Summary
Genomes are non-randomly arranged in the 3D space of the cell nucleus. Here we have developed HIPMap, a high-precision, high-throughput, automated fluorescent in situ hybridization imaging pipeline, for mapping of the spatial location of genome regions at large scale. High-throughput imaging position mapping (HIPMap) enabled an unbiased siRNA screen for factors involved in genome organization in human cells. We identify 50 cellular factors required for proper positioning of a set of functionally diverse genomic loci. Positioning factors include chromatin remodelers, histone modifiers and nuclear envelope and pore proteins. Components of the replication and post-replication chromatin re-assembly machinery are prominently represented amongst positioning factors and timely progression of cells through replication, but not mitosis, is required for correct gene positioning. Our results establish a method for the large scale mapping of genome locations and have led to the identification of a compendium of cellular factors involved in spatial genome organization.
Graphical Abstract

Introduction
Chromosomes and individual regions of the genome occupy preferential non-random positions inside the 3D space of the cell nucleus (Bickmore, 2013; Misteli, 2007). The position of genomic loci has been linked to numerous nuclear functions including transcription, replication, DNA repair, and chromosome translocations (Chiolo et al., 2011; Gilbert et al., 2010; Roix et al., 2003; Takizawa et al., 2008). The non-randomness of genome architecture can be measured by the proximity of a gene locus to the nuclear periphery, to nuclear structures such as the nucleolus or transcription centers, or by the proximity of a locus to another genomic region (Branco and Pombo, 2006; Chubb et al., 2002; Roix et al., 2003; Thomson et al., 2004; Zhang et al., 2012).
The spatial position of a genomic locus is routinely determined using fluorescence in situ hybridization (FISH) which allows physical mapping of a genomic region relative to a defined landmark (Speicher and Carter, 2005; Wei et al., 2013). DNA FISH has been used extensively to visualize the position of a locus and to document changes in positioning that occur during physiological and pathological processes (Ferrai et al., 2010; Meaburn et al., 2007b; Takizawa et al., 2008), such as the relocation of gene loci during differentiation (Hewitt et al., 2004; Kosak et al., 2002; Williams et al., 2006) or the proximity of translocation-prone genome regions in 3D space (Hakim et al., 2012; Mathas et al., 2009; Misteli and Soutoglou, 2009). The development of chromosome conformation capture techniques such as 3C, 4C, and Hi-C, which allow mapping of intra- and inter-chromosomal interactions at the scale of entire genomes, has further highlighted the non-randomness of higher genome organization and has revealed several novel principles of organization including the existence of functionally and structurally defined genomic sub-domains (de Wit and de Laat, 2012; Dixon et al., 2012; Lieberman-Aiden et al., 2009).
While the non-randomness of genome organization in the cell nucleus is well established, and some factors involved in shaping global higher-order chromatin structure such as CTCF, cohesin and Mediator have been identified (Botta et al., 2010; Ling et al., 2006; Phillips and Corces, 2009; Sofueva et al., 2013; Vogelmann et al., 2011; Zhao et al., 2006), the molecular machinery that determines the location of a gene or genome region in the 3D space of the nucleus are largely unknown. Physical mapping methods identified genome regions preferentially associated with the nuclear lamina, pointing towards a role for nuclear lamins in retaining genome regions at the nuclear periphery, and thus determining their spatial location (Guelen et al., 2008; Meuleman et al., 2013; Peric-Hupkes et al., 2010; Pickersgill et al., 2006). Furthermore, a genetic screen using a reporter gene in C. elegans identified histone methyltransferases and the H3K9me3 modification as determinants of peripheral localization (Towbin et al., 2012).
While DNA FISH can be performed in high-throughput format (Joyce et al., 2012), the systematic identification of molecular determinants of genome positioning has been hampered by the fact that spatial gene mapping by either imaging or chromosome conformation capture technology have not been amenable to implementation at a high-throughput scale and are thus not well suited for use in screening approaches. To overcome this limitation, we describe here the development of HIPMap (High-throughput Imaging Position Mapping), a fully automated FISH-based imaging pipeline to quantitatively determine the position of multiple endogenous loci in the nucleus of mammalian cells with high accuracy and high throughput. We use HIPMap in combination with siRNA screening to discover human genome positioning factors in an unbiased, large-scale fashion. We identify 50 cellular factors, most of them previously not implicated in genome organization, which affect positioning of a set of functionally diverse human genes. Our results provide insights into the mechanism by which genes are positioned in the cell nucleus and they represent a method for large-scale 3D gene mapping which will be applicable to the study of a wide variety of aspects of nuclear organization in diverse cellular systems.
Results
HIPMap: High-throughput imaging-based mapping of genome positions
In order to identify factors involved in spatial genome organization we developed an imaging-based method for the high-throughput, quantitative mapping of the spatial location of a genomic region in the mammalian cell nucleus. High-throughput imaging position mapping (HIPMap) is a robust, high-resolution imaging approach that quantitatively measures the spatial position of genome regions with high precision at a large scale. HIPMap is based on a streamlined FISH protocol optimized for use in a 384-well format enabling visualization of multiple endogenous gene loci in thousands of cells and several hundred samples, enabling accurate spatial gene mapping in large sample sets. The approach uses fluorescently labeled FISH probes in a fully automated liquid-handling FISH protocol, automated 3D image acquisition using confocal high-throughput microscopy, and a high-content image analysis pipeline (Figures 1A–1C; see Experimental Procedures). The custom designed analysis pipeline includes image and statistical analyses to quantitatively map the distribution profile of a gene locus on a single-cell basis with high accuracy and statistical power.
