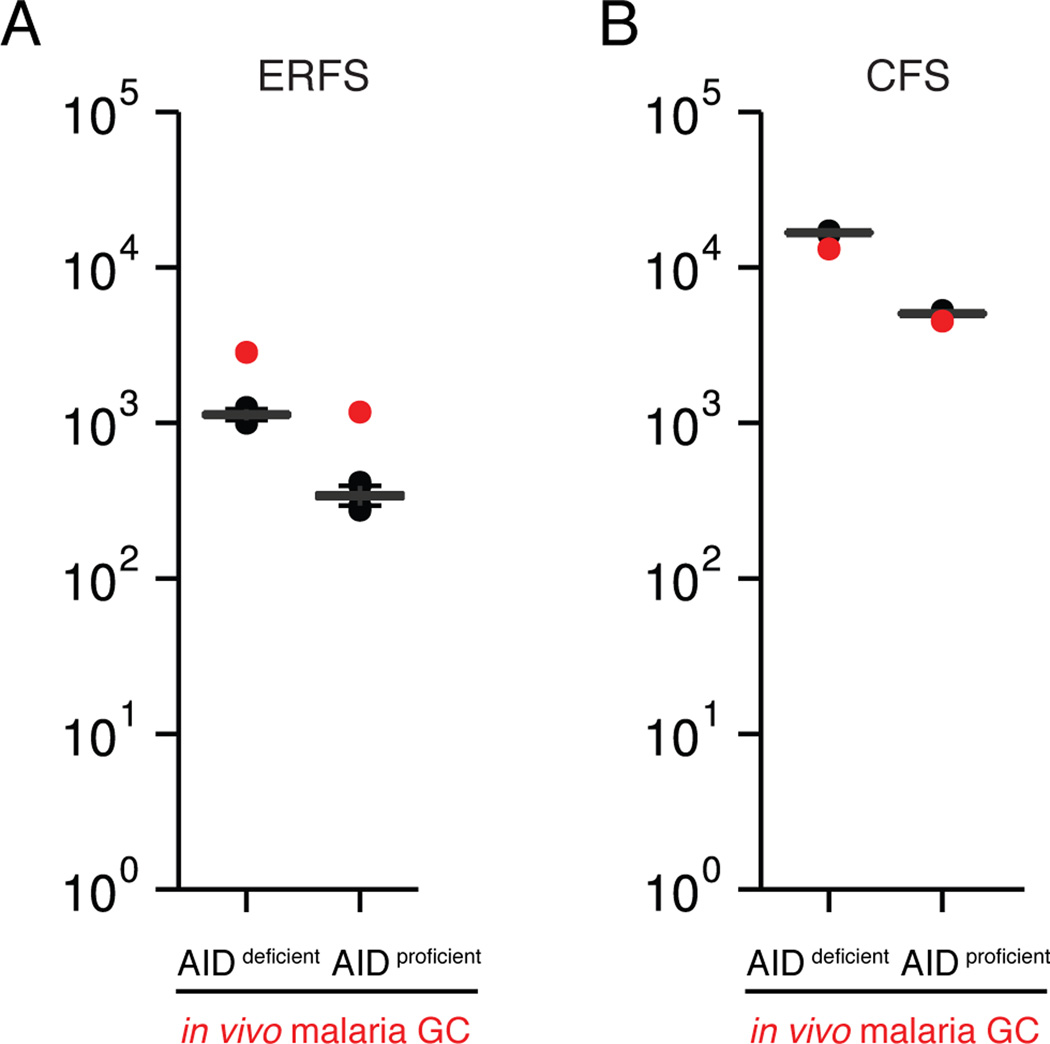
Early replication fragile sites (ERFS) are a set of genomic regions prone to breakage during DNA replication (Barlow et al., 2013). To assess the contribution of DNA replication to malaria-induced rearrangements, we analyzed their occurrence within hotspots of viral integration at ERFS (M.J. and I.T.S., unpublished data and (Barlow et al., 2013)). The fraction of translocations in ERFS was similar in the presence and absence of AID (1.5% and 2% of total, respectively), and significantly enriched over the random distribution determined by Monte-Carlo simulation (Figure 3A). ERFS translocations preferred genic regions and more highly transcribed genes (Figure S3). In contrast, no enrichment was observed within common fragile sites (CFS), which are late-replicating regions that preferentially break during mitosis (Figure 3B; (Helmrich et al., 2006)). We conclude that in malaria GC B cells DNA damage preferentially occurs at ERFS, irrespective of AID.
Figure 3. AID-independent DNA damage in regions of DNA replication-associated fragility.
A- Observed number of chromosome rearrangements within hotspots of viral integration at early replication fragile sites (ERFS, red dot), as compared to random Monte-Carlo simulation (p< 0.001 for both).
B- Number of chromosome rearrangements within common fragile sites (CFS, red dot), as compared to random.
See also Figure S3.
