Abstract
Background
Although some recent reports have shown that the expression level of miR-193a varied in different cancers, its role in hepatocellular carcinoma (HCC) remains unidentified. The aim of the current study was to validate the relationship between miR-193a-3p and clinicopathological characteristics in HCC patients.
Material/Methods
Expression of miR-193a-3p in 95 HCC cases and their corresponding peritumoral tissues (PT) was examined by using quantitative reverse transcription polymerase chain reaction (qRT-PCR). miR-193a-3p expression and its correlation with a variety of clinicopathological features and patient recurrence were analyzed.
Results
The relative level of miR-193a-3p was 3.2028±1.1951 in PT, significantly higher than its expression in HCC tissues (1.5941±0.7079, P<0.001). The area under the curve of underexpression of miR-193a-3p was 0.906 to distinguish HCC from normal liver (95% CI: 0.864–0.948, P<0.001). Expression of miR-193a-3p was negatively correlated to metastasis (r=−0.371, P=0.000), TNM (r=−0.321, P=0.002), respectively. Additionally, the recurrence time was 50.271±2.631 months for the low miR-193a-3p level group and 60.132±3.626 months for the high miR-193a-3p level group. However, no significant difference between them was found (chi-square=0.354, P=0.552).
Conclusions
MiR-193a-3p may be a tumor-suppressive miRNA which is down-regulated in HCC tissues. It could be regarded as a predictor for the deterioration of HCC patients.
MeSH Keywords: Carcinoma, Hepatocellular; MicroRNAs; Neoplasm Metastasis; Neoplasm Staging; Reverse Transcriptase Polymerase Chain Reaction
Background
Over the past decade the incidence of hepatocellular carcinoma (HCC) has doubled. In the Asia-Pacific region, 500 000 new cases occurred every year, with an estimated 360 000 deaths in Far East countries only [1]. China alone had more than 60% of the total HCC cases, most of which were due to chronic hepatitis B virus (HBV) infection [2–4]. In Japan, hepatitis C virus-(HCV-) related HCC made up 70% of all cases. In the US, where the 5-year survival rate remained under 12%, the incidence of HCC has tripled during the last 2 decades. In the European countries, because of the increased incidence of HCV, the occurrence of HCC has also increased [5,6]. Results from imageological examination, pathological biopsy, and serum AFP level are regarded as diagnostic criteria. Recently, the role of serum AFP level as an indicator for HCC has been shown to be less useful than previously assumed [7] and in most cases, the patients had late-stage HCC when diagnosed [8,9]. Only with early detection of the disease can we maximize clinical benefits while minimizing the toxicity and cost of nonsurgical treatments [10,11]. Therefore, there is a compelling need to find robust new diagnostic biomarkers.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that regulate the expression of protein-coding genes in a sequence-specific manner, including the genes controlling the DNA methylation state of the genome [12]. They can trigger the translational repression, deadenylation, and degradation of mRNA through binding to part of complementary sites within the 3′-untranslated region (UTR) of target messenger RNAs (mRNAs) [13]. Accumulating evidence shows that miRNAs controlled widely ranging physiological and pathological processes, including carcinogenesis and tumor progression [14]. Additionally, studies also found that miRNAs are involved in the invasion and metastasis of variety of cancers, including HCC [14,15]. Aberrant miRNA expression is also regarded as a promising biomarker for cancer detection.
Previous studies have shown that miR-193a repressed expression of certain oncogenic factors, including CDK6, c-kit, E2F6, and E-cadherin, which could enhance tumor invasiveness and angiogenesis [16–18]. Studies have also indicated that miR-193a exhibited significantly down-regulated expression in non-small cell lung cancer (NSCLC), and that they were associated with proliferation, metastasis, and apoptosis [19,20]. In epithelial ovarian cancer cells, miR-193a-3p plays a tumor-suppressive role [21], and in the neo-adjuvant chemotherapy (NACT) setting of ovarian cancer, miR-193a-5p is significantly over-expressed. In breast cancer, miR-193a-3p modulates the response of tumor cells to estrogen and anti-endocrine therapy, and up-regulated miR-193a-3p exaggerates the effects of estradiol and tamoxifen on estrogen receptor (ER)-positive breast cancer cells [22]. In colorectal cancer (CRC) patients, low expression of miR-193a-3p is correlated with poor prognosis and lymph node metastasis [23]. As for HCC, a study based on HCC cell lines and in vivo study of nude mice showed that miR-193a-3p regulated the 5-fluorouracil (5-FU) resistance of HCC, whose transcription could be repressed by the hypermethylated promoter state [17]. Another study verified the molecular interaction between miR-193a-3p and urokinase-type plasminogen activator (uPA) mRNA target. The study evaluated the expression of miR-193a-3p in HCC tumoral tissues from 39 patients and concluded that miR-193a-3p was down-regulated in HCC compared to the peritumoral tissues (PT) [24]. However, no study has assessed the correlation of miR-193a-3p expression between HCC and matched PT along with its clinicopathological significance in HCC patients. Thus, in order to investigate the relationship between expression of miR-193a-3p and clinicopathological features of HCC, especially its metastasis and recurrence, we quantified miR-193a-3p expression in HCC tissues and their corresponding PT.