Figure 1. A high-throughput imaging-based method to map gene positioning (HIPMap).
A. HIPMap outline. Cells are cultured in 384-well imaging plates and FISH is carried out in a fully automated fashion using directly labeled BAC probes, followed by automated image acquisition using high-throughput microscopy. Image analysis by Acapella segments the nucleus border and detects FISH signals. Normalized radial distances from the nuclear border are measured. Distance measurements distributions are plotted as histograms and/or estimated density curves.
B. Representative maximal projections of images acquired in three channels. FISH signals are automatically detected inside the nucleus ROI (see Experimental Procedures for details). Scale bar 10μm.
C. Density curves for normalized radial distance distributions for the indicated loci. For each locus the distance of at least 600 FISH spots from the nucleus periphery was determined.
D. Detection of radial position shift by siRNA silencing of lamins. Histograms of the normalized radial distance distributions of control non-targeting siRNA (green) compared to LMNA/C/B1 knock-down (purple) for the indicated locus. Distributions were generated from at least 600 FISH spots per samples. P<1e-16 for all 3 loci using the two sample KS test. Lines represent estimated density. See additional information in Experimental Procedures and Figure S1.
As proof of principle for the use of HIPMap to accurately determine spatial gene positions, we mapped the radial nuclear position of three diverse genome loci in hTERT immortalized CRL-1474 human skin fibroblasts (Fernandez et al., 2014; Scaffidi and Misteli, 2011). LADF is a lamina associated domain located on chromosome 5q35.2, devoid of ORFs and previously reported to localize to the nuclear periphery and to interact with the nuclear lamina (Guelen et al., 2008) (Figures 1B, 1C); COL1A1 is localized on chromosome 17q21.33 and is one of the most actively transcribed genes in CRL-1474 cells (Fernandez et al., 2014); OR5H1 is an olfactory receptor (OR) cluster on chromosome 3q11.2 which contains a set of silenced OR genes. Using fluorescently labeled FISH probes, the three loci were simultaneously visualized by high-throughput FISH. Z-stacks of 500–1,000 cells per well were acquired in four channels and analyzed using dedicated image and data analysis pipelines. The analysis pipeline detects the nuclear border with high accuracy using the DAPI channel (Figure S1A) and identifies FISH spots based on local maxima of the respective probe fluorophores in the other three channels simultaneously in maximal projections of z-stacks (Figures 1B and S1A; see Experimental Procedures for details). As expected, two FISH spots were detected in the majority of cells (60–80%) and the false-positive (FISH spots detected by image analysis but not visually) and false–negative (FISH spots detected visually but not by image analysis) detection rates were between 0–9%, as measured by comparing manual vs. software-based spot detection (Figures S1B, S1C). To determine the spatial position of a gene locus in the cell nucleus, the radial distance of the center of each measured FISH spot from the nuclear border was determined. To eliminate shape and size effects, nuclei were normalized using distance transformation and the normalized radial distance was measured as described in (Nandy et al., 2012) (nucleus periphery = 0; nucleus center = 1). (Figure S1A; see Experimental Procedures). All single-allele distance measurements were combined to generate a position distribution graph (Figure 1C). The minimally required sample size to achieve high statistical fidelity was determined to be ~600 FISH signals per well by computational simulation of 10,000 comparisons of variable population sizes generated by random sub-sampling (KS test; p < 0.005; Figure S1D).
As expected, when analyzed by HIPMap, we found LADF to localize strongly to the nuclear periphery with a median normalized radial distance from the nuclear edge of 0.2 ± 0.01 (SD, Figure 1C). In contrast, the highly expressed COL1A1 gene preferentially localized to a more central location with a median radial distance of 0.53 ± 0.01, and the OR5H1 locus assumed an intermediate position with a median radial distance of 0.39 ± 0.03 (Figure 1C). As quality control measures, the well-to-well variability of all three loci was minimal as indicated by similar distributions in multiple independent replicate wells and highly reproducible distance distributions in replicate experiments (p > 0.05, Figure S1E). The radial distance distributions of the three loci were conserved between CRL-1474 cells and human prostate PC3 cells, IMR90 and MRC-5 lung fibroblasts (Figure S1F). In HeLa cervical cancer cells the distribution of both LADF and OR5H1 was shifted towards the interior and periphery, respectively, compared to the other cell lines (Figure S1F), possibly due to abundant numerical and structural chromosome abnormalities in this cell line.
To determine whether HIPMap was sufficiently sensitive to detect changes in nuclear position of a specific endogenous locus, and to test whether HIPMap could be combined with siRNA strategies, we concomitantly knocked down the nuclear lamina genes LMNA/C and LMNB1 and examined the position of the peripheral LADF locus which is associated with the nuclear lamina (Guelen et al., 2008). RNAi knockdown of LMNA/C and LMNB1 for 72 hours resulted in elimination of >90% of the LaminA and LaminB1 mRNA (Figure S1G) and caused a concomitant shift in the position of LADF towards the center of the nucleus in CRL-1474, HeLa and IMR90 cells when compared with cells transfected with a negative, non-targeting siRNA (Figures 1D, S1E and S1H, p<1e-16, KS test). Similarly, COL1A1 and OR5H1 also underwent, albeit smaller, repositioning towards the periphery (Figures 1D and S1E; p<1e-16, KS test). Repositioning of these loci was not due to changes in nuclear size or shape during siRNA knockdown, since the mean nuclear area and the mean nuclear width to length ratio did not change between negative control and siLMNA/C/B1 transfected cells (Figures S2B, S2C). We conclude that HIPMap is a robust method for mapping the spatial position of genome regions and, in combination with siRNA approaches, has the potential to identify cellular determinants that affect gene positioning.