AID damages the genome of malaria GC B cells
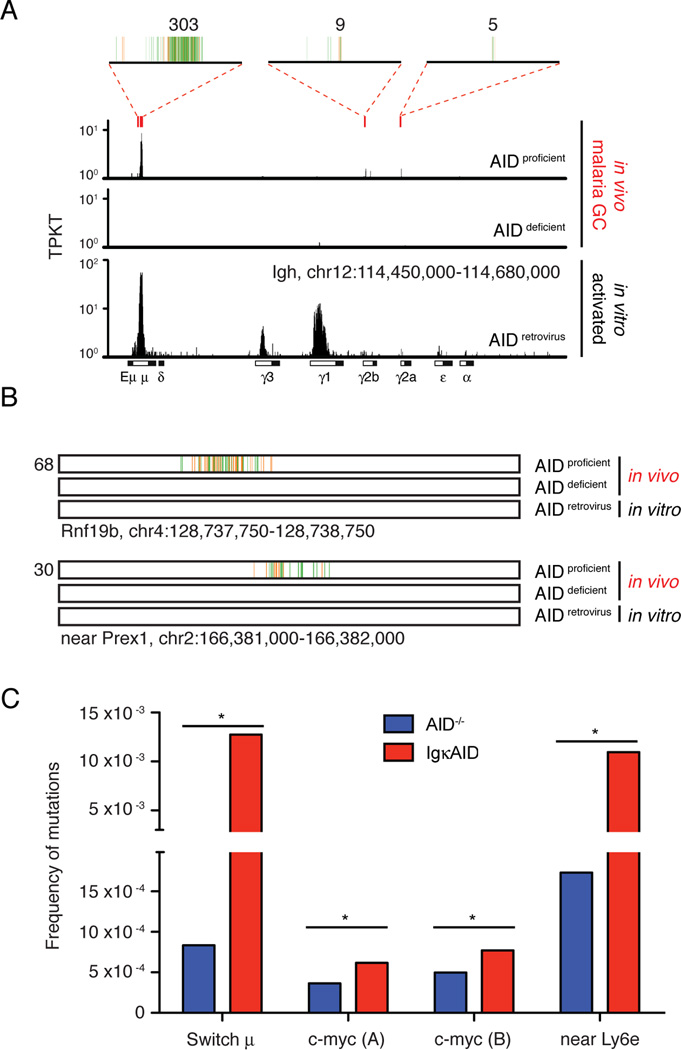
To determine whether AID destabilizes specific regions in the GC B cell genome we compared AID deficient and proficient TC-seq libraries for local accumulations of translocations using stringent criteria (Wang et al., 2014). As expected, hotspots of AID-dependent translocation were observed at the Igh locus, with accumulations involving the switch regions Sμ, Sγ2b, and Sγ2a (Figure 4A and Table S1). In contrast, AID translocations from B cells activated in vitro formed hotspots in all switch regions, and particularly at Sμ, Sγ1 and Sγ3 (Figure 4A). An additional 15 hotspots were identified in non-Ig regions, and off-target AID-dependent mutational activity was confirmed at c-myc and near Ly6e (Table S1 and Figures 4B, 4C, and S4A). In agreement with previous reports, AID translocations were enriched in genic regions and favored more highly transcribed genes (Figures S4B and S4C; (Klein et al., 2011; Wang et al., 2014)). Moreover, none of the AID translocation off targets identified in vivo was shared with those from the in vitro dataset (Table S1 and (Klein et al., 2011)). All together, translocations within AID-dependent hotspots including the Igh locus comprised only 0.7% of total. We conclude that AID is not limited to, but primarily destabilizes Ig regions in GC B cells.
Figure 4. AID contributes to Plasmodium induced DNA damage.
A- Translocations at the physiologic AID target Igh. Red rectangles indicate the location of AID hotspots, and 4kb regions at these hotspots are magnified on top, where vertical lines each represent a unique translocation event. Numbers on top indicate the total of translocations at each hotspot region. Rearrangements obtained in cultured B cells retrovirally expressing AID are shown for comparison (Klein et al., 2011). TPKT is the normalized number of translocations per kilobase per thousand translocations in the library.
B- Examples of non-Igh hotspots of AID-dependent translocation induced by malaria. One kb region is shown, with each vertical line representing a unique translocation. Numbers on the left indicate the total of translocations.
C- Mutational analysis of malaria GC B cells DNA by MutPE-seq. For c-myc, two adjacent regions in intron 1 were analyzed. P value is *<0.000001 for all (one-tailed Student’s T-test). See also Table S1 and Figure S4.
AID promotes late stage lymphomas
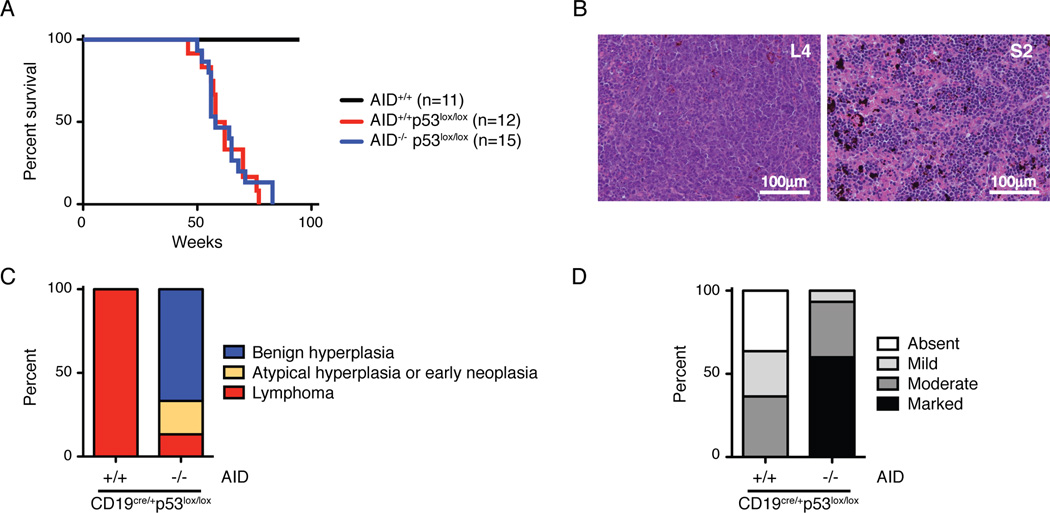
In addition to deregulated c-myc, BLs bear frequent mutation of p53 (Gaidano et al., 1991; Love et al., 2012). To model this phenomenon we produced mice that carry a B cell specific deficiency in p53 (CD19cre/+p53lox/lox) and are either proficient or deficient for AID. B lymphocyte development was not significantly altered in these mice despite efficient p53 deletion (Figure S5). To mimic the chronic exposure in areas of endemic malaria, the two cohorts were repeatedly infected with Pc and monitored for disease. Mice developed lymphomas in an AID dependent manner (Figure 5). Malaria infection was not required for lymphoma development but altered the phenotype to favor more mature B cell lymphomas (Figure 6).