Material and Methods
Patients and tissue samples
There were 95 HCC patients in total recruited to the current study. Among them, 75 were males and 20 were females, the mean age was 52 years old (range 29–82 years old). All patients were admitted to the First Affiliated Hospital, Guangxi Medical University from March 2010 to December 2011. We retrospectively evaluated the formalin-fixed, paraffin-embedded (FFPE) cancer tissues and corresponding PT. We also collected their clinicopathological parameters such as age, sex, tumor differentiation, tumor diameter, number of tumor nodes, metastasis, TNM stages, status of portal vein tumor embolus, vaso-invasion, capsular infiltration, and cirrhosis. In addition, enzyme-linked immunosorbent assay (ELISA) was applied to measure the expression of AFP in sera and immunohistochemistry was used to detect the level of metadherin (MTDH), nm23, p53, p21, vascular endothelial growth factor (VEGF), and microvessel density (MVD) calculated with CD34 in cancer tissues. All these features are recorded in Table 1. The current study was approved by the Ethics Committee of First Affiliated Hospital, Guangxi Medical University, China. Informed consent was achieved from all patients participating in the study, conducted according to the Helsinki Declaration. All samples were re-examined by 2 independent pathologists (Fanghui Ren and Gang Chen).
Table 1.
Relationship between the expression of miR-193a-3p and clinicopathological parameters in HCC.
| Clinicopathological parameters | N | miR-193a-3p relevant expression(2−ΔCq) | |||
|---|---|---|---|---|---|
| Mean ±SD | t | P | |||
| Tissue | Adjacent liver | 95 | 3.2028±1.1951 | −11.288 | <0.001 |
| HCC | 95 | 1.5941±0.7079 | |||
| Age | <50 | 46 | 1.6802±0.7575 | 1.151 | 0.253 |
| ≥50 | 49 | 1.5133±0.6555 | |||
| Gender | Male | 75 | 1.5035±0.5747 | −1.814 | 0.083 |
| Female | 20 | 1.9340±1.0193 | |||
| Differentiation | High | 6 | 1.6900±0.7780 | F=0.063a | 0.939 |
| Moderate | 60 | 1.5820±0.7234 | |||
| Poor | 29 | 1.5993±0.6851 | |||
| Size | <5 cm | 77 | 1.4006±0.3158 | 2.082 | 0.041 |
| ≥5 cm | 18 | 1.6394±0.7659 | |||
| Tumor nodes | Single | 52 | 1.7544±0.8745 | 2.673 | 0.009 |
| Multiple | 43 | 1.4002±0.3502 | |||
| Metastasis | − | 46 | 1.9241±0.8428 | 4.803 | 0.000 |
| + | 49 | 1.2843±0.3364 | |||
| Clinical TNM stage | I–II | 22 | 2.2718±1.0485 | 3.866 | 0.001 |
| III–IV | 73 | 1.3899±0.3886 | |||
| Portal vein tumor embolus | − | 63 | 1.6910±0.8202 | 2.400 | 0.018 |
| + | 32 | 1.4034±0.3428 | |||
| Vaso-invasion | − | 59 | 1.7178±0.8066 | 2.535 | 0.013 |
| + | 36 | 1.3914±0.4470 | |||
| Tumor capsular infiltration | With complete capsule | 45 | 1.7291±0.7708 | 1.784 | 0.078 |
| Infiltration or no capsule | 50 | 1.4726±0.6294 | |||
| HCV | − | 63 | 1.6976±0.7932 | 2.420 | 0.017 |
| + | 32 | 1.3903±0.4433 | |||
| HBV | − | 17 | 1.5453±0.6738 | −0.312 | 0.756 |
| + | 78 | 1.6047±0.7189 | |||
| AFP | − | 41 | 1.5978±0.6774 | 0.326b | 0.745 |
| + | 38 | 1.6511±0.7723 | |||
| Cirrhosis | − | 50 | 1.6630±0.9080 | −1.038 | 0.303 |
| + | 45 | 1.5176±0.3755 | |||
| MTDH | − | 38 | 1.6166±0.7713 | 0.996c | 0.322 |
| + | 51 | 1.4792±0.5295 | |||
| nm23 | − | 20 | 2.0550±1.0977 | 2.314 | 0.031 |
| + | 75 | 1.4712±0.5045 | |||
| p53 | − | 40 | 1.7810±0.8596 | 2.092 | 0.041 |
| + | 55 | 1.4582±0.5422 | |||