Identification of genome positioning determinants by RNAi screening
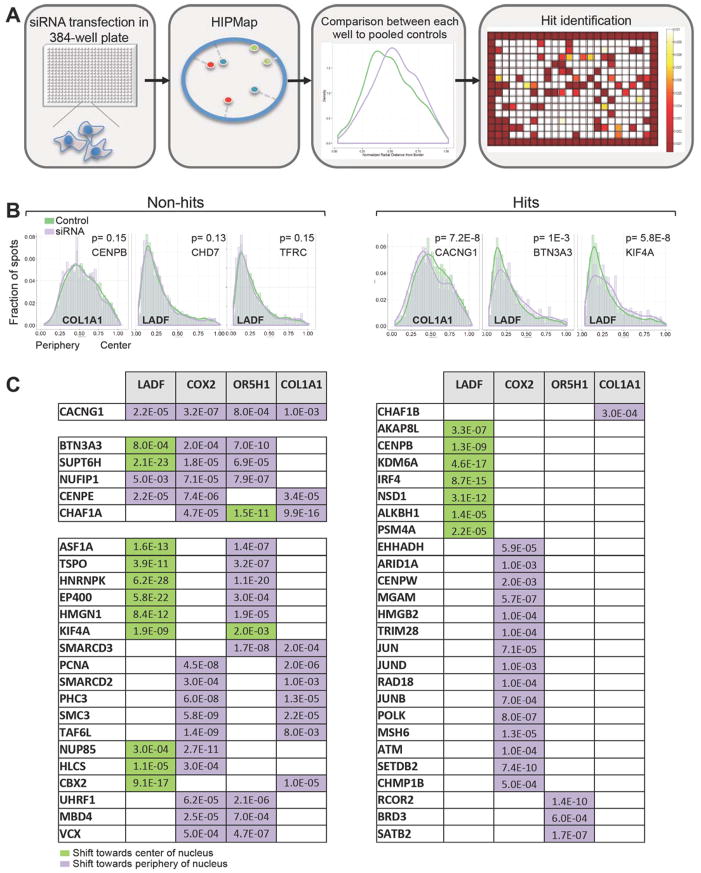
We applied HIPMap to conduct an siRNA screen to systematically identify determinants of genome positioning in human cells. CRL-1474 cells were reverse transfected in 384-well plates with a library of siRNA oligos targeting 669 nuclear genes (Figure 2A). The library contained siRNAs against annotated chromatin-binding proteins, nuclear structural proteins, and nuclear envelope- and lamina-associated proteins. Following knock-down for 72 hours, the position of LADF, OR5H1 and COL1A1 in each siRNA-treated well was determined using HIPMap. The distribution of each locus in the presence of each siRNA was statistically compared to that of six pooled, non-targeting siRNA wells using the KS-test as previously described (Meaburn et al., 2009; Meaburn and Misteli, 2008) (Figure 2A; see Experimental Procedures for details). The screen was conducted in biological duplicates using an siRNA pool targeting LMNA/LMNB1 as a positive control and non-targeting siRNA as negative control on each replicate plate (Figure 2A). Using a p value cut-off of p<2e-3, 135 of 669 (20%) targeted genes were identified as hits based on their ability to re-position at least one of the three target loci (Figures 2B, 2C and Table S1). The top 65 hits based on the most significant p values (p <1e-4) and the largest difference between sample and control distributions were validated in a secondary screen using an oligo siRNA pool of distinct chemistry and sequence to rule out possible siRNA off-target effects. A fourth locus, COX2, located at 1q31.1, is weakly expressed in CRL-1474 cells and has a peripheral localization, was also included in the secondary screen to expand the scope of tested loci. The validation screen identified 50 hits (77% confirmation rate) (Figure 2C). Several controls confirmed that the observed repositioning events were specific and were not due to indirect, global effects on nuclear organization upon knockdown of target proteins: 1) independent biological replicates yielded similar p values, 2) gene repositioning in replicates occurred to the same extent and in the same direction along the radial axis (Figure S2F), 3) positive hits did not reduce cell number, ruling out the possibility that they were identified due to effects on cell viability or cell cycle progression (Figures S2A, S2E, S2G), with the exception of, as expected, replication-related proteins (Figure S2E), 4) positive hits did not correlate with DNA damage, as measured by percentage of γH2AX positive cells (Figure S2D), 5) no effects of the hit siRNAs on nuclear size and shape as indicated by the mean nuclear area and the mean width/length ratio were detected (Figures S2B, S2C) and finally, there was no correlation with global transcription activity (Figures S2I, S2J) nor ROS production (Figure S2H). We conclude that the combination of HIPMap with RNAi screening allows the unbiased identification of cellular factors involved in determining the spatial position of endogenous genes in human cells.
Figure 2. Identification of genome positioning determinants by RNAi screening.
A. HIPMap-based siRNA screen outline. Reverse transfection of siRNA was conducted in 384- well plates and locus position was measured by HIPMap. P-values for each well were generated by pairwise comparison of the distribution of each sample to a negative control distribution generated by pooling together 6 wells containing non-targeting siRNA using the KS test. Hits were defined as siRNAs with a p value <2e-3..
B. Histograms of representative distributions of hits and non-hits. Distributions represent data from >600 FISH spots. Values represent one of two experimental replicates. Lines indicate the estimated density.