Figure 5. P53 suppresses and AID promotes Plasmodium induced lymphoma.
A- Survival of Plasmodium infected mice. All mice are also CD19cre/+.
B- Spleen histology of Plasmodium infected CD19cre/+p53lox/lox mice. L4 is AID proficient lymphoma and S2 is AID deficient benign B cell hyperplasia with marked extramedullary hematopoiesis.
C- Lymphoma versus benign hyperplasia in Plasmodium infected mice. Lymphoid tissues were evaluated by histology, immunohistochemistry, and flow cytometry. “Benign hyperplasia” indicates mice with splenomegaly but normal B cell distribution, with B220+ cells confined to follicular areas. “Atypical hyperplasia or early neoplasia” denotes splenomegaly and B220+ cells expanding into the periarteriolar lymphoid sheats (PALS). “Lymphoma” defines abnormal lymphoid tissue architecture and/or dissemination to multiple organs.
D- Extramedullary hematopoiesis in Plasmodium infected mice. Spleen sections were evaluated for the degree of extramedullary hematopoiesis.
See also Figure S5 and Table S2.
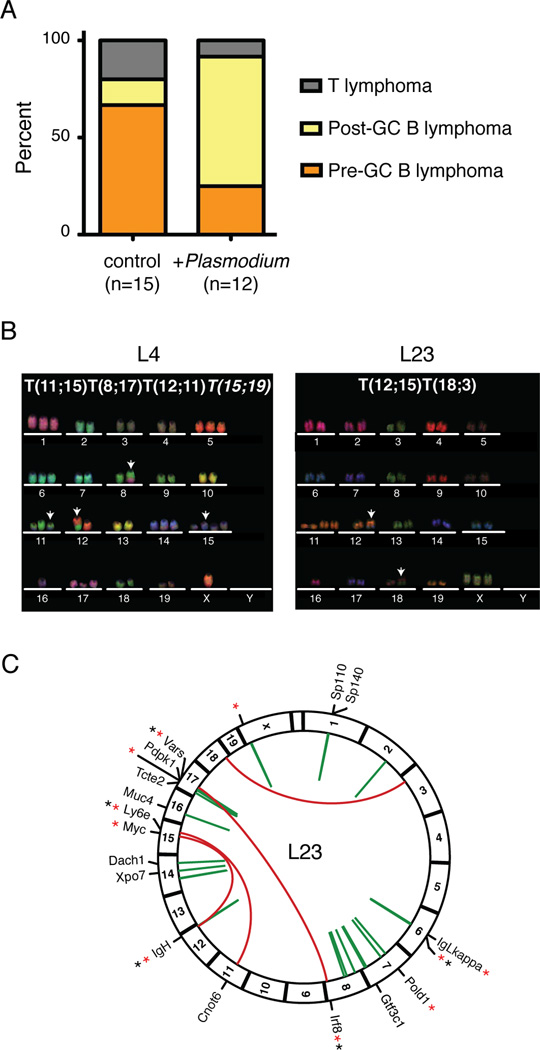
Figure 6. Genomic rearrangements in Plasmodium induced lymphomas.
A- Distribution of lymphoma phenotypes in Plasmodium-infected and control uninfected CD19cre/+p53lox/lox mice.
B- Representative M-FISH images of metaphases from Plasmodium-induced CD19cre/+p53lox/lox lymphomas. Arrows point to chromosomes with detectable translocations.
C- Circos diagram of the L23 genome. Red arches represent interchromosomal rearrangements, and green arches intrachromosomal ones. For genic rearrangements, the name of the gene is indicated. Asterisks indicate if the recombined site is a known AID target (red, (Hakim et al., 2012; Klein et al., 2011)), or within hotspots of viral integration at ERFS (black, M.J. and I.T.S., unpublished data).
See also Figures S6, S7 and Tables S3, S4, and S5.
Whereas none of eleven wild type mice became ill after infection, the two p53 deficient groups succumbed with a median of 60 weeks for CD19cre/+p53lox/lox and 58 weeks for CD19cre/+p53lox/lox AID−/− (p = 0.8 with the Mantel-Cox test; Figure 5A). However, all of twelve CD19cre/+p53lox/lox mice developed lymphoma while only 5 out of 15 CD19cre/+p53lox/lox AID−/− mice had evidence of lymphoma (33%, p < 0.0004 with Fisher’s exact test; Figures 5B and 5C). The majority of the CD19cre/+p53lox/loxAID−/− mice (67%) presented with splenomegaly associated with marked extramedullary hematopoiesis but no histologic evidence of cancer (Figures 5B, 5C, and 5D). Consistent with lack of transformation, splenocytes from these mice were unable to expand in vitro (n = 5), as opposed to 3 out of 4 control lymphomas (observation period of 3 weeks). Moreover, lymphoma developed only in 1 out of 4 immunodeficient mice, in which CD19cre/+ p53lox/lox AID−/− splenocytes from malaria infected mice had been transplanted (Figure S5D). Finally, all of the CD19cre/+ p53lox/lox AID−/− mice analyzed showed persistent parasitemia (n=5), low red blood cell counts, decreased hemoglobin levels, and increased mean cell volume in peripheral blood (Figure S5E). All together, these findings are consistent with the majority of CD19cre/+p53lox/loxAID−/− mice succumbing to a condition characterized by reactive extramedullary hematopoiesis in response to ongoing malaria infection. In contrast, all AID sufficient mice developed lymphoma.