| p21 | − | 62 | 1.6216±0.7549 | 0.517 | 0.606 |
| + | 33 | 1.5424±0.6177 | |||
| VEGF | − | 25 | 1.8120±0.9747 | 1.431 | 0.163 |
| + | 70 | 1.5163±0.5735 | |||
| Ki-67 LI | Low | 47 | 1.7004±0.8314 | 1.451 | 0.151 |
| High | 48 | 1.4900±0.5508 | |||
| MVD | Low | 47 | 1.6670±0.8120 | 0.993 | 0.323 |
| High | 48 | 1.5227±0.5887 | |||
ANOVA analysis was applied to assess the difference of miR-193a-3p among differentiation grading;
16 cases missed the detection of AFP;
6 cases lacked the measurement of MTDH.
N – number; AFP – alpha fetal protein; MTDH – metadherin; VEGF – vascular endothelial growth factor; LI – labeling index; MVD – microvessel density.
qRT-PCR
Isolation and normalization of RNA were performed as described before [25–27]. Reverse transcription (RT) and qPCR kits were used to examine the expression of miR-193a-3p, as reported previously [25–28]. A combinatorial reference of RUN6B and RUN48 was used for the detection of miR-193a-3p expression [29,30]. The primers for miR-193a-3p (Applied Biosystems Cat. No. 4427975-0000223: AACUGGCCUACAAAGUCCCAGU), RNU6B (Applied Biosystems Cat. No. 4427975-001093: CGCAA GGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU) and RNU48 (Applied Biosystems Cat. No. 4427975-001006: GAU GACCCCAGGUAACUCUGAGUGUGUCGCUGAUGCCAUCAC CGCAGCGCUCUGACC) were in TaqMan® MicroRNA Assays (4427975, Applied Biosystems, Life Technologies, Grand Island, NY 14072, USA). TaqMan® MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Life Technologies Grand Island, NY 14072 USA) also provided reverse primers, which were used in the RT process with a total volume of 10 μl. Applied Biosystems PCR7900 was used to perform the real-time qPCR for detection of miRNA. The expression of miR-193a-3p in the FFPE experiments was calculated with the formula 2−Δcq [25,26,29,30].
Statistical analysis
SPSS 20.0 (Munich, Germany) was used for the analysis of statistics. Student’s t test was performed to investigate the significance of alteration between 2 groups. As we divided differentiation into 3 groups, 1-way analysis of variance (ANOVA) test was performed to distinguish its significance of alteration. Spearman correlation was used to study the relationship between miR-193a-3p expression and clinicopathological parameters. Receiver operating characteristic (ROC) curve was drawn to test the effectiveness of miR-193a-3p when distinguishing HCC from PT. The relationship between miR-193a-3p and recurrence was analyzed with Kaplan-Meier (K-M) method. The log-rank test was performed to compare the recurrence time between different groups of miR-193a-3p. P-value less than 0.05 (2-tailed test) was statistically significant.
Results
Characteristics of the patients in current study
The clinicopathological parameters of the 95 HCC patients are enumerated in Table 1. All patients received no local ablative therapy, for example, percutaneous ethanol injection (PEI), radiofrequency ablation (RFA), and no transcatheter arterial chemoembolization (TACE) or transarterial radioembolization (TARE) except liver resection. Tumor sizes varied from 1 to 11 cm with a mean size of 6.4 cm. The follow-up time ranged from 2.68 to 68 months (mean time: 33.13±1.71 months). Among the 70 HCC patients available to the follow-up, 11 were censored or dead at the end of follow-up and the rest had recurrent tumors.