C. List of 50 high-confidence hits identified following primary- and validation-screen. Hits were defined as p values <2e-3 in both screens. See additional information in Experimental Procedures, Figure S2 and Tables S1 and S2.
Functional classification of genome positioning determinants
In order to characterize mechanisms of genome positioning, the 50 identified factors were analyzed for their functional properties. The majority (26/50) of hits affected only a single locus, suggesting that repositioning was not due to pleiotropic genome reorganization. 18 hits affected two loci, five hits affected three loci and only one hit affected all four loci (Figures 2C and 3A). The most enriched gene ontology groups compared to their representation in the siRNA library were centromere proteins (54 fold enrichment), chromatin remodeling (13 fold), nuclear envelope components (4 fold) and DNA repair and replication factors (3 fold) (Figure 3B).
Figure 3. Functional classification of genome positioning determinants.
A. Venn diagram representing the number of validated repositioning factors according to the affected locus.
B. Classification of hits into functional groups. Hits were grouped according to their assigned function and plotted according to the number of affected loci.
C. Bar graphs representing the relative contribution of each nuclear pathway to the re-positioning of the indicated locus.
D. Box plots of normalized radial distances from the nuclear border of the indicated locus following siRNA treatment. Dashed line indicates the median of the control distribution. Boxes show the 25th, 50th (median) and 75th percentile of the distributions and whiskers extend up to 1.5* inter-quantile range, outliers are shown as dots. Asterisks indicate hits identified in the screen using KS test with a p<2e-3. The same data is presented in Figure S3A as distribution curves.
E. mRNA expression, as measured by qRT-PCR, of COL1A1 and COX2 following siRNA transfection of the indicated gene or combination of genes for 72 hours. Expression is normalized to hTBP. All expression ratios are relative to non-targeting siRNA control (siNT). Values represent averages from 2 experiments ± SD.
See additional information in Figure S3.
All tested loci were affected by multiple, functionally diverse positioning factors, indicating that the location of a given locus is influenced by multiple pathways. As expected, the positioning of the peripheral LADF locus was strongly affected by nuclear envelope proteins (24% of all LADF hits). LADF was generally re-positioned towards the center of the nucleus regardless of the gene that was knocked-down (Figure 3D), suggesting that the identified factors are responsible for peripheral attachment and that its peripheral positioning is the result of an active process. Re-positioning of LADF towards the center was further confirmed by loss of co-localization between LADF and the nuclear lamina following knockdown of several hits as indicated by a reduction of more than 50% of cells containing two lamina-associated LADF loci (Figure S3B). Other hit classes that affected LADF included centromeric proteins and histone modifiers such as KDM6A and SUPT6H, which promote H3K27me3 demethylation (Wang et al., 2013). In contrast, the predominantly internally located COL1A1 locus was re-positioned towards a more peripheral location by all identified hits (Figure 3D). The active COL1A1 locus was mainly affected by chromatin remodeling and polycomb complex factors, both of which did not have a significant effect on the transcriptionally inactive loci tested (Figure 3C). OR5H1 re-positioned in both directions, depending on the specific hit and was mostly affected by nuclear envelope and mRNA processing factors (Figures 3C and 3D).
We identified several shared features of repositioning factors acting on genome regions with common properties. The peripherally located LADF, COX2 and the OR5H1 were affected by a set of factors (Figures 2C, 3A) enriched in nuclear envelope components, which combined represented 37% of the hits affecting these loci, including NUP85, AKAP8L and CACNG1. Other factors that affected the positioning of peripheral loci were histone modifiers and RNA binding proteins. A separate set of factors affected the position of the internal COL1A1 locus (Figure 3C), including several replication and chromatin remodeling factors such as PCNA, CHAF1A and SMARCD3.
Distinct sets of factors were found to affect the position of expressed genes when compared to inactive genome regions (Figure 3A). Positioning of the two expressed loci COX2 and COL1A1 was affected by several transcription factors and chromatin remodelers including TAF6L and SMARCD2. In contrast, the transcriptionally silent regions LADF and OR5H1 were most strongly affected by chromatin organizers such as HMGN1 and EP400. These results identify a set of molecular determinants of gene positioning and demonstrate selectivity of their effects on related subsets of genome regions.
Two of the genome regions we tested in the screen contain expressed genes: COL1A1 is highly expressed and COX2 is expressed at a low level (Fernandez et al., 2014). Gene repositioning of these genes in response to knockdown was uncorrelated with activity. For COL1A1, two out of four tested hits (SMC3 and SMARCD2) affected its position, but not activity, demonstrating that the two processes can be uncoupled (Figure 3E). Similarly, for COX2, knock-down of SETDB2 or CHAF1A and CHAF1B re-positioned the locus, but had no effect on its expression (Figure 3E). On the other hand, some hits affected both positioning and expression, for example CHAF1A and CHAF1B for COL1A1 and SMC3 and SMARCD2 for COX2 (Figure 3E). Changes in expression upon knockdown of these factors was not due to a global effect on the transcriptional machinery, since they did not affect expression of some of the test genes, nor of GAPDH or hTBP housekeeping genes (Figure S5F). These results demonstrate that gene positioning can be uncoupled from transcriptional activity.