To determine whether AID is required to control malaria, we inoculated AID−/− mice and monitored parasitemia over time. Regardless of p53, AID deficiency did not alter the acute infection, but AID was required to quench parasitemia in the chronic phase (Figures S5F). In agreement with this finding, AID deficiency was associated with anemia, splenomegaly, extramedullary hematopoiesis, and reduced survival (Figures S5G, S5H, S5I, and data not shown). We conclude that AID promotes Plasmodium-induced lymphomagenesis, but it is required to control chronic malaria infection.
Phenotype and karyotype of Plasmodium-induced lymphoma
We used flow cytometry to characterize the phenotype of lymphomas arising in AID sufficient CD19cre/+p53lox/lox mice. Infected and uninfected mice developed lymphoma with similar kinetics (median of 60 weeks for infected and 63 weeks for uninfected; p = 0.07 with the Mantel-Cox test; Figure S6A). However, malaria infected mice developed B cell lymphomas, a significant majority of which showed a more mature phenotype consistent with GC origin (72.7%, n = 11, Ig-switched or CD138+), while most uninfected controls developed pre-GC lymphomas (83.3%, n = 12; p < 0.01 with Fisher’s exact test; Figures 6A and S6B, and Tables 1, S2 and S3). As a control, we repeatedly immunized a cohort of AID sufficient CD19cre/+ p53lox/lox mice with sheep red blood cells (SRBC), a well-characterized antigen that induces long-lived germinal centers. Survival was similar in SRBC immunized mice (median of 68 weeks; p > 0.2 with the Mantel-Cox test; Figure S6A). However, in contrast to malaria, only a minority of SRBC associated lymphomas displayed a post-GC phenotype (p < 0.03 with Fisher’s exact test; Figure S5C and Table S4). We conclude that Plasmodium infection, unlike SRBC immunization, causes a shift towards a more mature, post-GC lymphoma phenotype.
Malaria-associated Burkitt’s lymphoma bears characteristic chromosome translocation of c-myc to Ig genes (Adams et al., 1983; Dalla-Favera et al., 1982; Hamlyn and Rabbitts, 1983; Taub et al., 1982). To determine whether Plasmodium-induced lymphoma in this model was associated with translocations, we performed multicolor-FISH (M-FISH) analysis of tumor metaphase spreads. We found that all of the informative lymphomas (n=7) contained clonal translocations, four tumors displayed multiple translocations (L4, L23, L24, L62) and one (L62) had a complex translocation involving three chromosomes (Figure 6B and Table 1). Lymphoma L23 had a T(12;15) between chromosomes 12 and 15, similar to the c-myc/Igh translocation found in pristane-induced plasmacytoma (Figure 6B; (Potter and Wiener, 1992)). To examine the L23 lymphoma at higher resolution, we sequenced its genome (see Experimental Procedures and Figure S7). Bioinformatic analysis confirmed the presence of a c-myc/Igh translocation at T(12;15), as well as the expected deletions at the p53 locus and at the physiologically rearranged Igh and Igκ loci (Figure 6C and data not shown). Sequence analysis also identified the breakpoint of T(18;3) and revealed additional intra- and interchromosomal rearrangements, which were not detected by M-FISH (Figures 6C, S7B, S7C and Table S5). Notably, 5 out of 8 interchromosomal breakpoints involved known AID targets (Figure 6C). We conclude that L23 carries numerous chromosome rearrangements, including the c-myc/Igh translocation characteristic of human Burkitt’s lymphoma, and that AID shapes this lymphoma’s genome.
Table 1. Karyotype and phenotype of lymphomas in P. chabaudi infected CD19cre/+ p53lox/lox mice.
Translocations are clonal if present in all metaphases. In italic are sporadic translocations and bolded are clonal, reciprocal translocations. At least 20 metaphases analyzed for each tumor.
| Tumor ID | Karyotype | Phenotype | S | MLN | sbLN | oLN | L |
|---|---|---|---|---|---|---|---|
| L21 | ND | CD3+ | + | − | − | − | + |
| L4 | T(11;15) T(8;17) T(12;11) T(15;19) | CD19+ IgM− IgD− IgK+ CD138− CD38+/− FAS− GL7− | + | + | + | + | − |
| L9 | no metaphases | CD19+ IgM− IgD− IgK+ CD138− CD38+ FAS+ GL7− | + | + | + | + | − |
| L23 | T(12;15) T(18;3) | CD19− IgM− IgD− IgK− CD138+ CD38− FAS-GL7dim | + | − | − | − | + |
| L24 | T(3;11) T(18;1) | CD19+ IgM− IgD− IgK+ CD138+ CD38+ FASdim GL7− | + | + | + | + | − |
| L33 | T(2;16) | CD19+ IgM+ IgD− IgK+ CD138+ CD38+ FAS+ GL7− | + | + | + | + | − |
| L52 | no metaphases | CD19+ IgM− IgD− IgK+ CD138− CD38+/− FAS− GL7+ | + | − | − | + | − |
| L62 | T(8;13) T(3;17;3;4) | CD19+ IgM+ IgD− IgK+ CD138+ CD38+ FAS+ GL7dim | + | + | + | + | − |
| L68 | ND | CD19− IgM− IgD− IgK− CD138+ CD38+ FASdim GL7− | − | + | + | + | − |
| L31 | T(2;10) | CD19+ IgM+ IgD+ IgK+ CD138− CD38+ FASdim GL7− | + | + | + | + | + |
| L39 | T(7;13) | CD19+ IgM+ IgD− IgK+ CD138− CD38+ FAS+ GL7+ | + | − | − | + | + |
| L69 | ND | CD19+ IgM+ IgD+ IgK+ CD138− CD38+ FAS− GL7− | + | − | − | − | − |
Macroscopic tumor dissemination at necropsy: S = spleen; MLN = mesenterial lymph nodes; sbLN = submandibular lymph nodes; oLN = axillary, thoracal or inguinal (other) lymph nodes; L = liver or other metastasis. ND is not determined. See also Figure S6 and Table S2.
