SUMMARY
Over thirty years ago, ZTP (5-amino-4-imidazole carboxamide riboside 5'-triphosphate), a modified purine biosynthetic intermediate, was proposed to signal 10-formyl-tetrahydrofolate (10f-THF) deficiency in bacteria. However, the mechanisms by which this putative alarmone or its precursor ZMP (5-aminoimidazole-4-carboxamide ribonucleotide, also known as AICAR) brings about any metabolic changes remain unexplained. Herein we report the existence of a widespread riboswitch class that is most commonly associated with genes related to de novo purine biosynthesis and one carbon metabolism. Biochemical data confirms that members of this riboswitch class selectively bind ZMP and ZTP with nanomolar affinity, while strongly rejecting numerous natural analogs. Indeed, increases in the ZMP/ZTP pool, caused by folate stress in bacterial cells, trigger changes in the expression of a reporter gene fused to representative ZTP riboswitches in vivo. The wide distribution of this riboswitch class suggests that ZMP/ZTP signaling is important for species in numerous bacterial lineages.
INTRODUCTION
Tetrahydrofolate (THF) and its various natural derivatives are essential cofactors involved in one-carbon metabolism (Schirch, 1998) in all cells. Among the biosynthetic pathways that employ folate-derived one-carbon units are those that generate purines, thymidine-5´-monophosphate, methionine, and glycine (Suh et al., 2001). Given the central role of various THF derivatives in these essential anabolic pathways, folate metabolism is often the target of anti-cancer and antibiotic agents (Kohanski et al., 2010; Longo-Sorbello and Bertino, 2001). Among different forms of folate, 10f-THF donates formyl groups to form the C2 and C8 atoms in the purine heterocycle during de novo purine biosynthesis. Given that purine production is a high-flux pathway in rapidly replicating cells, a robust supply of 10f-THF must be available. Therefore, the genes coding for folate biosynthesis and modification are expected to be tightly regulated.
Many bacteria rely on riboswitches (Serganov and Nudler, 2013) to sense and respond to changing levels of THF or its single-carbon derivatives (Ames et al., 2010). Riboswitches are structured noncoding RNA domains most commonly found in the 5´-untranslated regions (UTRs) of bacterial mRNAs where they control gene expression by selectively binding to cognate small molecule or ion ligands. Typically, riboswitches are comprised of a highly conserved ligand-binding aptamer and a poorly conserved expression platform that interfaces with one or more components of the cell’s gene expression systems. Ligand binding in the aptamer domain usually induces conformational changes in the adjoining expression platform, thereby exerting control over expression of the downstream coding sequences.
Given the broad ligand specificity of members of the known THF riboswitch class (Ames et al., 2010; Trausch et al., 2011), these RNAs would not be useful for selectively detecting 10f-THF in the presence of an abundance of other THF derivatives. If cells need to respond specifically to changing levels of 10f-THF, they must employ other more selective RNA- or protein-based sensors for this compound, or they must make use of an indirect method for determining the availability of this important formyl-carrying coenzyme.
More than three decades ago, ZTP (Figure 1A), a 5´-triphosphorylated derivative of the purine biosynthetic intermediate called AICAR (or ZMP), was proposed to act as a bacterial “alarmone” to signal 10f-THF deficiency (Bochner and Ames, 1982). An alarmone is loosely defined as a modified biosynthetic intermediate that signals metabolic imbalance (Stephens et al., 1975). The hypothesis that ZTP signals 10f-THF deficiency was based on the observation that both ZMP and ZTP significantly accumulate during folate depletion in Salmonella typhimurium. However, this hypothesis was called into question by a subsequent study that failed to find evidence for a ZTP-controlled regulon in Escherichia coli (Rohlman and Matthews, 1990). Indeed, there has since been no demonstration of receptors for ZTP or of its ability to influence gene expression.
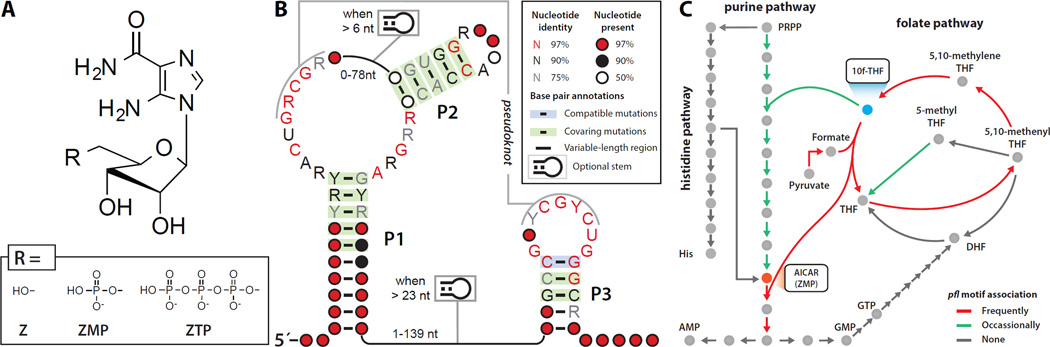
Figure 1. pfl Motif RNAs are Widespread and Commonly Associated with Genes for de novo Purine Biosynthesis and One-carbon Metabolism.
(A) Chemical structures of Z, ZMP, and ZTP.
(B) Consensus sequence and secondary structure model of pfl motif RNAs based on over 2000 representatives from bacteria.
(C) Simplified diagram of pathways relevant to the genes commonly associated with pfl RNAs.
We previously used a bioinformatics pipeline to identify the pfl motif (Figure 1B), which is a candidate riboswitch class that is most commonly associated with genes involved in de novo purine biosynthesis and one-carbon metabolism (Weinberg et al., 2010) (Figure 1C). If experimentally confirmed, the pfl motif would represent one of the most abundant and widespread riboswitch classes in bacteria, and we have speculated that its ligand likely controls fundamental and underappreciated aspects of microbial biology (Breaker, 2011; Meyer et al., 2011). Herein we demonstrate that pfl RNAs function as high-affinity receptors for ZMP and ZTP. Moreover we present evidence that these RNAs function as riboswitches to monitor the Z nucleotide pool and regulate the expression of genes whose protein products are important for maintaining an adequate supply of 10f-THF during de novo purine synthesis. Our findings resolve the mystery regarding the proposed role for ZTP and demonstrate that an off-path biosynthetic derivative can act as an alarmone to signal metabolic insufficiency.
RESULTS
A Conserved RNA Motif Associated with One-carbon Metabolism and Purine Biosynthesis
Several years ago, in an effort to identify riboswitch candidates and other structural regulatory RNA motifs, we identified an RNA motif termed pfl (Weinberg et al., 2010). Since our original discovery, more than 2000 examples of this RNA class (Supplemental Data) have been identified, and an updated consensus sequence and secondary structure model has been generated (Figure 1B). The wide phylogenetic distribution of this highly conserved RNA suggests that the motif is of ancient origin and that the ligand is likely of fundamental importance to a great diversity of organisms (Figure S1A). However, the ligand for this “orphan” riboswitch candidate has remained undiscovered.
The expanded collection of pfl representatives reinforces our original speculation that pfl motif RNAs bear hallmarks indicative of riboswitch function. For instance, the pfl motif has strikingly conserved sequence and structural features despite wide phylogenetic distribution, which is typical of many validated riboswitch classes. The predicted secondary structure based on nucleotide complementarity and on sequence covariation includes three essential base-paired substructures (P1, P2, and P3), and two additional base-paired elements that are present in some representatives. In addition to highly conserved nucleotides that reside within the junction between P1 and P2 (J1/2) and in the loop of P3 (L3), there also exists a highly conserved pseudoknot between J1/2 and L3.