DNA replication is a determinant of genome positioning
A prominent group of genome positioning factors affecting all tested loci included several DNA replication-associated proteins, particularly post-replication histone chaperones. In addition, several replication-associated DNA repair factors were identified as hits. The presence of replication-associated factors prompted us to explore in more detail the role of replication in gene positioning. The histone chaperones CHAF1A, CHAF1B and ASF1A, all involved in post-replication chromatin assembly, the PCNA replication sliding clamp, and the replication-associated mismatch repair protein MSH6 and the translesion DNA polymerase POLK were all identified as prominent hits in the screen. Knockdown of any one of these proteins led to repositioning of at least one target locus (Figures 4A, 4B and Table S3). Similar effects were observed when combinations of replication factors were knocked down (Figure 4B).
Figure 4. DNA replication is a determinant of genome positioning.
A. Histograms of representative distributions of loci following knock-down of the indicated gene. Distributions represent data from >600 FISH spots. Values represent one of three experimental replicates. Lines indicate estimated density.
B. Statistical testing of distributions using the KS test. P values are calculated by comparing the pooled negative controls distribution to the distribution in cells knocked down for the indicated gene or combination of genes. Values represent one of three independent experimental replicates.
C. Representative distributions of three loci comparing control (green) and siRNA transfected cells (purple) in cycling cells or cells treated with 2mM thymidine and the indicated siRNA for 72 hours. Values represent one of three experimental replicates.
D, E. P value heat maps comparing cycling cells to G1/S arrested cells for 3 different indicated loci. siRNA silencing of replication factors (D) or chromatin and structural proteins (E). Values represent one of three independent experimental replicates.
The primary and the secondary siRNA screens that led to the identification of replication-related proteins as genome positioning factors were performed in asynchronous cycling cells. To test whether DNA replication is required for re-positioning in response to knock-down, we arrested CRL-1474 cells at the G1/S boundary. To obtain a highly synchronous population, cellular quiescence was induced by growing cells at high density for 72 hours (Figure S4A), resulting in growth arrest of > 90% of cells in the population as measured by negative Ki67 and CyclinA staining (Figures S4B and S4C). Quiescence itself did not lead to significant re-positioning of all tested loci (Figure S4E). Quiescent cells were released into normal media or into media containing 2mM thymidine to prevent entry into S-phase and concomitantly transfected with siRNA (Figure S4A). Transfection efficiency and the extent of the siRNA knock-down were similar in cycling and arrested cells (Figure S4D). In contrast to control cycling cells released into normal media, in which re-positioning was observed following knock-down of replication-related factors, G1/S arrested cells showed no change in position of all tested loci (Figures 4C and 4D). Importantly, the requirement for S-phase progression for repositioning was not limited to replication-associated genome positioning factors. Knockdown of non-replication proteins such as NUP85 and AKAP8L, which resulted in significant repositioning in cycling cells, had no effect on the position of all assessed target genes in G1/S arrested cells (Figure 4E). We concluded that progression through S-phase is required for proper genome positioning by the identified factors, regardless of their direct involvement in DNA replication.
To directly test whether replication per se, rather than the absence of positioning factors during replication, contributes to gene positioning, we grew CRL-1474 cells under conditions of slowed replication by treatment for 24 hours with low-dose hydroxyurea (HU, 200μM). Under these conditions, S-phase progression was impaired, as indicated by diminished progression of cells into G2/M and accumulation of cells in G1 and S-phases (Figure S5A). In low-dose HU treated cells, LADF, COX2 and COL1A1 loci underwent significant re-positioning (Figure 5A). LADF was re-positioned towards the center of the nucleus (p= 5.7e-5) and COX2 and COL1A1 were re-positioned towards the periphery (p= 1.6e-7, p=2.6e-4, respectively). The extent and direction of re-positioning was similar to knock-down of replication-associated proteins (Figure 5A). Re-positioning was not due to the accumulation of cells in G1- or S-phase since gene positions were indistinguishable in the various cell cycle phases in normally cycling cells (Figure S5E) in agreement with mapping by Hi-C (Naumova et al., 2013; Sofueva et al., 2013). The observed repositioning was not due to DNA damage or DNA damage response (DDR) signaling caused by HU treatment, since no significant re-positioning of LADF, COL1A1 and COX2 was observed after treatment of cells with a high dose of HU (4mM) or etoposide for up to 4 hours to induce extensive DNA damage (Figures 5B and S5B–S5D). These observations demonstrate that timely progression of DNA replication is required for proper gene positioning.
Figure 5. Normal progression through replication is a required for accurate positioning.
A. Distributions of untreated (green) or hydroxyurea treated cells (200μM, 24 hours, purple) for the indicated locus. P values were calculated using KS test. Values represent one of three experimental replicates.
B. P value heat map comparing untreated cells to cells treated with the indicated DNA damaging agent for up to 4 hours for 3 different loci. Values represent one of two experimental replicates.
C. mRNA expression of COL1A1 and COX2 following knock-down of the indicated gene or set of genes for 72 hours in thymidine arrested cells. Values are normalized to hTBP. All expression ratios are relative to non-targeting siRNA control (siNT). Values represent averages from two experiments ± SD. See additional information in Experimental Procedures and Figure S5.
Interestingly, siRNA knockdown of several candidate positioning factors which affected both the localization and the transcriptional activity of their target loci in cycling cells, did not affect positioning nor expression in arrested cells (Figure 5C). For example, knock-down of LMNA/C and LMNB1 which increased expression of both COL1A1 and COX2 in cycling cells and repositioned these loci (Figure 3E), had no effect on their expression or re-localization in arrested cells. Similarly, knock-down of SMARCD2 changed the expression and position of COX2 in cycling cells, but had no effect on either in arrested cells. These observations suggest that the effect on activity in cycling cells was due to repositioning and not merely a consequence of factor knockdown on transcription of the target gene (Figure 5C).