Discussion
Burkitt’s lymphoma, a mature B cell lymphoma, occurs sporadically, but at higher incidence among immunodeficient individuals and in equatorial areas endemic for malaria (Cesarman, 2014; Magrath, 2012; Molyneux et al., 2012). In addition to malaria infection, most individuals developing eBL are also infected by EBV, a virus that encodes oncogenic proteins that can promote lymphomagenesis (Cesarman, 2014; Kutok and Wang, 2006; Magrath, 2012; Mesri et al., 2014; Molyneux et al., 2012; Zhang et al., 2012). Thus EBV is thought to play a direct role in the transformation process. In contrast, malaria’s contribution to lymphoma has remained elusive. We found that Plasmodium does not alter the incidence of cancers arising in B cells that carry a homozygous deletion of p53. Thus, Plasmodium infection alone does not induce cancer, and hence the requirement for EBV and additional transforming events. Instead, the parasitic infection alters lymphoma phenotype to favor more mature B cell lymphoma by inducing chronic GCs containing AID expressing B cells.
Mature B cell lymphomas, including eBL, are frequently associated with hallmark oncogenic translocations and display phenotypic and molecular characteristics of GC B cells. However, whether translocations are acquired within germinal centers or at earlier developmental stages is not clear (Shaffer et al., 2002). Supporting the notion that GCs are a lymphomagenic environment, we document widespread chromosome rearrangements occurring within malaria GCs. This is consistent with the high levels of AID and DNA replication in these rapidly proliferating cells (Gitlin et al., 2014; Robbiani and Nussenzweig, 2013; Victora and Nussenzweig, 2011). Early replication fragile sites (ERFS) map to sites of early DNA replication and are particularly vulnerable to fork collapse. ERFS DNA is CG-rich, and mostly euchromatic (Barlow et al., 2013). Translocations were significantly enriched at ERFS but not at Common Fragile Sites (CFS). This finding was unanticipated given that CFS had been previously implicated in the genesis of lymphomagenic rearrangements (Chesi et al., 1998; Kameoka et al., 2004).
AID was not absolutely required for malaria-associated lymphomagenesis since a small number of the AID−/− p53 deficient mice also developed lymphoma. Moreover, only a fraction of rearrangements found in malaria GC B cells in vivo could be attributed to AID. Nevertheless AID strongly promoted lymphomagenesis possibly because in addition to causing translocations (Chiarle et al., 2011; Klein et al., 2011; Ramiro et al., 2004; Robbiani et al., 2008; Robbiani et al., 2009) it can mutate cancer genes directly (Hakim et al., 2012; Liu et al., 2008; Pasqualucci et al., 2001).
In the absence of Plasmodium superinfection EBV is associated with B cell cancers other than eBL. For example, EBV is commonly detected in plasmablastic and Hodgkin’s lymphoma, and approximately one third of immunodeficiency-associated diffuse large B cell lymphoma is EBV positive (Cesarman, 2014). Moreover, when immune surveillance is impaired, the EBV gene LMP1 alone directly promotes B cell transformation (Zhang et al., 2012). Thus EBV is oncogenic in B cells, but alone cannot account for the specific phenotype of eBL. Similarly, mutations in p53 and other cancer genes are not exclusive for eBL, as they occur broadly in B cell malignancy (O'Shea et al., 2008; Young et al., 2008). Instead, the distinctive phenotype of eBL is specifically associated with malaria. Essentially all eBLs bear c-myc translocation to Ig regions, and AID is accountable for the DNA lesions at both sites leading to their translocation (Robbiani et al., 2008). By eliciting protracted, high levels of AID in GCs, malaria acts as a disease modifier, and predisposes B cells to acquire c-myc translocation at a stage in B cell development that mirrors the molecular and phenotypic characteristics of eBL.
Experimental Procedures
Mice and parasites
All experiments were performed in agreement with protocols approved by the Rockefeller University Institutional Animal Care and Use Committee. Mutant mice used in this study include: CD19cre/cre (Cd19tm1(cre)Cgn; (Rickert et al., 1997)), p53lox/lox (Trp53tm1Brn; (Marino et al., 2000)), AID−/− (Muramatsu et al., 2000), AIDGFP (Crouch et al., 2007), IgHI-96k/+ (Bothmer et al., 2010), MycI/I and IgHI/I (Robbiani et al., 2008), IgκAID (Robbiani et al., 2009). The ROSAAIDer and ROSAerISCEI alleles were generated in house by gene targeting into the ROSA26 locus of C2J embryonic stem cells, as previously described (Srinivas et al., 2001), and the loxP-flanked transcriptional stop was removed from the germline by crossing to EIIAcre mice (B6.FVB-TgN(EIIa-Cre)C5379Lmgd; (Lakso et al., 1996)). With the exception of the immunodeficient NRG strain (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ; (Pearson et al., 2008)), mice used in this study were either generated in C57Bl/6 background or backcrossed into it for at least 11 generations. Plasmodium chabaudi chabaudi ASS (Pc; MRA-429; (Carter and Walliker, 1975; Peters and Robinson, 2000)) was obtained through the MR4 as part of the BEI Resources Repository, NIAID, NIH.