Another hallmark of riboswitches is a strong association with a set of genes related to the cognate ligand of the RNA. Therefore, examination of the genes putatively controlled by candidate riboswitches often provides valuable clues to the identity of the ligand. Members of the pfl motif class of RNAs most commonly precede genes involved in de novo purine biosynthesis or in the synthesis of 10f-THF (Figure 1C, Figure S1B). Specifically, pfl motif RNAs are predominantly associated with genes for the bifunctional enzyme PurH (AICAR formyltransferase/IMP cyclohydrolase), Fhs or Fthfs (formate tetrahydrofolate ligase), Pfl (pyruvate formate lyase), GlyA (glycine hydroxymethyltransferase), and the bifunctional enzyme FolD (5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylenetetrahydrofolate cyclohydrolase).
In addition to gene contexts, the architectures of expression platforms can also help identify the cognate ligand. Analysis of pfl motif RNAs suggests that they are typically genetic ON switches, and therefore a build-up of a certain compound likely triggers the activation of genes controlled by this orphan riboswitch candidate. During de novo purine synthesis, 10f-THF is required for key formylation reactions. Thus, we hypothesized that during 10f-THF deficiency, a build-up of ZMP or its derivative ZTP (Figure 1A) might be sensed by pfl motif RNAs and subsequently trigger expression of downstream genes to maintain adequate level of 10f-THF (Weinberg et al., 2010).
The types of genes associated with pfl motif RNAs are consistent with this hypothesis. For example, 10f-THF can be produced from THF and serine by GlyA and FolD. In anaerobes, formate can be produced from pyruvate by Pfl (Becker et al., 1999) or transported by formate transporters, such as FocA (Sawers, 2005). 10f-THF is subsequently produced from THF and formate by Fhs. Indeed, it has been reported that, in Streptococcus thermophilus, Pfl is highly expressed during simultaneous starvation for formate and purines, but repressed when either formate or purines are provided (Derzelle et al., 2005).
ZMP could also accumulate as a result of inadequate production of PurH, which is commonly found to be associated with pfl motif RNAs. Notably, genes in de novo folate biosynthesis are not associated with pfl motif RNAs, which suggests that these RNAs are more likely to be involved in maintaining an adequate 10f-THF level and folate pool balance rather than maintaining the global amount of THF and its various derivatives. This distinction is further supported by the existence of a separate riboswitch class that senses THF and controls genes directly involved in folate biosynthesis (Ames et al., 2010).
Representative pfl Motif RNAs Bind the Purine Biosynthetic Intermediate ZMP
Previous efforts to identify the ligand for the pfl riboswitch candidate were carried out using a 92-nt pfl RNA construct (92 Cac) derived from the 5´-UTR of the purH gene from Clostridium acetobutylicum (Weinberg et al., 2010) (Figure S2A). We used a method called in-line probing to map and quantify any ligand-induced structural changes in the putative riboswitch aptamer by monitoring spontaneous RNA cleavage patterns (Soukup and Breaker, 1999). Using this method, many compounds related to purine and folate metabolic pathways were previously tested, including ZMP, without any indication of ligand-induced RNA structure modulation.
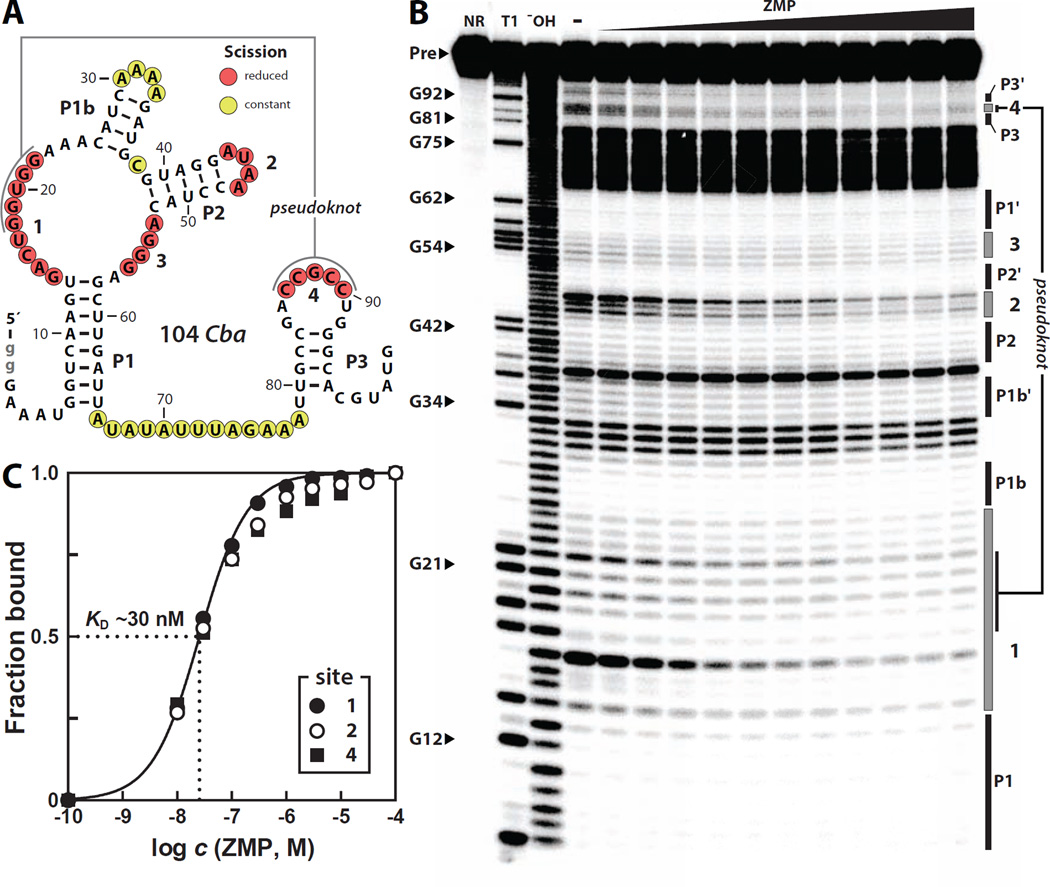
Upon re-examining the 92 Cac construct, however, we noticed that the P1 and P2 stems of this RNA do not properly base-pair under our assay conditions (Figure S2B). We hypothesized that the previous negative results might have been due to the failure of this particular RNA construct to properly fold to form the binding pocket required to recognize its cognate ligand. To avoid this possible problem, we synthesized and tested several additional pfl motif RNA constructs, including a 104-nucleotide RNA (104 Cba) that encompasses the pfl motif from Clostridium bartlettii (Figure 2A). Unlike 92 Cac, 104 Cba exhibits a pattern of spontaneous RNA phosphodiester cleavage that is consistent with the secondary structure model. Specifically, all the internucleotide linkages residing within the predicted stems exhibit strong resistance to spontaneous cleavage, whereas most regions predicted to be unpaired exhibit robust spontaneous cleavage (Figure 2B).
Figure 2. Recognition of ZMP by a pfl Motif RNA.
(A) Sequence and secondary structure model for a pfl RNA called 104 Cba from Clostridium bartlettii. Lowercase nucleotides in gray were added to facilitate transcription. Nucleotides undergoing structural stabilization upon the addition of ZMP are annotated with red circles while positions remaining unstructured are annotated with yellow circles. Data are derived from the inline probing results in B.
(B) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of 5´ 32P-labeled 104 Cba with various concentrations of ZMP. NR, T1, and −OH designate no reaction, partial digestion with RNase T1, and partial digestion with alkali, respectively.
(C) Plot of the fraction of RNA bound to ligand, as inferred from the normalized fraction of spontaneous RNA cleavage versus the logarithm of the concentration of ZMP. The most robust regions of modulation (sties 1, 2 and 4) were used to create the plot. The solid line represents a theoretical binding curve expected for a one-to-one interaction between ligand and RNA with a KD of 30 nM.
Upon addition of ZMP, the most highly conserved nucleotides undergo structural stabilization (Figure 2B), including those that form the predicted pseudoknot (Figure 1B). Ligand-induced structural changes in phylogenetically conserved regions are typical of many known riboswitches. Therefore our results suggest that many of the conserved nucleotides that undergo ZMP-mediated structural stabilization are critical for forming the molecular architecture needed for ZMP binding. Representatives of this RNA class from other phyla, including 118 Har from the β-proteobacterium Hermininiimonas arsenicoxydans (Figure S3) and 92 Ske from the Actinobacterium Sanguibacter keddieii (Figure S4), also exhibit proper folding of all the predicted secondary structures under in-line probing conditions and undergo structural modulation in the presence of ZMP. The apparent dissociation constant (KD) of 104 Cba RNA and ZMP is estimated to be ~30 nM by quantifying the extent of spontaneous cleavage at several nucleotide positions over a range of ZMP concentrations (Figure 2C).
