Abstract
Ten-eleven translocation (Tet) genes encode for a family of hydroxymethylase enzymes involved in regulating DNA methylation dynamics. Tet1 is highly expressed in mouse embryonic stem cells (ESCs) where it plays a critical role the pluripotency maintenance. Tet1 is also involved in cell reprogramming events and in cancer progression. Although the functional role of Tet1 has been largely studied, its regulation is poorly understood. Here we show that Tet1 gene is regulated, both in mouse and human ESCs, by the stemness specific factors Oct3/4, Nanog and by Myc. Thus Tet1 is integrated in the pluripotency transcriptional network of ESCs. We found that Tet1 is switched off by cell proliferation in adult cells and tissues with a consequent genome-wide reduction of 5hmC, which is more evident in hypermethylated regions and promoters. Tet1 downmodulation is mediated by the Polycomb repressive complex 2 (PRC2) through H3K27me3 histone mark deposition. This study expands the knowledge about Tet1 involvement in stemness circuits in ESCs and provides evidence for a transcriptional relationship between Tet1 and PRC2 in adult proliferating cells improving our understanding of the crosstalk between the epigenetic events mediated by these factors.
INTRODUCTION
DNA methylation is the most important epigenetics event involved in mammalian development and cancer growth (1–6). Recently, several papers reported that the demethylation mechanism involves TET family genes, in particular Tet1 member (7–11). Tet1 is a DNA hydroxylases that can convert 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) through three consecutive oxidation reactions (7,12–14).
The functional role of Tet1 has been largely studied in embryonic stem cells (ESCs) and during early embryonic development where its principal functions are to maintain hypomethylated developmental and housekeeping gene promoters and to contribute to the genome DNA demethylation taking place during primordial germinal cells specification (15–22). Several works demonstrated that Tet1 is of crucial importance during dedifferentiation of adult cells in reprogramming experiments thus it plays an essential role in the stemness circuits in mammalian (23–28). In adult tissues Tet1 downregulation has been found to be implied in tumor initiation and progression (29–32).
If on one hand the Tet1 biological and molecular function has been thoroughly studied, on the other hand its regulation in mammalian cells remains almost completely unknown. Recent works demonstrated that Tet1 can undergo post-transcriptional and post-translational negative modulation, respectively by microRNA (miR-22) and cellular proteolytic system (calpain) (31,33). The understanding of Tet1 transcriptional regulation and its chromatin state as well as the identification of the transcription factors involved in its activation is essential to better delineate its biological role in stemness and during development.
In this study, we analyzed Tet1 regulatory elements in mouse and human cells. We identified the key players of its transcriptional regulation and its chromatin state and in different cellular systems.
MATERIALS AND METHODS
Cell culture condition
E14 mouse ES cells were cultured on feeder-free gelatin-coated plates in dulbecco's modified Eagle's medium (DMEM) high glucose medium (Invitrogen) supplemented with 15% fetal bovine serum (FBS) (Millipore), 0.1 mM non-essential amino acids (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 0.1 mM 2-mercaptoethanol, 1500U/ml Leukemia Inhibitory Factor (LIF) (Millipore), 25 U of penicillin/ml and 25 μg of streptomycin/ml. Mouse embryonic fibroblasts (MEFs) were derived from 13.5 d pregnant female mice and cultured in DMEM high glucose medium supplemented with 10% fetal calf serum (FCS). Mouse tissues were extracted from 8 weeks old age mice. 3T3 and 3T3-Myc fibroblast cells were cultured in growth medium (DMEM high glucose with 10% FCS). Human umbilical vein endothelial cells (HUVECs) were grown on gelatin-coated surfaces in M199 medium (Gibco) supplemented with 20% FBS, 50 U/ml penicillin-streptomycin, 10 U/ml heparin and 100 μg/ml brain extract and filtered through a 0.22-μm-pore-size sterile filter (Millipore). BGO1V human ES cells were purchased by Invitrogen and cultured on feeder free system in STEMPRO® hESC SFM (Invitrogen) following the manufacturing protocol. Human MCF10A cells were cultured in DMEM/F12 with 5% HS (horse serum) freshly supplemented with insulin, EGF, hydrocortisone and cholera toxin. All the other human cancer cells were cultured in Roswell Park Memorial Institute (RPMI) with 10% FCS. For differentiation in EBs, mouse and human ES cells were cultured in non-adherent 96-wells plates in α-MEM medium with 5% FCS and 10% of Serum Replacement (SR—Gibco). For experiments with Ezh2 inhibitor cells were treated with Dimethyl sulfoxide (DMSO) or 2 μM GSK343 (Sigma) for 48 h.
Animals
C57BL/6 mice (8–10 weeks old) were obtained from our mouse facility. BALB/c-nude (6 weeks old) were obtained from Janvier-labs. Housing and all experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of the University of Turin.