Mitosis is not required for establishment of genome positioning
Having identified normal progression through S-phase as a determinant of proper gene positioning, we asked whether passage through mitosis was also required to establish gene position. To test this hypothesis, cells were arrested for 72 hours at the G1/S boundary and simultaneously transfected with siRNA followed by release into normal media. Cells were fixed 6 hours after release from the G1/S block, prior to their entry into M-phase as assessed by cell cycle analysis and pospho-Ser10 H3 levels (Figures S6A–S6C) or, as a control, after 24 or 48h (Figure S6D). As expected, thymidine-arrested cells showed no repositioning of LADF, but significant re-positioning in response to LMNA/B1 knockdown was already observed six hours after release, when the majority of the cells had not entered mitosis yet (Figures 6A, 6B). Similarly, knockdown of CTCF resulted in gene repositioning prior to entry into mitosis (Figure 6B). A similar extent of repositioning was observed upon knockdown in cells released for 24h or 48hrs, indicating completion of the repositioning event prior to mitosis (Figure 6B). These results indicate that re-positioning does not require passage through mitosis and occurs prior to entry in M-phase.
Figure 6. Mitosis is not required to establish gene positioning.
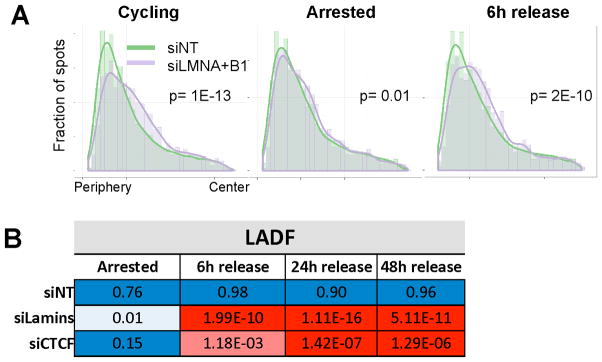
A. Radial distance distributions of control (green) and siRNA transfected cells (purple) for the LADF locus. For each treatment, non-targeting siRNA control (siNT) distributions are plotted together with knock-down of lamin distribution.
B. P value heat map comparing each treatment to the indicated knock-down and treatment. Data represent one of four experiments.
See additional information in Figure S6.
Discussion
We have developed HIPMap, an imaging pipeline for the accurate mapping of genome loci in a high-throughput fashion. The ability to quantitatively determine the position of endogenous gene loci in thousands of samples enabled us to perform an unbiased screen to identify cellular factors involved in determining the position of individual loci in the 3D space of the human cell nucleus.
Mapping gene positioning using HIPMap
HIPMap combines optimized, fully automated high-throughput FISH with high-resolution confocal microscopy and a robust image analysis platform for accurate determination of the position of FISH signals in the cell nucleus at a large scale. HIPMap has the ability to routinely image and measure hundreds of cells per sample in thousands of samples. The dense datasets generated by the large number of cells imaged and the small variability between replicates allows determination of gene positioning in the nucleus with high precision enabling reliable detection of even relatively small changes in positioning of gene loci, including repositioning events in subpopulations of cells. The method is based on single-cell analysis and provides detailed information on the variability of gene localization within the population. The limiting step in the throughput of the method is the imaging time, which depends on the chosen number of channels, number of z-stacks and the number of imaged fields. The primary siRNA screen described here was achieved by continuous automated imaging of 1512 wells over ~200 hours.
Using HIPMap as a screening tool to identify gene positioning factors
The high-throughput nature of HIPMap makes it suitable as a screening tool and has made possible the characterization of gene positioning factors in an unbiased discovery approach. Screening approaches to identify factors that determine the position of genome regions have in the past been severely limited by the difficulty in reliably detecting and measuring the position of genome regions by FISH at a large scale. In C. elegans the methyltransferases MET2 and MET25 were identified as key factors for sequestration of a transcriptionally repressed artificial GFP-tagged reporter to the nuclear periphery during embryonic development (Towbin et al., 2012). Furthermore, a high-throughput FISH screen in Drosophila cells identified factors involved in homologous chromosomes pairing in mitosis, but did not address interphase positioning (Joyce et al., 2012). Importantly, HIPMap detects endogenous loci, thus overcoming the limitation of screening approaches to artificial reporters and allows the analysis of genes with variable expression profiles and in a wide range of biologically relevant settings.
We used HIPMap to discover nuclear factors that determine the positioning of several endogenous loci of variable functional status ranging from a gene desert to a highly active locus. Reassuringly, we identified several factors that were previously reported to contribute to global genome organization such as SMC3, LMNA/C and SETDB2 (Guelen et al., 2008; Peric-Hupkes et al., 2010; Sofueva et al., 2013; Towbin et al., 2012; Zuin et al., 2014). In addition, we identified a set of novel factors not previously implicated in genome positioning. Importantly, most identified factors affected only a subset of target loci, demonstrating that their effects are not global. Conversely, we find that all tested loci are affected by multiple, functionally diverse positioning factors, indicating that the location of a locus is likely determined by the integrated action of multiple processes and molecular pathways, rather than by dedicated genome positioning machinery.