Using 104 Cba as a parent construct, we examined the importance of some of the highly conserved features of the aptamer to provide further support our consensus model. For example, mutation of a single highly conserved nucleotide (M1) (Figure 3A) results in the loss of the normal ZMP binding pattern (Figure 3B). A mutation that disrupts base-pairing in the P1 stem (M2) also results in a loss of binding activity, which can be reversed through a compensatory mutation (M3) that restores base pairing within P1. An additional series of mutational studies using 118 Har also provided similar results (Figure S5). Taken together, our results demonstrate that pfl motif RNAs are high-affinity aptamers for ZMP and that ligand binding stabilizes highly-conserved portions of this widespread riboswitch candidate.
Figure 3. In-line Probing Analysis of Aptamer Mutants.
(A) Sequence and secondary structure of wild-type (WT) and mutant 104 Cba RNA constructs used to evaluate the importance of highly conserved nucleotides and structural elements. Mutant constructs (M1–M3) carry nucleotide changes as indicated. Other annotations are as described in the legend to Figure 2A.
(B) PAGE analysis of in-line probing reactions with WT 104 Cba RNA or mutants M1, M2, and M3. Each 5´ 32P-labeled RNA construct was incubated in the absence or presence of 10 µM ZMP. Other annotations are as described in the legend to Figure 2B.
Molecular Recognition Characteristics of ZMP-binding RNAs
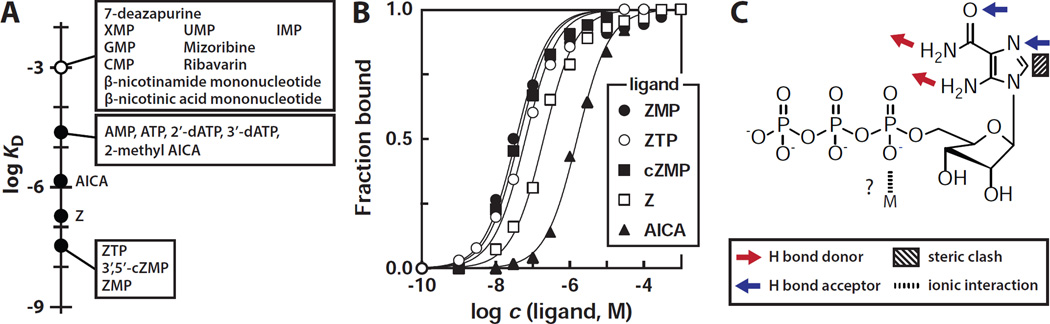
The molecular recognition characteristics of 104 Cba were further assessed by establishing the apparent KD values for various analogs that carry certain structural features in common with ZMP (Figure 4A, Figure S6). The nucleotide AMP (adenosine-5´-monophosphate) is bound by the RNA with an affinity that is about 1000-fold poorer than for ZMP. Similarly, the affinities for other natural nucleotide derivatives such as IMP (inosine-5´-monophosphate), GMP (guanosine-5´-monophosphate), and XMP (xanthosine-5´-monophosphate) are all at least 30,000-fold poorer. Therefore ZMP aptamers are likely to selectively bind their cognate ligand even in the presence of a large excess of natural nucleotide derivatives. Furthermore, the importance of the ribosyl group in ligand recognition is apparent because 5-amino-4-imidazolecarboxamide (AICA, or Z nucleobase), which lacks the ribosyl group entirely, is bound by 104 Cba approximately 50-fold poorer (KD ~1.5 µM) than ZMP (Figure 4B). Similar results are observed with 118 Har (Figure S4D).
Figure 4. Molecular Recognition Characteristics of a ZMP/ZTP Aptamer.
(A) Summary of the apparent KD values for ZMP and various ZMP analogs with 104 Cba as established by in-line probing (Figure 2, Figure S7 and data not shown). The open circle indicates that no evidence for binding by the compounds indicated was observed at 1 mM.
(B) Plot of fraction of 104 Cba RNA bound to ligand versus the logarithm of the concentration of ZMP and its analogs. The lines represent theoretical binding curves expected for a one-to-one interaction between ligand and RNA.
(C) Schematic representation of the molecular recognition determinants of ZMP/ZTP binding by 104 Cba RNA. Predicted molecular recognition contacts are as indicated.
Binding affinities for various AICA analogs (Figure S6) were also established to determine important functional groups within the ligand, leading to the identification of key molecular features recognized by the 104 Cba aptamer (Figure 4C). The presence of a methyl group at the C2 position of the AICA decreases affinity by 50 fold, likely due to steric clash between the ligand and aptamer. The amino group of the carboxamide moiety of the ligand is likely to come into contact with the aptamer because methylation at this position causes a complete loss of binding affinity. Similarly, methylation at the N2 position reduces affinity by 1000 fold or more (KD >1 mM), suggesting that this chemical change also results in steric hindrance and/or disruption of hydrogen bond interactions with the aptamer.
1H-imidazole-4-carboxamide, which lacks the 5-amino group of AICA, also exhibits a loss of at least three orders of magnitude in affinity compared to AICA. Ribavirin, a ZMP analog that is a nucleoside inhibitor used to treat viral infections (Cameron and Castro, 2001), also lacks the 5-amino group and exhibits a decrease of at least five orders of magnitude in affinity. These data suggest that the 5-amino group is a critical determinant of ligand recognition for the aptamer.
Analogs with various 5´-phosphorylation states were evaluated to determine whether the aptamer could discriminate among derivatives with differing numbers of phosphate groups. Interestingly, both 104 Cba and 118 Har do not discriminate between ZMP and ZTP (Figure 4B, Figure S3D), indicating that the 5´-polyphosphate group is unlikely to serve as a critical molecular recognition determinant. Structural studies of TPP and FMN riboswitches have revealed that pyrophosphate and phosphate groups, respectively, form direct and Mg2+-mediated contacts with their riboswitch RNA receptors to increase ligand-binding affinity and specificity (Serganov and Patel, 2012). Given these precedents, it is somewhat surprising that similar contacts are unlikely to be exploited by this RNA class to discriminate among various Z riboside phosphorylation states. Regardless, pfl motif RNAs robustly bind to ZTP, demonstrating that this orphan riboswitch class could represent a widespread receptor and gene control element for this proposed alarmone.
Although Z riboside and 3', 5'-cZMP are bound strongly by 104 Cba, neither compound is known to be synthesized by cells, and therefore the high affinity exhibited by these compounds is likely to be irrelevant in vivo. If ZTP were to function as the exclusive alarmone for signaling 10f-THF deficiency, it might be expected that a riboswitch for this signal would more strongly exclude the ZMP molecules that directly accumulate during 10f-THF deficiency. ZTP production could occur if excess ZMP is pyrophosphorylated by the promiscuous action of the enzyme PRPP synthetase (Sabina et al., 1984) or by a dedicated unidentified enzyme. However, given the lack of discrimination between ZMP and ZTP, members of this RNA class might sense and respond to the combined ZMP/ZTP pool, rather than respond exclusively to ZTP.