Diagnostic value of miR-193a-3p in HCC
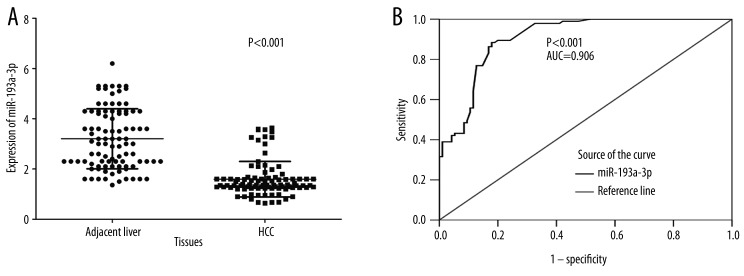
A significant difference in miR-193a-3p expression level was found between HCC tissues and the relevant PT. The relative level of miR-193a-3p was 1.5941±0.7079 in HCC tissues, which was clearly lower than that of the PT (3.2028±1.1951, P<0.001, Figure 1A). In addition, we performed ROC curve to prove the diagnostic role of miR-193a-3p in HCC. The area under curve (AUC) of miR-193a-3p was 0.906 (95% CI: 0.864–0.948, P<0.001, Figure 1B).
Figure 1.
Expression of miR-193a-3p in adjacent PT and HCC tissues. Quantitative real-time RT-PCR was performed to detect the expression of miR-193a-3p. (A) The difference of relevant miR-193a-3p expression between adjacent PT and HCC tissues. (B) ROC curve of miR-193a-3p expression to distinguish HCC from PT. The area under curve (AUC) of miR-193a-3p was 0.906 (95% CI: 0.864–0.948, P<0.001).
The relationship between miR-193a-3p level and various HCC clinicopathological parameters
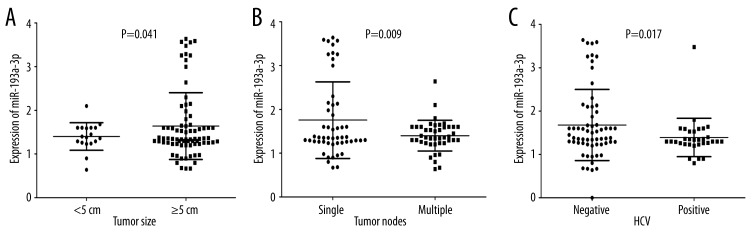
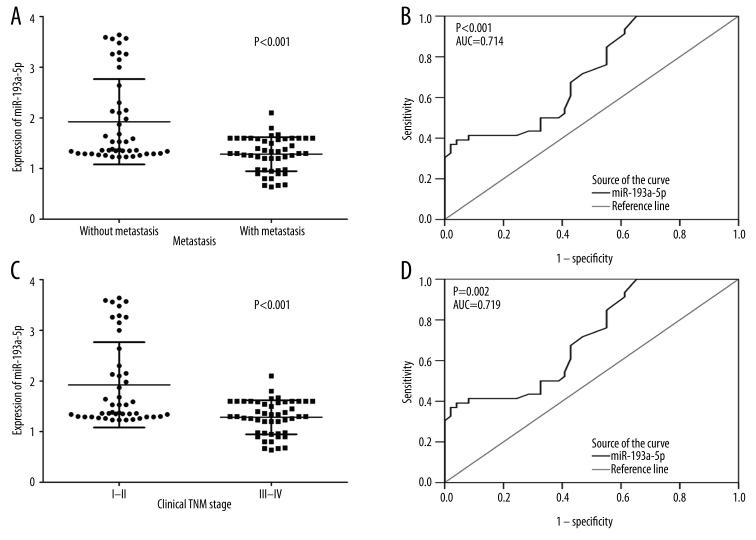
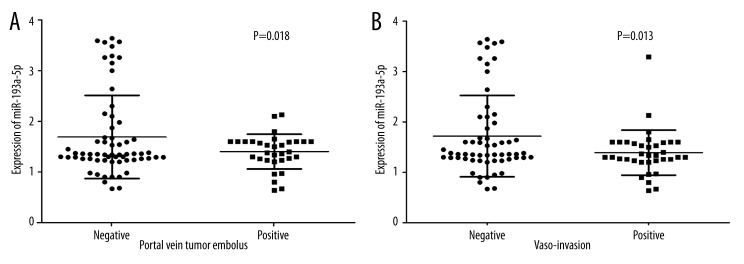
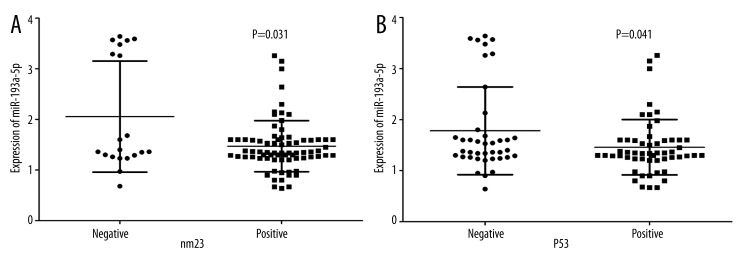
In HCC patients with tumor size was under 5 cm, their levels of miR-193a-3p (1.4006±0.3158) were clearly lower than those with tumor size larger than 5 cm (1.6394±0.7659, P=0.041, Figure 2A). The relative level of miR-193a-3p in HCC patients with multiple tumor nodes (1.4002±0.3502) was prominently lower than those with single tumor nodes (1.7544±0.8745, P=0.009, Figure 2B). In association with HCV infection, HCC patients with positive infection (1.3903±0.4433) had greater down-expression of miR-193a-3p than those without infection (1.6976±0.7932, P=0.017, Figure 2C). Compared with those without metastasis (1.9241±0.8428), the expression of miR-193a-3p was reduced in HCC patients with metastasis (1.2843±0.3364, P<0.001, Figure 3A). ROC curve showed an AUC of 0.714 (95% CI: 0.612–0.817, P<0.001, Figure 3B) to predict the status of metastasis. When compared to early-stage patients (I and II, 2.2718±1.0485), the relative level of miR-193a-3p in those with advanced stages (III and IV, 1.3899±0.3886, P<0.001, Figure 3C) markedly decreased. The AUC of ROC curve was 0.719 (95% CI: 0.590–0.848, P=0.002, Figure 3D) to evaluate the clinical TNM stage of HCC. In HCC patients who had portal vein tumor embolus, levels of miR-193a-3p (1.4034±0.3428) were prominently lower than those who had no embolus (1.6910±0.8202, P=0.018, Figure 4A). Additionally, the relative level of miR-193a-3p in HCC