Plasmodium infection
Parasites were maintained as frozen stocks and passaged in wild type mice. For the analysis of B cell responses, 6–12 weeks old mice were injected intraperitoneally (i.p.) with 105 Pc infected erythrocytes from a donor mouse. In tumor experiments, 6–8 week old mice were used for primary infection, and secondary and tertiary infections were performed at 8 weeks intervals using the same dose and route. Parasitemia was monitored by flow cytometry after staining 1 µl of blood with anti-Ter119 (eBiosciences) and Hoechst33342 (Invitrogen) for 30 minutes at room temperature, and confirmed by blood smear analysis.
Immunization with Sheep Red Blood Cells (SRBC)
One ml of SRBC (Colorado Serum Company) was diluted with 4ml PBS, and 0.2ml of the mix were injected intraperitoneally. Mice were immunized five times at 4 weeks intervals.
Flow cytometry
Single-cell suspensions of lymphatic or tumor tissues were stained after erythrocyte lysis with fluorophore-conjugated anti-mouse antibodies (from BD Pharmingen or eBiosciences) to detect: CD19 (clone 1D3), B220 (RA3–6B2), GL-7, FAS (Jo2), CD38 (90), CD86 (GL1), IgM (AF6.78,II/41), IgD (11–26c.2a), IgK (187.1), CD138 (281.2), CD43 (S7), CD3 (145-2C11), or biotin-conjugated antibody for CXCR4 (2B11) followed by streptavidin-APC. Samples were acquired on a FACSCalibur or Fortessa instrument (Becton Dickinson) and analyzed with FlowJo software.
Western Blot
Germinal center light and dark zone (LZ and DZ) B cells were purified from spleens of Pc infected mice at 3 weeks post inoculation. Upon magnetic enrichment with anti-CD19 microbeads (Miltenyi Biotech), B cells were stained with antibodies to sort DZ (B220+ CD38− GL7+ FAS+ CXCR4hi CD86lo) and LZ (B220+ CD38− GL7+ FAS+ CXCR4lo CD86hi) B cells with a FACSVantage SE with Diva option or FACSAria instruments (Becton Dickinson, >95% purity). Resting B lymphocytes from wild type and AID−/− mice were isolated from spleens by immunomagnetic depletion with anti-CD43 MicroBeads (Miltenyi Biotech) and cultured for four days in the presence of LPS (25 µg/ml) and IL-4 (5ng/ml). AID protein was detected as previously described (McBride et al., 2008).
Purification of malaria GC B cells and TC-seq library preparation
Three-to-five weeks after Plasmodium infection, MycI/I ROSAerISCEI/AIDer or MycI/I ROSAerISCEI/erISCEI AID−/− mice were injected intraperitoneally with 0.8mg Tamoxifen in corn oil, and euthanized 36 hours later. Upon ACK lysis, single-cell suspensions of splenocytes were incubated with biotinilated antibodies to mouse CD43 (clone S7), CD11c (N418), F4/80 (BMO), Gr-1 (RB6–8C5), and IgD (11–26c) for 20 minutes on ice (all from BD Pharmingen or eBiosciences). Upon washing, cells were incubated with anti-biotin microbeads and negatively depleted with magnetic columns (Miltenyi). This was followed by incubation with anti-CD19 microbeads and positive magnetic selection. GC B cells purity was monitored by flow cytometry and ranged between 88–95%. Two TC-seq libraries for each genotype were independently generated as previously described, with capture from the I-SceI site at c-myc (Klein et al., 2011), with the exception of using an improved linker (5’-GCA GCG GAT AAC AAT TTC ACA CAG GAC GTA CTG TGC GGC CGC T and 5’-/5Phos/GCG GCC GCA CAG TAC TTG ACT GAG CTT TA/3ddC/). Deep sequencing was with Illumina HiSeq, 100 cycles, paired-ends.
TC-seq and computational analysis
Pooled data from two independent libraries were analyzed as described (Klein et al., 2011; Wang et al., 2014), with minor modifications. Sequencing reads were first trimmed for quality with seqtk (error rate threshold of 0.01; Broad Institute). Reads containing primer sequences from the first PCR reaction were discarded from subsequent analysis. Finally, reads were mapped with SMALT (version 0.7.6; Sanger Institute), and only reads uniquely aligning at least 28 bps were included. To determine the enrichment at genic regions (Figure 2D), the following portions of the genome were excluded: 1Mb surrounding the I-SceI site, Igh, Igκ, and Igγ regions, 2kb-regions surrounding cryptic I-SceI sites (according to the consensus [TCA][AT]GGGATA[AC]CAGG[GCT][TC][ATC][AG][TAC]), AID-coding exons (likely representing retroviral insertions), and 3Mb at each centromere. As genic was considered the portion of DNA from −2kb of the most upstream transcription start site to the end of the last exon. For the role of transcription (Figure 2E), RNA-seq data (Hogenbirk et al., 2013) were mapped with STAR aligner v2.3.0.1 (Dobin et al., 2013) using the mouse genome (mm9) and removing multiple alignments. Transcripts were quantified and annotated using cufflinks v2.2.1 (Trapnell et al., 2013) and Ensembl annotation (release 66), and the following regions were excluded from downstream analysis: 1Mb surrounding the I-SceI site, 2kb-regions surrounding cryptic I-SceI sites, and AID-coding exons. For the analysis of enrichment within hotspots of viral integration at ERFS (M.J. and I.T.S., unpublished data) or CFS (Figure 3), and of hotspots of AID-dependent translocation (Table S1 and Figure 4), rearrangements within 1Mb surrounding the I-SceI site, within 2kb surrounding cryptic I-SceI sites, and within AID-coding exons were excluded from analysis. Translocation histograms (Figure 4A) were normalized for the size of the library and represented in log scale as TPKT (translocations per kilobase per thousand translocations in the library): TPKT = N/(B * S * 10−6), where: N is the number of translocations in each bin, B is the bin size in nucleotides (in this case B = 100), and S is the total number of translocations in the library. See also Extended Supplemental Procedures online.