Riboswitch-mediated Control of Gene Expression by ZMP and ZTP
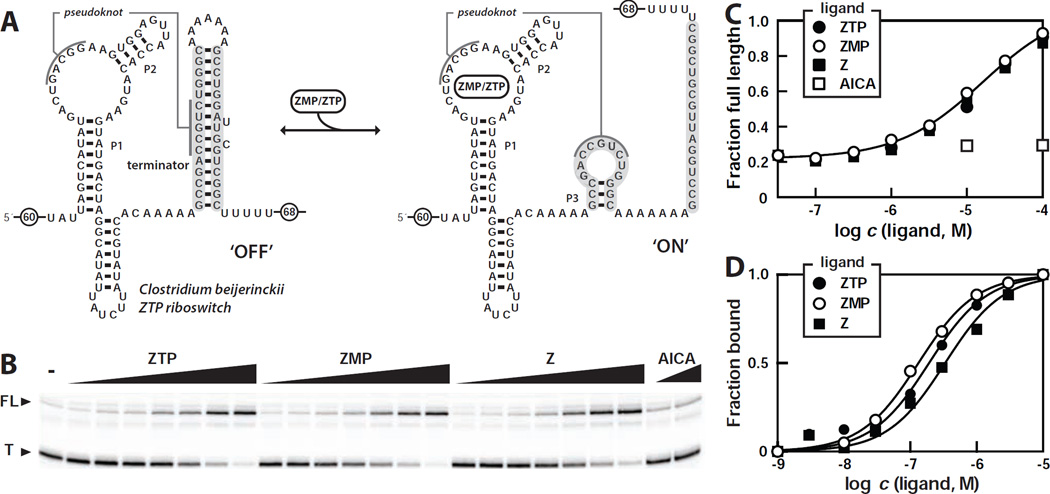
To determine whether members of this RNA class regulate gene expression in response to changing levels of ZMP or ZTP, we examined the transcription of a DNA template encompassing the sequence of a Clostridium beijerinckii pfl motif RNA with its natural terminator stem (Figure 5A). In single-round transcription assays lacking ligand (Figure 5B), the majority of RNAs generated are terminated within the run of U residues that are characteristic of intrinsic transcription terminator stems (Gusarov and Nudler, 1999; Yarnell and Roberts, 1999). Upon addition of increasing amounts of ZMP, the yield of full-length RNA transcripts increases while the amount of terminated transcripts decreases (Figure 5B, 5C). ZTP and Z induce a similar effect on the products of transcription compared to ZMP, whereas AICA fails to trigger a change in the yields of the two major transcription products.
Figure 5. The Nucleoside Z and the Nucleotides ZMP and ZTP Trigger Riboswitch-mediated Transcription Elongation.
(A) Sequence and secondary structure of a full-length ZTP riboswitch construct from C. beijerinckii used for transcription termination assays. In the absence of ligand (left), an intrinsic terminator stem (participating nucleotides shaded in gray) is expected to form and cause transcription termination. In the ligand-bound state (right), both P3 and the pseudoknot are expected to form, thereby blocking formation of the terminator stem and leading to transcription read-through. Encircled numbers indicate the presence of additional nucleotides from the natural sequence.
(B) Single-round in vitro transcription termination assays conducted in the absence of ligand (−) or in the presence of ZMP, ZTP, and Z ranging from 100 nM to 100 µM. AICA was tested at concentrations of 30 µM and 100 µM. T and FL denote terminated and full-length RNA transcripts, respectively.
(C) Plot of the fraction of full-length RNA product versus the logarithm of the concentration of ligands. Data are derived from B.
(D) Plot of fraction of RNA bound to ligand versus the logarithm of the concentration of ZMP, ZTP, or Z, as estimated from in-line probing assays with the riboswitch aptamer from C. bejerinckii (nucleotides 1 through 99 as depicted in A). The solid lines represent theoretical binding curves expected for one-to-one interactions between ligand and RNA.
These findings are consistent with our hypothesis that many ZMP/ZTP aptamers are riboswitches that activate gene expression upon ligand binding. Moreover, these results demonstrate that the RNA is not capable of distinguishing between ZTP, ZMP, and Z during in vitro transcription (Figure 5B, 5C), which matches our binding assay results with the aptamer from this same species as determined by in-line probing (Figure 5D). However, there is a ~50-fold shift in the dose-response curve between the KD values derived by in-line probing and the concentrations of ligand needed to trigger half-maximal transcription read-through. These results suggest that the RNA is operating as a kinetically-driven riboswitch, as has been observed for representatives of other riboswitch classes (Wickiser et al., 2005a; Wickiser et al., 2005b).
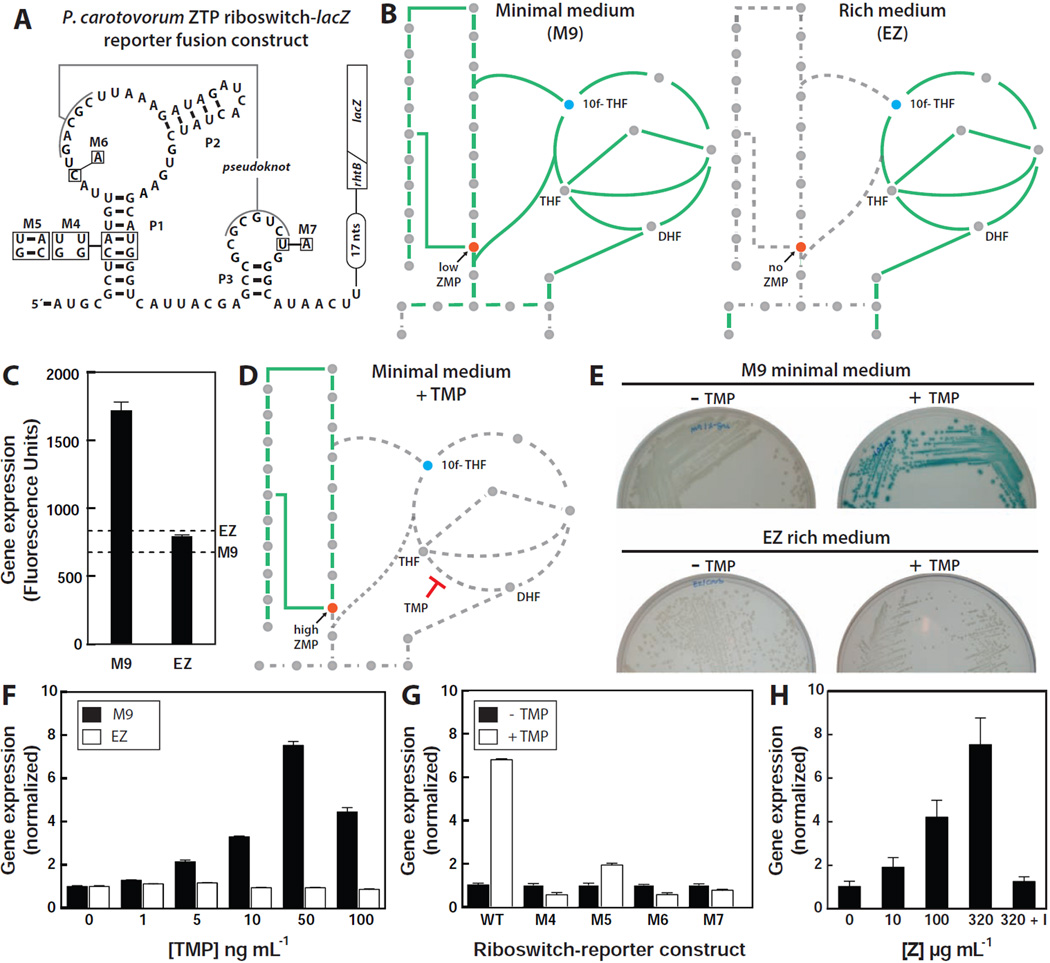
Gene control in vivo was evaluated by transforming E. coli BW25113 with a plasmid containing a translational fusion between a ZTP riboswitch from the γ-proteobacterium Pectobacterium carotovorum and a lacZ reporter gene (Figure 6A). During growth in minimal medium, the de novo purine biosynthetic pathway is used to produce purines, and therefore a basal level of the biosynthetic intermediate ZMP is expected to be present (Figure 6B, left). However, during growth in rich medium, the purine salvage pathway is preferentially utilized. Under these conditions, ZMP is expected to be absent, or present at an exceedingly low concentration (Figure 6B, right). The reporter strain was grown either in minimal or rich medium, and the levels of β-galactosidase activity were determined by using a fluorescence-based assay (Figure 6C). As expected, the riboswitch-reporter strain grown in M9 minimal medium exhibits modest expression, as fluorescence values are about 2-fold above background. By contrast, there is no measurable fluorescence above background from cells grown in EZ rich medium.