patients with vaso-invasion (1.3914±0.4470) was obviously lower than those without (1.7178±0.8066, P=0.013, Figure 4B). With respect to the correlation with the expression of nm23, HCC patients of nm23 positive (1.4712±0.5045) showed a noticeably lower level of miR-193a-3p than the negative one (2.0550±1.0977, P=0.031, Figure 5A). Moreover, miR-193a-3p in p53-positive HCC patients was down-regulated (1.4582±0.5422), compared with p53-negative HCC patients (1.7810±0.8596, P=0.041, Figure 5B). A further Spearman correlation test was performed and showed a negative correlation between miR-193a-3p and metastasis (r=−0.371, P<0.001), as well as between miR-193a-3p and clinical TNM stages (r=−0.321, P=0.002). However, no noteworthy association was observed between miR-193a-3p level and other clinicopathological features.
Figure 2.
The relationship between miR-193a-3p and tumor size, tumor nodes, and HCV infection in HCC. (A) The difference of relevant miR-193a-3p expression between tumor smaller than 5cm and tumor larger than 5cm, P=0.041. (B) The difference of relevant miR-193a-3p expression between single tumor node and multiple tumor nodes, P=0.009. (C) The difference of relevant miR-193a-3p expression between negative infection and positive infection of HCV, P=0.017.
Figure 3.
The relationship between miR-193a-3p and metastasis, clinical TNM stage in HCC. (A) The difference of relevant miR-193a-3p expression between patients without metastasis and with metastasis. P<0.001. (B) ROC curve of miR-193a-3p expression to distinguish whether there was metastasis for HCC patients. The area under curve (AUC) was 0.714 (95% CI: 0.612–0.817, P<0.001). (C) The difference of relevant miR-193a-3pexpression between early TNM stage and advanced TNM stage. P<0.001. (D) ROC curve of miR-193a-3p expression to distinguish early TNM stage from advanced TNM stage. The area under curve (AUC) was 0.719 (95% CI: 0.590–0.848, P=0.002).
Figure 4.
The relationship between miR-193a-3p and portal vein tumor embolus and vaso-invasion in HCC. (A) The difference of relevant miR-193a-3p expression between negative and positive portal vein tumor embolus. P=0.018. (B) The difference of relevant miR-193a-3p expression between no vaso-invasion and positive vaso-invasion. P=0.013.
Figure 5.
The relationship between miR-193a-3p and nm23, p53 in HCC. (A) The difference of relevant miR-193a-3p expression between negative and positive expression of nm23. P=0.031. (B) The difference of relevant miR-193a-3p expression between negative and positive expression of p53. P=0.041.