Various functional groups of positioning factors were identified. Our observation that loss of lamins results in internalization of a LAD suggests that the peripheral location of LADs is not a default localization, but that lamins actively tether these regions to the nuclear periphery. This interpretation is in line with the finding based on live cell observations that LADs are relatively dynamic and undergo periodic cycles of association- and dis-association with the nuclear lamina (Akhtar et al., 2013) and with the reported repositioning of genome regions in laminopathy patient cells (Meaburn et al., 2007a). In contrast, the position of transcriptionally active genome regions was more prominently affected by transcription factors and chromatin remodelers. However, we find that gene positioning is not tightly linked with gene activity, suggesting that these factors act in a transcription-independent fashion and that positioning and expression can be uncoupled. This conclusion is in line with the recent finding that chromatin decondensation, rather than transcriptional activation, is sufficient to reposition endogenous genes (Therizols et al., 2014). For some genes, however, repositioning was accompanied by a change in expression. Interestingly, in several instances where knockdown of positioning factors resulted in a change in gene expression, no such effect was seen in non-cycling cells, where re-positioning is suppressed, suggesting that positioning affects expression of these genes. Detailed characterization of each identified repositioning factor to determine their mode of action should shed light on whether distinct functional classes of genes are affected by different positioning mechanisms.
Replication is required for re-positioning
The results of our screen point to a significant role of replication in determining the position of genome regions. We find several components of the replication machinery and several DNA repair factors, which are active during replication, as prominent hits. Furthermore, interference with replication by drug treatment resulted in repositioning of several target loci, suggesting that the process of replication itself, rather than the individual replication-associated factors, determines gene positioning. Further evidence for this notion is that even non-replication related factors required progression of cells through S-phase to mediate their repositioning effects, strongly suggesting that replication and timely passage through S-phase is a major determinant of genome positioning. Considering that several of the replication-associated repositioning factors are involved in chromatin assembly, it is tempting to speculate that post-replication chromatin assembly, during which proper chromatin states are re-established and epigenetic modifications are transmitted to the daughter strands, is a critical contributor to establishing and maintaining gene position.
A major question is how spatial genome organization and gene positions are maintained in cells as they pass through mitosis. Hi-C analysis has recently demonstrated that the internal domain structure of chromosomes is lost during mitosis and re-established in early to mid G1 (Naumova et al., 2013). Previous analysis of entire chromosomes or large genome regions suggested that overall patterns of organization are partially maintained during cell division (Cvackova et al., 2009; Gerlich et al., 2003; Walter et al., 2003). Furthermore, tethering experiments have indicated a requirement for progression through mitosis for proper positioning of artificial chromatin arrays linked to the nuclear periphery via nuclear envelope proteins (Finlan et al., 2008; Kumaran and Spector, 2008; Reddy et al., 2008). In our analysis of endogenous gene loci, we find that gene positions are largely determined before cells reach mitosis. Single-cell analysis to track the location of individual genes through the cell cycle will be required to study positioning of genome regions in individual living cells.
Future Applications of HIPMap
Here, we applied HIPMap to generate a list of factors with a potential role in determining radial gene positioning in the human cell nucleus. The identification of candidate genome positioning factors now opens the door to the investigation of the precise mechanisms of each factor. In addition to screening approaches as described here, HIPMap will also be useful in numerous other applications. HIPMap is equally well suited to measure distances between gene loci or cellular landmarks, such as nuclear bodies, allowing interrogation of higher order chromatin organization and its relationship to nuclear features. Of particular relevance will be the use of HIPMap in determining the frequencies of Hi-C interactions to uncover how Hi-C signal strength relates to single cell interactions. This will enable to determine variability amongst interactions in individual cells in a population, and the analysis of combinatorial occurrence of multiple mapped interactions in a single cell nucleus (Williamson et al., 2014). Furthermore, HIPMap in combination with immunofluorescence staining will allow detection of FISH signals in a defined sub-population of cells identified by a particular marker, for example to mark cancer stem cells or a particular differentiation stage, thus allowing analysis of subpopulation-specific localization patterns. A combined approach of HIPMap and immunofluorescence may also be used for interrogation of correlations between expression level of a protein and locus positioning. Additionally, DNA FISH may be combined with RNA FISH in HIPMap to assess the effect of positioning on gene expression at the single allele level. We suggest that HIPMap, and related methods, will be useful tools to study the molecular basis of various aspects of genome architecture.
Experimental Procedures
High throughput FISH in 384-well plates
For high-throughput FISH, cells were plated in 384-well CellCarrier plates (Perkin-Elmer) at a concentration of 80 cells/μl (~2000 cells/well). Cells were plated automatically using a Multidrop Combi (Thermo Scientific) or manually. Cells were fixed in 4% PFA in PBS for 15 minutes, permeabilized in 0.5% Saponin (Sigma Aldrich) /0.5% Triton X100/PBS for 20 minutes at RT and incubated in 0.1N HCl for 15 minutes. Cells were kept in 50% formamide/2X SSC for at least 30 minutes at room temperature. A probe mix containing 60ng of fluorescently-labelled probe, 1mg human COT1 DNA (Invitrogen), and 20μg yeast tRNA (Ambion) was ethanol precipitated, and re-suspended in 10μl of hybridization buffer (10% dextran sulphate, 50% formamide, 2X SS, 1% Tween 20). Probe mix was then manually added to each well, denatured together with cells at 85°C for 7 minutes and left to hybridize at 37°C overnight. Excess probe was washed three times with each: 1X SSC and 0.1X SSC at 42°C for 5 minutes using an automated EL406 plate washer (Biotek). Cells were finally stained with DAPI in PBS (5ng/μl) before imaging.