Figure 6. ZMP/ZTP-induced Reporter Gene Expression Controlled by a Riboswitch Associated with Pectobacterium carotovorum rhtB.
(A) Sequence and secondary structure of the WT ZTP riboswitch associated with the rhtB gene of P. carotovorum and various mutants fused to a β-galactosidase reporter gene (lacZ). The diagonal line indicates the fusion of the first eight codons of rhtB to the ninth codon of lacZ. Other annotations are as described for Figure 3A.
(B) Schematic representation of the expected metabolic flux in minimal (left) and rich (right) medium. Solid green lines indicate pathways with flux whereas dashed gray line represents pathway with low or no flux.
(C) Plot of reporter gene activity for E. coli cells containing the riboswitch reporter construct and grown on minimal (M9) or rich (EZ) medium supplemented with X-gal (See Extended Experimental Procedures). Dashed lines represent the background measurements for M9 and EZ media in the absence of cells.
(D) Schematic representation of the expected metabolic flux in minimal medium containing trimethoprim (TMP). Other annotations are as described for B.
(E) Agar plate assays with E. coli cells containing the riboswitch reporter construct and grown under the conditions as indicated in media supplemented with X-gal.
(F) Plot of the relative reporter gene expression levels with increasing concentrations of TMP relative to no treatment either in minimal (M9) or rich (EZ) medium.
(G) Plot of the relative β-galactosidase activity in cells containing the WT or mutant riboswitch reporter constructs depicted in A in the absence or presence of 0.1 µg mL−1 TMP, as indicated, in minimal medium.
(H) Plot of the relative β-galactosidase activity of cells carrying the WT riboswitch reporter construct upon the addition of increasing concentrations of Z riboside in rich medium. “I” indicates addition of 1 mM 5´-amino-5´-deoxyadenosine, a selective inhibitor of adenosine kinase.
Many antifolates target de novo folate synthesis, which should lead to 10f-THF depletion and the subsequent build-up of ZMP and ZTP (Figure 6D). Indeed, several research groups have reported that treatment with antifolates results in rapid and dramatic ZMP (and in some cases ZTP) accumulation (Bochner and Ames, 1982; Chakraborty et al., 2013; Kwon et al., 2010). Therefore, we speculated that treatment with trimethoprim (TMP), a dihydrofolate reductase (DHFR) inhibitor (Dauber-Osguthorpe et al., 1988), would result in an increase in reporter gene expression when cells are grown in minimal medium. As expected, TMP treatment in minimal (but not rich) medium results in a high reporter gene expression (Figure 6E, Figure S7A), and the level of expression increases in a concentration-dependent manner (Figure 6F). Transformed strains carrying mutant riboswitch-reporter constructs that either disrupt the P1 stem (M4) (Figure 6A) or alter highly conserved nucleotides (M6 and M7) fail to exhibit detectable expression upon TMP treatment (Figure 6G, Figure S7B). In contrast, restoring base pairing within the P1 stem (M5) partially restores riboswitch function.
As noted above, ZMP should not be produced when cells are grown in rich medium, and the enzyme that utilizes ZMP to build purines (PurH) is not expected to be expressed (Figure 6B, right). Therefore any ZMP artificially introduced into cells should accumulate. Exogenous supplementation of Z riboside is expected to artificially elevate the concentration of ZMP in cells when grown in rich medium due to its uptake by adenosine transporters and subsequent phosphorylation by adenosine kinase (Fryer et al., 2002; Sabina et al., 1985). Indeed, exogenous addition of Z strongly induces the expression of β-galactosidase in cells carrying the riboswitch-reporter construct in a concentration-dependent manner (Figure 6H). However, when the reporter strain is grown with Z and a selective inhibitor of adenosine kinase (5´-amino-5´-deoxyadenosine) that precludes intracellular conversion of Z to ZMP (Wiesner et al., 1999), we observed only a low level of β-galactosidase expression. This latter result is somewhat surprising given that our in vitro binding and transcription termination data indicate that Z can trigger the riboswitch as well as ZMP or ZTP. However, if nucleosides are not phosphorylated to add a negative charge to the molecule, they cannot be efficiently retained within bacteria (Deutscher et al., 2006). Furthermore, non-phosphorylated ribonucleosides might also be degraded to yield other metabolites. These possible drains on the cellular concentration of Z might explain the low reporter gene expression observed.
Notably, the addition of Z to E. coli transformed with WT and with various mutant riboswitch reporter constructs (Figure S7C, Figures S7D) yields reporter gene expression levels similar to that observed upon the addition of TMP (Figure 6G). Therefore, the manipulation of ZMP concentrations by either Z supplementation or antibiotic treatment yields the same riboswitch responses, which is consistent with our hypothesis that pfl RNAs are riboswitches for ZMP and/or ZTP.
Finally, we measured β-galactosidase activities from the riboswitch reporter constructs in purH knockout (KO) E. coli cells. During growth in minimal medium, the purH KO strain is expected to accumulate ZMP since PurH is responsible for the conversion of ZMP to IMP. However, bacteria deficient in purH are unable to synthesize purines and therefore are unable to grow under strict minimal medium conditions (Figure S7E) (Jenkins et al., 2011). Therefore, we grew WT and purH KO E. coli strains in rich media before transferring and incubating them in minimal media overnight. Importantly, increased expression was observed in the purH KO strain relative to the WT strain (Figure S7F). Similarly, examination of Bacillus subtilis WT and purH KO strains harboring the Bacillus sp. SG-1 ZTP riboswitch reporter construct displayed an increased reporter gene expression in the purH KO strain compared to WT after growth for 24 hours in rich medium (Figure S7F). These results are again consistent with our hypothesis that ZTP riboswitch-mediated activation of gene expression occurs as a result of Z nucleotide accumulation.
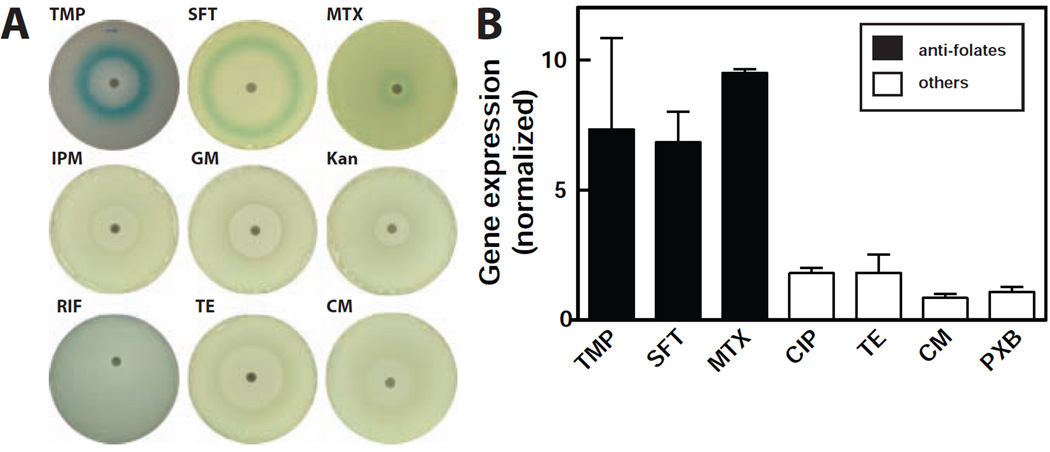
Given the sensitivity of ZTP riboswitches to the antifolate compound TMP, we sought to demonstrate that this effect is not simply due to general cellular distress that could be brought about by antibiotics that operate via other mechanisms. Using agar-diffusion (Figure 7A) and liquid-based (Figure 7B) assays, we treated the E. coli riboswitch reporter strain with other antifolates such as sulfathiazole and methotrexate. All three antifolate compounds trigger riboswitch-mediated gene expression as demonstrated by the formation of blue halos (Figure 7A). Laboratory strains of E. coli are strongly resistant to methotrexate, and therefore methotrexate did not prevent bacterial growth under our assay conditions (Kopytek et al., 2000). However, the compound is expected to retain its ability to block its DHFR target because resistance in E. coli is due to a drug pump mechanism rather than the presence of drug-resistant DHFR. Curiously, methotrexate fails to yield reporter gene expression in cells nearest the filter disk, which should contain the highest concentration of drug. One possible explanation is that these cells might undergo additional metabolic alterations that either reduce ZTP concentration or otherwise suppress reporter gene expression.