Role of miR-193a-3p expression in recurrence of HCC
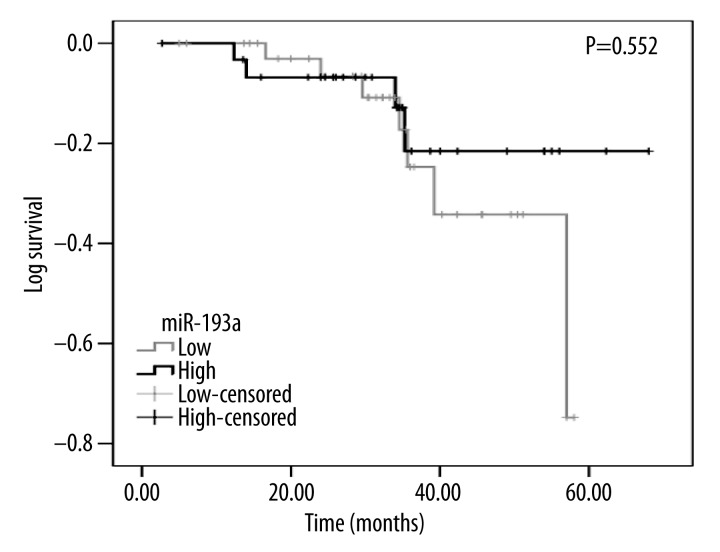
Out of the 95 patients participating in our study, 70 remained available in the follow-up, with an overall recurrence time of 57.095±2.876 months. Among the 70 patients, 38 had low miR-193a-3p expression (lower than the median level of 1.36), while 32 had high miR-193a-3p expression. Concerning time to recurrence, low miR-193a-3p expression group was 50.271±2.631 months, shorter than that in the high expression group (60.132±3.626 months). However, no marked alteration of recurrence time was achieved between the low and high miR-193a-3p groups (chi-square=0.354, P=0.552, Figure 6).
Figure 6.
Recurrence K-M curve between high and low level of miR-193a-3p expression.
Discussion
Only 2 studies have explored the role of miR-193a-3p in HCC. One was carried out by Ma et al. [17], and a pair of HCC cell lines that differ in chemoresistance to 5-fluoro-2,4(1 h, 3 h) pyrimidinedione (5-FU) were selected to perform both genome-wide analysis and mechanistic studies. They found that the expression level of miR-193a-3p was low in 5-FU-sensitive HCC cell line HepG2, while it was high in all other 5-FU-resistant cell lines: PLC, FOCUS, Hep3B, and BEL-7402. They also selected nude mice for in vivo study to investigate the potential mechanism, and found that repression of the miR-193a-3p level by antagomir technologies inhibited tumor growth and sensitized SMMC-7721 cells to 5-FU. miR-193a-3p regulated the 5-FU resistance of HCC by targeting SRSF2 and E2F1. The other study was presented by Salvi et al. [24], who aimed to understand the co-treatment of HCC cells with the combination of a molecular targeting drug (sorafenib) and miRNAs that target uPA. As a result, they validated that miR-193a-3p was a negative regulator of uPA in 2 HCC undifferentiated cell lines. Also, they evaluated the expression of miR-193a-3p in HCC tumoral tissues from 39 patients and found there was dysregulation of miR-193a-3p in tumoral tissues. However, the sample size was small (n=39) in the study of Salvi et al. [24]. In the present study, we examined the miR-193a-3p expression in 95 cases of HCC tissues and their corresponding PT. We found lower expression of miR-193a-3p in HCC tissues, which coincided exactly with the results of Salvi et al. [24]. The relevant level of miR-193a-3p was 1.5941±0.7079 in HCCs, which was significantly lower than that of the PT (3.2028±1.1951, P<0.001). The additional ROC curve demonstrated that miR-193a-3p had a valid diagnostic value for HCC, with the AUC of 0.906. Thus, we were strongly convinced that miR-193a-3p was a tumor-suppressive predictor in HCC.