M-FISH
To analyze chromosome rearrangements, metaphase spreads were obtained from primary tumor cells upon treatment with 100ng/ml colcemid for 1 hour and were hybridized with the 21× mouse probe cocktail (Metasystems). Imaging was on a Zeiss AxioImager M1, equipped with a motorized scanning stage, and Isis software was used for the analysis.
Deep-sequencing and computational analysis of lymphoma L23
Whole-genome sequencing and data analysis were performed as previously described (Jankovic et al., 2013). In brief, 5µg of genomic DNA were sonicated to an average fragment size of 500bps and processed according to the Paired-End (PE) Sample Preparation kit protocol (Illumina). Upon quality assessment by Bioanalyzer (Agilent), the library was subjected to paired-end sequencing. Data were analyzed for structural variation (SV) discovery using the following programs: Hydra-SV (Quinlan et al., 2010), BWA aligner (Li and Durbin, 2009), Novoalign, the intersectBed software from BEDtools suite (Quinlan and Hall, 2010), RepeatMasker and RefSeq track from the UCSC Genome Browser. In order to clear artifacts resulting from genetic variation, we subtracted SVs that were similarly found in the genomes of C57BL/6 and 129/Sv mice (Jankovic et al., 2013), or of 17 other inbred strains of laboratory mice (Keane et al., 2011). Further, SVs were removed from final analysis if any of the two reads matched to repeats for >50%, if deletions were <1kb, or if their Final Weighted Support value was <2. Circular plots of genomic rearrangements were generated using Circos software (Krzywinski et al., 2009).
Supplementary Material
Highlights.
Capture of chromosome translocations occurring in germinal center B cells, in vivo
AID is required for Plasmodium-associated B cell lymphoma
Chronic infection causes a shift towards a more mature lymphoma phenotype
Plasmodium-induced lymphomas bear chromosome translocations, including c-myc/Igh
Acknowledgments
We thank members of the lab for discussions and suggestions, the MR4 for providing the malaria parasites contributed by Drs. W. Peters, B. Robinson, and I. Landau, Dr. Frank Costantini for providing the ROSA26 targeting plasmids, and Dr. Rafael Casellas for AIDGFP mice. A particular thank you to Klara Velinzon and Yelena Shatalina for FACS sorting, and to Ariel Halper-Stromberg for help with figure 2. Moreover, we are thankful to Dr. Julie White (Laboratory of Comparative Pathology, Memorial Sloan-Kettering Cancer Center) for histopathological evaluation. TC-seq and lymphoma L23 sequencing data generated in this study have been deposited to the SRA database (SRP053308). This work was supported by grant 11-0022 from Worldwide Cancer Research (formerly known as Association for International Cancer Research), by the Fondazione Ettore e Valeria Rossi, and by NIH grant AI112602 to DFR, and in part by NIH grants AI037526 and AI072529 to MCN. AN and EC are supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research, and by a Department of Defense grant (BCRP DOD Idea Expansion Award, grant 11557134). NF is a Bennett Fellow of Leukaemia & Lymphoma Research. MCN is an HHMI Investigator. This manuscript is dedicated to Georg Klein on the occasion of his 90th birthday.
Footnotes
Extended Supplemental Procedures are provided in the Supplemental Information section available online.
The authors declare no competing financial interests.
References
- Achtman AH, Stephens R, Cadman ET, Harrison V, Langhorne J. Malaria-specific antibody responses and parasite persistence after infection of mice with Plasmodium chabaudi chabaudi. Parasite immunology. 2007;29:435–444. doi: 10.1111/j.1365-3024.2007.00960.x. [DOI] [PubMed] [Google Scholar]
- Adams JM, Gerondakis S, Webb E, Corcoran LM, Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983;80:1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkitt DP. Observations on the geography of malignant lymphoma. East African medical journal. 1961;38:511–514. [PubMed] [Google Scholar]
- Carter R, Walliker D. New observations on the malaria parasites of rodents of the Central African Republic - Plasmodium vinckei petteri subsp. nov. and Plasmodium chabaudi Landau, 1965. Ann Trop Med Parasitol. 1975;69:187–196. doi: 10.1080/00034983.1975.11687000. [DOI] [PubMed] [Google Scholar]
- Cesarman E. Gammaherpesviruses and lymphoproliferative disorders. Annual review of pathology. 2014;9:349–372. doi: 10.1146/annurev-pathol-012513-104656. [DOI] [PubMed] [Google Scholar]
- Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schrock E, Ried T, Kuehl WM. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, et al. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The lancet oncology. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epeldegui M, Vendrame E, Martinez-Maza O. HIV-associated immune dysfunction and viral infection: role in the pathogenesis of AIDS-related lymphoma. Immunologic research. 2010;48:72–83. doi: 10.1007/s12026-010-8168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, Magrath IT, Knowles DM, Dalla-Favera R. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014 doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlyn PH, Rabbitts TH. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 1983;304:135–139. doi: 10.1038/304135a0. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome research. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenbirk MA, Heideman MR, Velds A, van den Berk PC, Kerkhoven RM, van Steensel B, Jacobs H. Differential programming of B cells in AID deficient mice. PloS one. 2013;8:e69815. doi: 10.1371/journal.pone.0069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Debets JM, Faleiro R, Karunarathne DS, Liu XQ, Lineburg KE, Poh CM, Grotenbreg GM, Hill GR, MacDonald KP, Good MF, et al. PD-1 dependent exhaustion of CD8+ T cells drives chronic malaria. Cell reports. 2013;5:1204–1213. doi: 10.1016/j.celrep.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Feldhahn N, Oliveira TY, Silva IT, Kieffer-Kwon KR, Yamane A, Resch W, Klein I, Robbiani DF, Casellas R, et al. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Mol Cell. 2013;49:623–631. doi: 10.1016/j.molcel.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz S, Potter M, Rabkin CS. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes, chromosomes & cancer. 2003;36:211–223. doi: 10.1002/gcc.10178. [DOI] [PubMed] [Google Scholar]