Figure 7. Antibiotic Agents that Target Folate Biosynthesis Trigger ZTP Riboswitch-mediated Gene Expression.
(A) Agar diffusion assays of the E. coli WT riboswitch reporter strain described in Figure 6A. Blank discs were spotted with 5 µL of either 1 µg mL−1 TMP, 400 µg mL−1 sulfathiazole (SFT), 45 mg mL−1 methotrexate (MTX), 4 µg mL−1 kanamycin (Kan) or 5 µg mL−1 chloramphenicol (CM), Disks infused with gentamycin (GE), rifamycin (RIF), tetracycline (TE), and imipenem (IPM) were purchased from BD. All plates contained M9 agar medium with X-gal.
(B) Plots of the relative β-galactosidase activity of the E. coli WT riboswitch reporter strain grown with various antibiotics as indicated. Concentrations tested are approximately one half log unit below the minimum inhibitory concentration (MIC) for each antibiotic, with the exception of methotrexate, which was tested at 100 µg mL−1
In solution (Figure 7B), TMP and sulfathiazole were tested at a concentration just below their MIC, where folate distress should be strong but not lethal. Again, methotrexate was tested at increasing concentrations only up to a maximum of 100 µg mL−1 due to solubility concerns. Although methotrexate at this concentration does not inhibit growth, robust reporter gene expression is observed.
By contrast, the Z nucleotide pool is not expected to increase in cells exposed to antibiotic classes that target pathways other than folate biosynthesis. Indeed, treatment with other classes of antibiotics had no effect on reporter gene expression. These observations suggest that ZTP riboswitches could be exploited to establish the mechanisms of novel compounds that disrupt folate metabolism and influence the Z nucleotide pool.
DISCUSSION
Identifying the ligands for novel candidate riboswitch classes is likely to become increasingly difficult for various reasons. For example, some riboswitch candidates are associated with genes whose proteins they encode have unknown functions, and therefore no clues regarding ligand identity can be derived from gene associations. Some newfound riboswitch candidates are very rare, and these might respond to ligands that are not widespread in biology. Rare riboswitch classes also can yield imperfect sequence and structural consensus models, and therefore constructs chosen for testing might miss key structural features or carry accessory sequences that interfere with aptamer function. Moreover, experimental validation of common riboswitch candidates can also be frustrated by constructs that misfold. For example, our earlier efforts to observe reporter gene regulation with a pfl motif construct upon TMP manipulation of folate levels were not successful (Meyer et al., 2011). Since the pfl RNA for this previous reporter construct was derived from the same C. acetobutylicum RNA that was used to generate the misfolded 92 Cac, and the host bacterium was B. subtilis, the riboswitch might not have been able to fold properly in its surrogate cell. To overcome such riboswitch validation problems, ligand candidates should be tested with multiple different constructs using various biochemical and genetic analyses.
We previously noted that the cognate ligands for the most abundant and widespread riboswitch classes are likely to control fundamental and perhaps underappreciated aspects of biology (Breaker, 2011; Meyer et al., 2011; Nelson et al., 2013). To date, ZTP riboswitches rank as the 14th most common class among ~35 validated classes. Other riboswitch classes that are similar in abundance respond to fundamental ligands such as coenzymes, amino acids and common bacterial signaling compounds. Indeed, ZMP has long been known as a de novo purine biosynthesis intermediate, but its role in gene regulation has remained largely unknown. Intriguingly ZTP, the pyrophosphorylated derivative of ZMP, was discovered more than 30 years ago and its possible importance in signaling 10f-THF deficiency was proposed (Bochner and Ames, 1982). However, no mechanism for this regulation had been described.
Our findings demonstrate that the pfl motif RNAs represent a widespread class of receptors for the Z nucleotide pool. Members of this riboswitch class directly and selectively bind to ZMP and ZTP, and are capable of strongly discriminating against other nucleotides and nucleotide derivatives. We also provide in vitro and in vivo evidence for riboswitch-mediated gene expression via ZMP or ZTP binding. In addition to earlier reports (Bochner and Ames, 1982; Rohlman and Matthews, 1990), more recent studies using metabolomics approach have clearly established that ZMP rapidly accumulates upon treatment with antifolates (Chakraborty et al., 2013; Kwon et al., 2010; Wei et al., 2011). Our riboswitch-reporter constructs yield gene expression results that are consistent with these reports, and also demonstrate that the accumulation of ZMP directly activates the expression of genes that counter 10f-THF insufficiency. It is quite common for some species to lack a member of a particular riboswitch class for a ligand, but instead sense the ligand with a member of another riboswitch class (e.g., SAM, preQ1, c-di-GMP) or with a protein factor (Breaker, 2011). Although organisms such as E. coli lack representatives of this ZTP riboswitch class, it is possible that ZTP signaling regulates the same biological processes via other receptors that have yet to be discovered.
ZTP riboswitch binding data suggests that the 5´ phosphate group of the ribosyl moiety is not critical for ligand recognition. Perhaps there has been no selective pressure for cells to discriminate between ZMP and ZTP, since both compounds are elevated in concentration when cells are deficient in 10f-THF. Moreover, chemically-similar phosphorylated nucleotides, such as AMP and ATP, are present at high concentrations in cells under normal conditions. Therefore, a riboswitch class that cannot easily discriminate against ZMP and ZTP because it lacks molecular recognition contacts with phosphates might be more capable of avoiding unwanted activation by other phosphorylated nucleotides that are more abundant.
Our results suggest that ZMP and ZTP as a pool can trigger changes in gene expression in response to 10f-THF deficiency. Given that this riboswitch class cannot distinguish between ZMP and ZTP, our findings do not resolve the question of whether ZTP is being made as a dedicated alarmone. It is possible that, in some organisms, the ability of the riboswitch to sense ZMP and ZTP with equal affinity allows the cell to tolerate ZTP production as the unavoidable action of a promiscuous enzyme, but still account for both ZMP and ZTP when indirectly monitoring for 10f-THF deficiency. However, we cannot rule out the possibility that there exist ZTP-specific receptors that might enable responses that are exclusive to this alarmone.
The majority of genes regulated by ZTP riboswitches are directly involved in either de novo purine biosynthesis or in the generation of one-carbon units associated with folate coenzymes. However, there are a number of other genes controlled by this riboswitch class (Supplemental Data). We speculate that most of these other genes are also related to de novo purine and 10f-THF biosynthesis. For example, in Bacillales, some ZTP riboswitches are found to control rpiB, which encodes ribulose-5-phosphate isomerase. RpiB may help harness the end-product of the oxidative phase of the pentose phosphate pathway to generate PRPP, which is the starting product for de novo purine biosynthesis. In three instances where the ZTP riboswitch is associated with rpiB, the expression platform architecture appears consistent with a genetic ‘OFF’ switch, wherein a strong intrinsic transcription terminator stem can form apart from the ZTP aptamer domain (data not shown). In these instances, ZTP accumulation presumably reduces expression of a gene that otherwise would unnecessarily promote the production of more purines. In several species of Proteobacteria, ZTP riboswitches are found to control the putative threonine transport gene, rhtB. In these organisms, threonine likely enters the one-carbon metabolism pathway via glycine (Jungermann et al., 1970; Locasale, 2013).