Image acquisition and analysis
Cells were imaged in 384-well plates (Perkin Elmer Cell Carrier) on the Opera QEHS (PerkinElmer, Waltham, MA) confocal high-throughput imaging system using a 40X water objective lens (NA 0.9), 12- bit 1.3 Mb CCD cameras and with camera pixel binning of 2. Image stacks of 6 images at steps of 1.2 μm were acquired. Under these imaging conditions the pixel size was 320nm. At least 63 randomly sampled fields were imaged per well containing a total of >250 cells. All image analysis steps were performed using Acapella 2.0 (PerkinElemer, Waltham, MA). First, images from the same field of view and channel were maximally projected. Then, nuclei were segmented using the DAPI channel. Nucleus border was increased by one pixel to allow proper identification of very peripheral FISH signals (Figure S1A). The resulting Nucleus ROI was used as the search region for the FISH spot detection algorithm in the Alexa488, Alexa 568 and Cy5 channels, respectively. The nucleus ROI was then subdivided in 1-pixel wide equidistant, concentric regions. The normalized radial distance of each Nucleus ROI pixel was then measured by dividing each absolute radial distance value by the per-cell maximum radial distance value. The nucleus border assumes a normalized value of 0, whereas the nucleus center has a normalized value of 1. The normalized absolute radial position of the FISH signal was calculated at the spot center pixel. All acquired single-spot level data were exported as text files and further analyzed by using either MATLAB (R2014b, TheMathworks) or R (http://www.R-project.org/).
Statistical analysis
For the primary screen analysis, empirical cumulative distribution functions (ECDF) for the normalized radial distance of each FISH spot from the border were generated as described for each well in a 384- well plate (Meaburn et al., 2009; Meaburn and Misteli, 2008). Data from all FISH spots from negative control wells (6 wells/plate for primary screen, 12 wells/plate in validation screen) were combined to generate a single ECDF that was used in a pairwise comparisons to each assay well in a plate using the two-sample Kolmogorov-Smirnov (KS) test (Mitchison, 2005; Perlman et al., 2004; Smellie et al., 2006). For validation screen results, at least two out of three replicates had a significant p value. For the analysis of the validation screen results, the distribution histograms, estimated density curves and boxplots were generated using R and the ggplot2 graphics package. Gene ontology analysis was performed using DAVID functional annotation tools.
Supplementary Material
Highlights.
Development of a method for accurate mapping of gene position in the 3D nuclear space at large scale
siRNA screen identifies nuclear factors involved in genome organization
Replication is a critical step in determining spatial gene positioning
Acknowledgments
We thank Karen Meaburn for insightful suggestions for developing hiFISH, members of the Misteli laboratory for helpful discussions and comments, Laurent Ozbun from the NCI HiTIF for technical assistance with the screen and Murali Palangat for help with polymerase II activity assays. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research.
Footnotes
Author contribution
SS, TM designed the study; SS, performed most experiments; NS conducted HU treatment experiments; TCV, GP, SS analyzed the data; TCV, GP developed scripts for image and data analysis; SS, TM wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annual review of genomics and human genetics. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Molecular systems biology. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS biology. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Current biology: CB. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Cvackova Z, Masata M, Stanek D, Fidlerova H, Raska I. Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. Journal of structural biology. 2009;165:107–117. doi: 10.1016/j.jsb.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes & development. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P, Scaffidi P, Markert E, Lee JH, Rane S, Misteli T. Transformation resistance in a premature aging disorder identifies a tumor-protective function of BRD4. Cell reports. 2014;9:248–260. doi: 10.1016/j.celrep.2014.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Gene positioning. Cold Spring Harbor perspectives in biology. 2010;2:a000588. doi: 10.1101/cshperspect.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS genetics. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harbor symposia on quantitative biology. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. European journal of immunology. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- Joyce EF, Williams BR, Xie T, Wu CT. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS genetics. 2012;8:e1002667. doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. The Journal of cell biology. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5831–5836. doi: 10.1073/pnas.0900912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging cell. 2007a;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. The Journal of cell biology. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Locus-specific and activity-independent gene repositioning during early tumorigenesis. The Journal of cell biology. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Seminars in cancer biology. 2007b;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome research. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nature reviews Molecular cell biology. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Small-molecule screening and profiling by using automated microscopy. Chembiochem: a European journal of chemical biology. 2005;6:33–39. doi: 10.1002/cbic.200400272. [DOI] [PubMed] [Google Scholar]
- Nandy K, Gudla PR, Amundsen R, Meaburn KJ, Misteli T, Lockett SJ. Automatic segmentation and supervised learning-based selection of nuclei in cancer tissue images. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2012;81:743–754. doi: 10.1002/cyto.a.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature genetics. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nature genetics. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. In vitro generation of human cells with cancer stem cell properties. Nature cell biology. 2011;13:1051–1061. doi: 10.1038/ncb2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smellie A, Wilson CJ, Ng SC. Visualization and interpretation of high content screening data. Journal of chemical information and modeling. 2006;46:201–207. doi: 10.1021/ci050404g. [DOI] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO journal. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nature reviews Genetics. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Current biology: CB. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus. 2011;2:358–369. doi: 10.4161/nucl.2.5.17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. The Journal of cell biology. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. The EMBO journal. 2013;32:1075–1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Huang D, Gao F, Chang WH, An W, Coetzee GA, Wang K, Lu W. Biological implications and regulatory mechanisms of long-range chromosomal interactions. The Journal of biological chemistry. 2013;288:22369–22377. doi: 10.1074/jbc.R113.485292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. Journal of cell science. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, Paquette D, Dostie J, Bickmore WA. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes & development. 2014;28:2778–2791. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.