No previous study has focussed on the relationship between expression of miR-193a-3p and clinicopathological parameters of HCC. We primarily hypothesized that miR-193a-3p expression was responsible for the deterioration of HCC for the following reasons: 1). There was a significant negative correlation between miR-193a-3p and metastasis as well as TNM stages and these 2 indexes indicate poor prognosis of HCC. The AUC of miR-193a-3p in metastasis and TNM was 0.714 and 0.719, respectively, which suggested its role in diagnosis and prognosis of speculation. 2). MiR-193a-3p was expressed at low levels in HCC patients whose tumors were smaller than 5 cm. It was also repressed in those patients with multiple tumor nodes compared to those with single node. 3). As HCC developed, there appeared portal vein tumor embolus and vaso-invasion, and miR-193a-3p was found to be down-regulated along with them. 4). In nm23-positive patients, level of miR-193a-3p decreased compared to nm23-negative patients, as is also true in p53-positive patients. MiR-193a-3p level was lower than that in p53-negative patients. Studies have already suggested that nm23 with pro-oncogenic potential was involved in hepatocarcinogenesis [31], and that its expression in tumor tissues was correlated with metastasis and survival in HCC, which may also be an indicator for prognosis [32]. Studies also showed that overexpression of p53 contributed to the hepatocarcinogenesis and more advanced stage of HCC [33]. As we found there was significant difference of miR-193a-3p expression in nm23 as well as p53-positive and -negative HCC patients in the current study, miR-193a-3p may be a pro-oncogenic potential indicator, as nm23 and p53 are in HCC.
From the K-M curve, we observed a tendency that the recurrence of patients with low miR-193a-3p level was shorter than those with a high level, although there was no significant difference. This suggested that miR-193a-3p could be a predictor of prognosis and survival. However, studies with larger sample sizes are needed to test the significant difference between miR-193a-3p and recurrence, as well as the relationship between miR-193a-3p and overall survival of HCC.
The 95 patients recruited into the current study received no additional treatment, and their tumor tissues were compared to their own paired PT, which was a guarantee of minimized individual differences and better comparability. The most noteworthy point is that we primarily identified relationship between miR-193a-3p expression and clinicopathological features, especially its relationship with metastasis and TNM stages.
However, the present study has a few limitations. Firstly, the sample size in our study was still small, which limited its corroborative role in drawing the conclusion of the impact of miR-193a-3p in patients with HCC. Secondly, we only used the method of qRT-PCR in our current study to evaluate the expression of miR-193a-3p, and fluorescence in situ hybridization (FISH) could be another option to detect the expression level of miRNA. Thirdly, we only analyzed the recurrence-free survival (RFS) of patients rather than overall survival (OS), which may not be sufficient to reflect the value of miR-193a-3p in predicting the prognosis of HCC. Fourthly, this study was limited to exploring the relationship between miR-193a-3p and its clinicopathological features in HCC tissues, not the correlation of miR-193a-3p with the expression profiling of its potential targeting genes. Furthermore, it would be better to identify its function in vivo and in vitro at a further step and discover new target genes of miR-193a-3p.
Conclusions
Along with previous studies, the current observations confirm that miR-193a-3p is a tumor-suppressive miRNA that plays an essential role in the tumorigenesis and deterioration of HCC. It could also be regarded as a predictor of prognosis. Cohorts with larger sample sizes and further in vitro and in vivo studies are needed define the relevant mechanism of miR-193a-3p in HCC.
Footnotes
Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors have no declared conflicts of interest.
Ethical considerations
The current study was approved by the Ethics Committee of First Affiliated Hospital, Guangxi Medical University, China. Related research was conducted in accordance with the Helsinki Declaration.
Source of support: The study was partly supported by the Fund of Guangxi Natural Scientific Research (No. 2013GXNSFBA019191), Guangxi Provincial Health Bureau Scientific Research Project (Z2014054), Youth Science Foundation of Guangxi Medical University (GXMUYSF201311), Guangxi University Science and Technology Research Projects (LX2014075), and the Fund of National Natural Science Foundation of China (NSFC 81360327)
References
- 1.Lin CL, Kao JH. Risk stratification for hepatitis B virus related hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:10–17. doi: 10.1111/jgh.12010. [DOI] [PubMed] [Google Scholar]
- 2.Liang T, Chen EQ, Tang H. Hepatitis B virus gene mutations and hepatocarcinogenesis. Asian Pac J Cancer. 2013;14:4509–13. doi: 10.7314/apjcp.2013.14.8.4509. [DOI] [PubMed] [Google Scholar]