Other genes associated with ZTP riboswitches are not so easily linked to purine or folate biosynthesis. In Burkholderiales, there are a number of instances where ZTP riboswitches control the glyA-nrdR transcriptional unit. NrdR is annotated as a negative regulator of ribonucleotide reductase (rnr) (Borovok et al., 2004; Rodionov and Gelfand, 2005), which converts RNA monomers into DNA monomers. If the annotations for these nrdR genes are correct, then this suggests a possible role for ZTP in the regulation of 2´-deoxyribonucleotide monomer production for the synthesis of DNA.
Intriguingly, both ZMP and 5´-phosphoribosyl-4-succinocarboxamide-5-aminoimidazole ribonucleotide (SAICAR, the precursor to ZMP) have been proposed to upregulate de novo purine biosynthesis in Saccharomyces cerevisiae (Rebora et al., 2005). In yeast, the interaction of two transcription factors Bas1p and Pho2p are required to promote expression of de novo purine biosynthetic genes (Daignan-Fornier and Fink, 1992). Based on genetic evidence, it was proposed that this interaction is promoted by SAICAR and ZMP. It has also been suggested that, in S. cerevisiae, ZMP controls phosphate metabolism (Pinson et al., 2009). Although we do not see an association between ZTP riboswitches and genes involved in phosphate metabolism in bacteria, it is possible that the regulatory networks have differentiated through evolution (Babu et al., 2004).
Z riboside-mediated cellular physiology has been studied in depth in mammalian cells. It is known that Z riboside is phosphorylated to ZMP by adenosine kinase and acts an agonist of AMPK, a master regulator of metabolism (Sullivan et al., 1994). Its administration to eukaryotes has shown promising results in treating cardiovascular diseases, malignant hyperthermia, and leukemia (Daignan-Fornier and Pinson, 2012; Kuznetsov et al., 2011; Lanner et al., 2012). Z riboside has also been shown to increase endurance in sedentary mice (Narkar et al., 2008), which has in turn lead to its use as a performance-enhancing drug among professional athletes. ZMP- or ZTP-mediated folate pool balance may also occur in eukaryotes, including in humans and other mammals. Despite the importance of ZMP and possibly ZTP in eukaryotes, we have not observed any examples of ZTP riboswitches in this domain of life. Perhaps other classes of RNA or protein factors are involved in Z-dependent regulation in eukaryotes.
Riboswitches that regulate genes in biosynthetic pathways typically sense an end-product and control genes by feedback control (Breaker, 2011). However, monitoring a biosynthetic intermediate at a key node could also relay rich information regarding metabolic status of a cell. In that regard, ZMP seems to be an ideal metabolite to be monitored, as it lies at the node of three interconnected biochemical pathways for purines, folates and histidine. Interestingly, the ykkC RNA motif, another widespread and long-standing orphan riboswitch, is commonly associated with de novo purine biosynthesis and nitrogen metabolism (Barrick et al., 2004; Meyer et al., 2011). Although its ligand remains unidentified, our current study highlights the possibility that perhaps ykkC motif RNAs bind to yet another biosynthetic intermediate at a critical node.
The widespread phylogenetic distribution of this riboswitch class suggests an ancient origin, and perhaps the original members of this motif existed in the RNA World (Breaker, 2009, 2012). Intriguingly, ZTP riboswitches join a growing list of ligand-binding RNA switches that sense and respond to compounds such as coenzymes, nucleosides and nucleoside derivatives, and other fundamental metabolites that have been proposed to have originated in the RNA World (White, 1976; Woese, 1967). Each of the three pathways closest to ZMP and ZTP production are likely to have arisen before the emergence of proteins. Specifically, purine biosynthesis would have been necessary as RNA World entities evolved beyond their most primitive states. Folate compounds originate from the purine nucleotide GTP, and the THF derivative 10f-THF has become an essential coenzyme for purine biosynthesis. Also, histidine is the only amino acid whose synthesis begins with a nucleotide (ATP), and the biosynthetic pathways for both these compounds generate ZMP.
Additionally, ZTP riboswitches join a list of riboswitches that recognize RNA-derived signaling molecules, such as c-di-GMP (Lee et al., 2010; Sudarsan et al., 2008), c-di-AMP (Nelson et al., 2013), and c-AMP-GMP (Nelson et al., submitted; Kellenberger et al., submitted). These findings suggest that regulatory systems comprised exclusively of RNA polymers and small RNA-derived signals likely existed in the RNA World, and demonstrate that they persist in modern cells amongst the more typical protein-mediated signaling systems. This makes the apparent lack of riboswitches for other RNA-derived signaling molecules, such as cAMP, cGMP, and (p)ppGpp all the more puzzling.
Given that modern riboswitch classes that recognize THF and ZMP/ZTP exist, it seems reasonable to speculate that a ribozyme or ribozyme complex once existed that carried both 10f-THF and ZMP binding sites. This architecture could have been exploited to catalyze a one-carbon transfer reaction as part of the purine biosynthetic pathway. Interestingly, in modern cells, there are a number of examples where THF and ZTP aptamers are found in a tandem arrangement with independent expression platforms (Ames et al., 2010), and such arrangements with other riboswitches are known to act as two-input logic gates (Sudarsan et al., 2006). Although these modern RNAs are almost certainly present to integrate regulation of the purine and THF biosynthetic pathways, their existence provides a precedent for RNAs that bind a folate derivative and ZMP in the same RNA polynucleotide.
Regardless of the origin of ZTP riboswitches, constructs based on this riboswitch class might be useful for drug discovery. Novel antibiotics classes are greatly needed even though the current commercial landscape is not conducive to drug discovery in this area (Payne et al., 2007). While target-based screening has made advances in modern drug discovery, some drug developers have noted challenges associated with this approach compared to phenotypic-based screening (Swinney and Anthony, 2011). Moreover, there is renewed interest in phenotypic screening among the scientific community (Zheng et al., 2013). However, one of the main challenges of phenotypic screening is target identification. Our result suggests that ZTP riboswitches fused to reporter genes could be exploited for phenotypic screens wherein compounds that target folate biosynthesis by novel mechanisms could be discovered.
EXPERIMENTAL PROCEDURES
Procedures used for in vitro transcription assays, preparation of RNA oligonucleotides, bacterial strains and growth conditions, design of reporter gene constructs, liquid-based β-galactosidase assays, and RNA homology search are described in the Extended Experimental Procedures.
In-line Probing
In-line probing experiments were conducted as previously described (Soukup and Breaker, 1999). Briefly, a trace amount (~5 nM) of 5´ 32P-labeled RNA was incubated at 25°C for approximately 40 hours in the presence of 100 mM KCl, 50 mM Tris-HCl (pH 8.3 at 23°C), and 20 mM MgCl2 and the desired ligand. RNA spontaneous cleavage products were resolved by denaturing 10% PAGE and imaged with a PhosphorImager (Molecular Dynamics). ImageQuant 5.1 was used to establish band intensities. Values for the apparent dissociation constants (KD) were determined as previously described (Baker et al., 2012).
Single-round in vitro Transcription Termination Assays
A DNA template for in vitro transcription, consisting of the native promoter, predicted riboswitch aptamer and terminator stem and corresponding to coordinates NC_009617.1/1211871-1212120, was constructed via PCR of Clostridium beijerinckii spores. An additional 70 nucleotides following the predicted transcription terminator were included to aid in resolution of the terminated and full-length products. For more detailed protocol, see Extended Experimental Procedures.