- Kameoka Y, Tagawa H, Tsuzuki S, Karnan S, Ota A, Suguro M, Suzuki R, Yamaguchi M, Morishima Y, Nakamura S, et al. Contig array CGH at 3p14.2 points to the FRA3B/FHIT common fragile region as the target gene in diffuse large B-cell lymphoma. Oncogene. 2004;23:9148–9154. doi: 10.1038/sj.onc.1208136. [DOI] [PubMed] [Google Scholar]
- Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nature reviews Immunology. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Koduru PR, Raju K, Vadmal V, Menezes G, Shah S, Susin M, Kolitz J, Broome JD. Correlation between mutation in P53, p53 expression, cytogenetics, histologic type, and survival in patients with B-cell non-Hodgkin's lymphoma. Blood. 1997;90:4078–4091. [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. The New England journal of medicine. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- Kutok JL, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annual review of pathology. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrath I. The pathogenesis of Burkitt's lymphoma. Advances in cancer research. 1990;55:133–270. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- Magrath I. Epidemiology: clues to the pathogenesis of Burkitt lymphoma. British journal of haematology. 2012;156:744–756. doi: 10.1111/j.1365-2141.2011.09013.x. [DOI] [PubMed] [Google Scholar]
- Marcucci F, Mele A. Hepatitis viruses and non-Hodgkin lymphoma: epidemiology, mechanisms of tumorigenesis, and therapeutic opportunities. Blood. 2011;117:1792–1798. doi: 10.1182/blood-2010-06-275818. [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Feitelson MA, Munger K. Human Viral Oncogenesis: A Cancer Hallmarks Analysis. Cell host & microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux EM, Rochford R, Griffin B, Newton R, Jackson G, Menon G, Harrison CJ, Israels T, Bailey S. Burkitt's lymphoma. Lancet. 2012;379:1234–1244. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- O'Shea D, O'Riain C, Taylor C, Waters R, Carlotti E, Macdougall F, Gribben J, Rosenwald A, Ott G, Rimsza LM, et al. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood. 2008;112:3126–3129. doi: 10.1182/blood-2008-05-154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Pearson T, Shultz LD, Miller D, King M, Laning J, Fodor W, Cuthbert A, Burzenski L, Gott B, Lyons B, et al. Non-obese diabetic-recombination activating gene-1 (NOD-Rag1 null) interleukin (IL)-2 receptor common gamma chain (IL2r gamma null) null mice: a radioresistant model for human lymphohaematopoietic engraftment. Clinical and experimental immunology. 2008;154:270–284. doi: 10.1111/j.1365-2249.2008.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W, Robinson BL. The chemotherapy of rodent malaria. LVIII. Drug combinations to impede the selection of drug resistance, Part. 2: The new generation--artemisinin or artesunate with long-acting blood schizontocides. Ann Trop Med Parasitol. 2000;94:23–35. doi: 10.1080/00034980057581. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Potter M, Wiener F. Plasmacytomagenesis in mice: model of neoplastic development dependent upon chromosomal translocations. Carcinogenesis. 1992;13:1681–1697. doi: 10.1093/carcin/13.10.1681. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Clark RA, Sokolova S, Leibowitz ML, Zhang Y, Hurles ME, Mell JC, Hall IM. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome research. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annual review of pathology. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- Saha A, Robertson ES. Epstein-Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:3056–3063. doi: 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen A, Sauerwein RW. How malaria modulates memory: activation and dysregulation of B cells in Plasmodium infection. Trends in parasitology. 2013;29:252–262. doi: 10.1016/j.pt.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nature reviews Immunology. 2002;2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal Centers. Annu Rev Immunol. 2011 doi: 10.1146/annurev-immunol-120419-022408. [DOI] [PubMed] [Google Scholar]
- Wang Q, Oliveira T, Jankovic M, Silva IT, Hakim O, Yao K, Gazumyan A, Mayer CT, Pavri R, Casellas R, et al. Epigenetic targeting of activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1420575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KH, Leroy K, Moller MB, Colleoni GW, Sanchez-Beato M, Kerbauy FR, Haioun C, Eickhoff JC, Young AH, Gaulard P, et al. Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B-cell lymphoma: an international collaborative study. Blood. 2008;112:3088–3098. doi: 10.1182/blood-2008-01-129783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kracker S, Yasuda T, Casola S, Vanneman M, Homig-Holzel C, Wang Z, Derudder E, Li S, Chakraborty T, et al. Immune surveillance and therapy of lymphomas driven by Epstein-Barr virus protein LMP1 in a mouse model. Cell. 2012;148:739–751. doi: 10.1016/j.cell.2011.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucca E, Bertoni F, Roggero E, Cavalli F. The gastric marginal zone B-cell lymphoma of MALT type. Blood. 2000;96:410–419. [PubMed] [Google Scholar]
- Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.