- 3.Ayub A, Ashfaq UA, Haque A. HBV induced HCC: major risk factors from genetic to molecular level. Biomed Res Int. 2013;2013:810461. doi: 10.1155/2013/810461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robotin MC, George J. Community-based hepatitis B screening: what works? Hepatol Int. 2014;8:478–92. doi: 10.1007/s12072-014-9562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davila JA, Kramer JR, Duan Z, et al. Referral and receipt of treatment for hepatocellular carcinoma in United States veterans: effect of patient and nonpatient factors. Hepatology. 2013;57:1858–68. doi: 10.1002/hep.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh JJ, Uemura M. Hepatocellular carcinoma. N Engl J Med. 2012;366:92. doi: 10.1056/NEJMc1112501. author reply 92–93. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Xie L, Yang WS, et al. Risk factors of hepatocellular carcinoma – current status and perspectives. Asian Pac J Cancer. 2012;13:743–52. doi: 10.7314/apjcp.2012.13.3.743. [DOI] [PubMed] [Google Scholar]
- 8.Yeh YT, Chang CW, Wei RJ, Wang SN. Progesterone and related compounds in hepatocellular carcinoma: basic and clinical aspects. Biomed Res Int. 2013;2013:290575. doi: 10.1155/2013/290575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han MS, Moon KS, Lee KH, et al. Brain metastasis from hepatocellular carcinoma: the role of surgery as a prognostic factor. BMC Cancer. 2013;13:567. doi: 10.1186/1471-2407-13-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuang L, Zeng X, Yang Z, Meng Z. Effect and safety of interferon for hepatocellular carcinoma: a systematic review and meta-analysis. PLoS One. 2013;8:e61361. doi: 10.1371/journal.pone.0061361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang P, Hu JH, Cheng ZG, et al. Efficacy and safety of bevacizumab for the treatment of advanced hepatocellular carcinoma: a systematic review of phase II trials. PLoS One. 2012;7:e49717. doi: 10.1371/journal.pone.0049717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi M, Han W, Spivack SD. A quantitative method to identify microRNAs targeting a messenger RNA using a 3’UTR RNA affinity technique. Anal Biochem. 2013;443:1–12. doi: 10.1016/j.ab.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie W, Rasko JE. Refining microRNA target predictions: sorting the wheat from the chaff. Biochem Biophys Res Commun. 2014;445:780–84. doi: 10.1016/j.bbrc.2014.01.181. [DOI] [PubMed] [Google Scholar]
- 14.Di Leva G, Briskin D, Croce CM. MicroRNA in cancer: new hopes for antineoplastic chemotherapy. Ups J Med Sci. 2012;117:202–16. doi: 10.3109/03009734.2012.660551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Lu H, Wang X, Jin H. MicroRNAs in hepatocellular carcinoma: regulation, function, and clinical implications. Scientific World Journal. 2013;2013:924206. doi: 10.1155/2013/924206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao XN, Lin J, Li YH, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30:3416–28. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 17.Ma K, He Y, Zhang H, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287:5639–49. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iliopoulos D, Rotem A, Struhl K. Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011;71:5144–53. doi: 10.1158/0008-5472.CAN-11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34(4):413–23. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Yang B, Han L, et al. Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol Biochem. 2013;32:1707–19. doi: 10.1159/000356605. [DOI] [PubMed] [Google Scholar]
- 21.Nakano H, Yamada Y, Miyazawa T, Yoshida T. Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int J Oncol. 2013;42:1875–82. doi: 10.3892/ijo.2013.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillai MM, Gillen AE, Yamamoto TM, et al. HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res Treat. 2014;146:85–97. doi: 10.1007/s10549-014-3004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang P, Ji DB, Han HB, et al. Downregulation of miR-193a-5p correlates with lymph node metastasis and poor prognosis in colorectal cancer. World J Gastroenterol. 2014;20:12241–48. doi: 10.3748/wjg.v20.i34.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvi A, Conde I, Abeni E, et al. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer. 2013;12:162. doi: 10.1186/1476-4598-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Kronenberger P, Teugels E, et al. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Med. 2012;10:28. doi: 10.1186/1741-7015-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen G, Umelo IA, Lv S, et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G, Kronenberger P, Teugels E, De Greve J. Influence of RT-qPCR primer position on EGFR interference efficacy in lung cancer cells. Biol Proced Online. 2011;13:1. doi: 10.1186/1480-9222-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MJ, Xu DY, Li H, et al. Pro-oncogenic potential of NM23-H2 in hepatocellular carcinoma. Exp Mol Med. 2012;44:214–24. doi: 10.3858/emm.2012.44.3.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An R, Meng J, Shi Q, et al. Expressions of nucleoside diphosphate kinase (nm23) in tumor tissues are related with metastasis and length of survival of patients with hepatocellular carcinoma. Biomed Environ Sci. 2010;23:267–72. doi: 10.1016/S0895-3988(10)60062-1. [DOI] [PubMed] [Google Scholar]
- 33.Cioca A, Cimpean A, Ceausu R, et al. Crosstalk between EGFR and p53 in hepatocellular carcinoma. Asian Pac J Cancer. 2014;15:8069–73. doi: 10.7314/apjcp.2014.15.19.8069. [DOI] [PubMed] [Google Scholar]