DNA constructs and shRNA
shRNA against murine c-Myc and N-Myc were previously described in (34). shRNA against human c-Myc (shRNA#1: TRCN0000174055, shRNA#2: TRCN0000039640), Nanog (shRNA#1: TRCN0000075334, shRNA#2: TRCN0000075336) and Suz12 (TRCN0000123889) were purchased from Open Biosystem. All the other shRNA for was cloned into PLKO vector as described in Public TRC Portal (http://www.broadinstitute.org/rnai/public/). Oligonucleotides sequences is provided in Supplementary Table S1. pAAV-EF1a-HA-hTet1CD-WPRE-PolyA and pAAV-EF1a-HA-hTet1CDmu-WPRE-PolyA were purchased from Addgene (plasmid 39454 and 39455). pLVX-Tight-Puro Vector was purchased from Clontech (plasmid S4934 and S4932). Catalytic domain of TET1 was subcloned into pLVX-Tight-Puro-Vector.
Transfection and transduction
Transfection of 3T3 cells and human/mouse ESCs was performed using Lipofectamine™ 2000 Transfection Reagent in according to manufacturing protocol using equal amount of each plasmid in multiple transfections. To obtain 3T3-Myc stable line, transfected cells were plated as single cell, cultured for 2 weeks in growth medium with Puromycin 1 μg/ml and then drug resistant clones were picked, grown and analyzed. MEFs and HUVECs were transduced with 10 μl of concentrated third generation lentivirus prepared using Lenti-XTM Lentiviral Expression Systems (Clontech).
Antibodies
The antibodies were purchased from Millipore (anti-Tet1, anti-H3K27me3, anti-H3K4me3), Abcam (anti-ssDNA, anti-Tet2), Roche (anti-HA-12CA5), Cell Signaling (anti-EZH2) Sigma-Aldrich (anti-β-actin), SantaCruz (anti-PCNA-sc56; anti-cMYC sc-764; anti-mOCT3/4 sc-5279; anti-N-MYC sc56729; anti-E2F1;anti-Stat3 sc-482; anti-hNANOG sc-33759; anti-hOCT3/4 sc-9081, anti-p16 sc-1661, anti-p21 sc-397), Bethyl (anti-mNanog) and Active Motif (anti-5mC, anti-5hmC).
Partial hepatectomy
Eight to ten-week old C57 black 6 (C57BL/6) male mice were fasted overnight. The next morning, the mice were anesthetized with Zoletil/Rompun mix and the median and left lateral lobes of the liver were ligated at their stem and excised. Food was re-introduced 6–8 h after surgery. The animals were sacrificed by cervical dislocation at the indicated time points following surgery. The Institutional Animal Care and Use Committee of the University of Turin approved all experiments.
Cell growth and cell cycle analysis
For cell growth assay, 5 × 104 cells were plated in 35 mm wells and counted at the indicated time point using Scepter™ Automated Cell Counter (Millipore). For FACS cell cycle analysis, the cells were stained with propidium iodide (PI) solution (0.1% Triton X-100, 200 μg/ml RNase, 20 μg/ml PI in PBS) for 30 min at room temperature. EdU incorporation FACS analysis was performed by using Click-iT® EdU Flow Cytometry Cell Proliferation Assay (Invitrogen) in according to manufacturing protocol by stimulating MEF cells for 3 h with 5 mM EdU. Acquisition was performed using Becton Dickinson FACS Canto and analysis was done with FACS FlowJo Software.
Colony assay
Colony forming assay survival was defined as the ability of the cells to maintain their clonogenic capacity and form colonies. Briefly, after transfection, cells were trypsinized, counted and seeded for colony formation in 100 mm dishes at 1000 cells/dish. After 14 days incubation, the colonies were stained with crystal violet and manually counted.
Protein extraction and western blotting
For total cell extracts, cells were resuspended in F-buffer (10 mM Tris–HCl pH 7.0, 50 mM NaCl, 30 mM Na-pyrophosphate, 50 mM NaF, 1% Triton X-100, anti-proteases) and sonicated for three pulses. Extracts were quantified using bicinchoninic acid (BCA) assay (BCA protein assay kit; catalog no. 23225; Pierce) and were run on sodium dodecyl sulphate-polyacrylamide gels at different percentages, transferred to nitrocellulose membranes and incubated with specific primary antibodies overnight.