Agar Diffusion Assays
A diluted overnight culture of the E. coli strain carrying the riboswitch-reporter fusion plasmid was spread onto an agar plate containing carbenicillin (100 µg mL−1) and X-gal (80 µg mL−1). A single BBL™ Sensi-Disc™ Susceptibility Test Disc (BD) was placed on each plate and a specific antibiotic was added to the disc (5 µL of either 1 µg mL−1 trimethoprim, 400 µg mL−1 sulfathiazole, 5 µg mL−1 chloramphenicol, or 4 µg mL−1 kanamycin). Disks pre-impregnated with imipenem, tetracycline, rifampicin, and gentamycin antibiotic were purchased (BD). Bacteria were grown on M9 minimal agar medium supplemented with carbenicillin and X-gal. The plates were incubated overnight at 37°C.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Adam Roth, Dr. Narasimihan Sudarsan, Dr. Zasha Weinberg, Dr. Kenneth Blount, Shira Stav, and other members of the Breaker laboratory for helpful discussions. We also thank N. Carriero and R. Bjornson for their help in using the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02). This work was supported by grants from the National Institutes of Health (GM022778 and DE022340) to R.R.B and by the Howard Hughes Medical Institute. P.B.K was supported by Gruber Foundation, Davenport Hooker Memorial Fellowship, and NIH Genetics Training Grant (5T32GM007499).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication., As a service to our customers we are providing this early version of the manuscript., The manuscript will undergo copyediting, typesetting, and review of the resulting proof, before it is published in its final citable form. Please note that during the production, process errors may be discovered which could affect the content, and all legal disclaimers, that apply to the journal pertain.
Supplemental Information includes Extended Experimental Procedures, seven figures, one table, and one date file.
AUTHOR CONTRIBUTIONS P.B.K., J.W.N., and R.R.B. designed experiments for the biochemical and genetic analyses of riboswitch constructs. P.B.K. performed bioinformatics searches, designed genetic reporter constructs, and created the reporter strains. P.B.K. and J.W.N. conducted in-line probing assays and conducted genetic reporter analyses. J.W.N. performed in vitro transcription assays. All authors contributed to discussions regarding the data and their interpretation. P.B.K. and R.R.B. wrote the manuscript with assistance from all authors.
REFERENCES
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Biol. 2010;17:681–685. doi: 10.1016/j.chembiol.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Fritz-Wolf K, Kabsch W, Knappe J, Schultz S, Volker Wagner AF. Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat Struct Biol. 1999;6:969–975. doi: 10.1038/13341. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Ames BN. ZTP (5-amino 4-imidazole carboxamide riboside 5'-triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell. 1982;29:929–937. doi: 10.1016/0092-8674(82)90455-x. [DOI] [PubMed] [Google Scholar]
- Borovok I, Gorovitz B, Yanku M, Schreiber R, Gust B, Chater K, Aharonowitz Y, Cohen G. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol Microbiol. 2004;54:1022–1035. doi: 10.1111/j.1365-2958.2004.04325.x. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future microbiology. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA world. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Castro C. The mechanism of action of ribavirin: lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr Opin Infect Dis. 2001;14:757–764. doi: 10.1097/00001432-200112000-00015. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Gruber T, Barry CE, 3rd, Boshoff HI, Rhee KY. Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science. 2013;339:88–91. doi: 10.1126/science.1228980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B, Fink GR. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci U S A. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B, Pinson B. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5'-Monophosphate (AICAR), a Highly Conserved Purine Intermediate with Multiple Effects. Metabolites. 2012;2:292–302. doi: 10.3390/metabo2020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductasetrimethoprim, a drug-receptor system. Proteins. 1988;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Derzelle S, Bolotin A, Mistou MY, Rul F. Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl Environ Microbiol. 2005;71:8597–8605. doi: 10.1128/AEM.71.12.8597-8605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer LG, Parbu-Patel A, Carling D. Protein kinase inhibitors block the stimulation of the AMP-activated protein kinase by 5-amino-4-imidazolecarboxamide riboside. FEBS Lett. 2002;531:189–192. doi: 10.1016/s0014-5793(02)03501-9. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Jenkins A, Cote C, Twenhafel N, Merkel T, Bozue J, Welkos S. Role of purine biosynthesis in Bacillus anthracis pathogenesis and virulence. Infect Immun. 2011;79:153–166. doi: 10.1128/IAI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungermann KA, Schmidt W, Kirchniawy FH, Rupprecht EH, Thauer RK. Glycine formation via threonine and serine aldolase. Its interrelation with the pyruvate formate lyase pathway of one-carbon unit synthesis in Clostridium kluyveri. Eur J Biochem. 1970;16:424–429. doi: 10.1111/j.1432-1033.1970.tb01097.x. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytek SJ, Dyer JC, Knapp GS, Hu JC. Resistance to methotrexate due to AcrAB-dependent export from Escherichia coli. Antimicrob Agents Chemother. 2000;44:3210–3212. doi: 10.1128/aac.44.11.3210-3212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov JN, Leclerc GJ, Leclerc GM, Barredo JC. AMPK and Akt determine apoptotic cell death following perturbations of one-carbon metabolism by regulating ER stress in acute lymphoblastic leukemia. Mol Cancer Ther. 2011;10:437–447. doi: 10.1158/1535-7163.MCT-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Higgins MB, Rabinowitz JD. Antifolate-induced depletion of intracellular glycine and purines inhibits thymineless death in E. coli. ACS Chem Biol. 2010;5:787–795. doi: 10.1021/cb100096f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner JT, Georgiou DK, Dagnino-Acosta A, Ainbinder A, Cheng Q, Joshi AD, Chen Z, Yarotskyy V, Oakes JM, Lee CS, et al. AICAR prevents heat-induced sudden death in RyR1 mutant mice independent of AMPK activation. Nat Med. 2012;18:244–251. doi: 10.1038/nm.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo-Sorbello GS, Bertino JR. Current understanding of methotrexate pharmacology and efficacy in acute leukemias. Use of newer antifolates in clinical trials. Haematologica. 2001;86:121–127. [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA biology. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013 doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson B, Vaur S, Sagot I, Coulpier F, Lemoine S, Daignan-Fornier B. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 2009;23:1399–1407. doi: 10.1101/gad.521809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebora K, Laloo B, Daignan-Fornier B. Revisiting purine-histidine cross-pathway regulation in Saccharomyces cerevisiae: a central role for a small molecule. Genetics. 2005;170:61–70. doi: 10.1534/genetics.104.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS. Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 2005;21:385–389. doi: 10.1016/j.tig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Rohlman CE, Matthews RG. Role of purine biosynthetic intermediates in response to folate stress in Escherichia coli. J Bacteriol. 1990;172:7200–7210. doi: 10.1128/jb.172.12.7200-7210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina RL, Holmes EW, Becker MA. The enzymatic synthesis of 5-amino-4-imidazolecarboxamide riboside triphosphate (ZTP) Science. 1984;223:1193–1195. doi: 10.1126/science.6199843. [DOI] [PubMed] [Google Scholar]
- Sabina RL, Patterson D, Holmes EW. 5-Amino-4-imidazolecarboxamide riboside (Z-riboside) metabolism in eukaryotic cells. J Biol Chem. 1985;260:6107–6114. [PubMed] [Google Scholar]
- Sawers RG. Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans. 2005;33:42–46. doi: 10.1042/BST0330042. [DOI] [PubMed] [Google Scholar]
- Schirch V. Mechanisms of folate-requiring enzymes in one-carbon metabolism. In: Sinnott M, editor. Comprehensive Biological Catalysis. Academic Press; 1998. pp. 211–252. [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annual review of biophysics. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JC, Artz SW, Ames BN. Guanosine 5'-diphosphate 3'-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc Natl Acad Sci U S A. 1975;72:4389–4393. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- Sullivan JE, Carey F, Carling D, Beri RK. Characterisation of 5'-AMP-activated protein kinase in human liver using specific peptide substrates and the effects of 5'-AMP analogues on enzyme activity. Biochem Biophys Res Commun. 1994;200:1551–1556. doi: 10.1006/bbrc.1994.1627. [DOI] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, Rubin EJ. Depletion of antibiotic targets has widely varying effects on growth. Proc Natl Acad Sci U S A. 2011;108:4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome biology. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HB., 3rd Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005a;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005b;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wiesner JB, Ugarkar BG, Castellino AJ, Barankiewicz J, Dumas DP, Gruber HE, Foster AC, Erion MD. Adenosine kinase inhibitors as a novel approach to anticonvulsant therapy. J Pharmacol Exp Ther. 1999;289:1669–1677. [PubMed] [Google Scholar]
- Woese CR. The genetic code; the molecular basis for genetic expression. New York: Harper & Row; 1967. [Google Scholar]
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18:1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.