Immunohistochemistry and immunofluorescence
For histology and immunostaining, organs were dissected and fixed in 2% Paraformaldehyde (PFA) overnight at 4°C, dehydrated and embedded in paraffin. Thick sections (5 μm thickness) were treated as previously described (35). Immunostainings were performed using the following primary antibodies: rabbit anti-PCNA (SantaCruz, sc56, 1:200), rabbit anti-5methyl and 5-hydroxymethyl cytosine (Active Motif, 1:200). Sections were then incubated with the appropriate fluorescently conjugated secondary antibodies (Alexa 488 or 546, Molecular Probes, 1:500) or for immunohistochemistry with secondary biotinylated antibodies (R&D Cell and Tissue Staining kit), followed by amplification with the high sensitivity streptavidin-horseradish peroxidase (HRP) conjugate and reaction with 3, 3'-diaminobenzidine (DAB). We assessed liver morphology based on hematoxylin and eosin-stained paraffin sections. The frequency of nuclear PCNA, 5mC and 5hmC staining (anti-PCNA antibody; SantaCruz, sc56) was determined by examination of at least three random 200X field.
DNA extraction and dot-blot analysis
Genomic DNA was extracted from cells using DNeasy Blood and Tissue kit (Qiagen). For dot-blot analysis, extracted genomic DNA was sonicated for 15 cycles to obtain 300 bp fragments, denatured with 0.4 M NaOH and incubated for 10 min a at 95°C prior to being spotted onto Hybond™-N+ (GE Healthcare). Membranes were saturated with 5% milk and incubated 16 h with the specific antibodies as described (36).
RNA extraction and RT-PCR analysis
Total RNA was extracted as previously described in (37) by using TRIzol reagent (Invitrogen). Real-time polymerase chain reaction (PCR) was performed using the SuperScript III Platinum One-Step Quantitative RT-PCR System (Invitrogen, cat.11732-020) following the manufacturer's instructions. For analysis of the hnRNA, RNA was treated with DNAse I for 2 h and then analyzed with specific primers on intronic sequences. For primers efficiency analysis, RT-PCR products were run on gel and purified by using QIAquick Gel Extraction Kit (Qiagen). Purified DNA was quantified by nanodrop and a three-fold standard curve was prepared by starting from 20 pg of DNA. qPCR of standard curve was performed in triplicate in the same conditions of RT-qPCR and CT values were used to calculate primer-specific amplification coefficient. Primers sequences are provided in Supplementary Table S2.
Chromatin immunoprecipitation
Each chromatin immunoprecipitation (ChIP) experiment was performed in at least three independent biological samples and performed as previously described described (38). Primers for ESCs were designed on E14 specific genome assembly (39). The sequences are provided in Supplementary Table S3.
RESULTS AND DISCUSSION
Tet1 expression is regulated by an ESC-specific promoter
To understand how Tet1 expression is regulated in mammals during development, we first analyzed the RNA-seq and H3K4me3 ChIP-seq data in mouse ESCs and MEFs to better define the Tet1 gene structure (40).
The promoter-specific H3K4me3 modification and RNA-seq indicate the presence of two transcriptional start sites (TSSs) in ESCs (Figure 1A) that give origin of two different Tet1 isoforms, which we named Tet1.1 and Tet1.2 bearing respectively a distal and a proximal promoter relatively to the annotated gene. The first exon of each isoform does not contain an open reading frame therefore both isoforms code for the same Tet1 protein. We designed different primers to discriminate between the two isoforms. Quantitative RT-PCR (RT-qPCR) shows that Tet1.1 is highly expressed in ESCs, its expression is rapidly reduced during EBs differentiation (Figure 1 and Supplementary Figure S1) and is almost undetectable in adult tissues, MEFs and NIH-3T3. Tet1.2 is expressed at a lower level in all samples, with a strong reduction in proliferating MEFs and NIH-3T3 (Figure 1B). These results suggest that the two promoters are regulated independently and that the Tet1.1 isoform can be controlled by an ESC-specific mechanism. The analysis of a meta-dataset of ChIP-seq experiments (41) revealed that, in ESCs, the Tet1.1 promoter (from −500 to +500 bp) is bound by the core ESC factors Oct3/4 and Nanog, and by Myc, Mycn, Stat3, and E2f1 transcriptional factors known to be required for ESC self-renewal (34,42–46). Inspection of Tet1.1 promoter sequence showed an enrichment of the DNA binding motifs for these factors around the TSS (Supplementary Figure S1A). By ChIP experiments, we confirmed the binding of these factors on the TSS of the Tet1.1 promoter using as negative controls the TSS of the annotated Tet1 and the Tet1.2 promoter (Figure 1C). To analyze the effect of these factors on the transcription driven by the Tet1.1 promoter we cultured ESCs without LIF which promotes ESC differentiation with the downmodulation of these nuclear factors trough a Stat3 dependent mechanism (34,46–48). LIF withdrawal induced a significant reduction of the Tet1.1 mRNA level, which could be due to the reduction of the pluripotency factors without affected the transcriptional regulation of the Tet1.2 isoform (Figure 1D). To further investigate the direct contribution of each ESC-specific factor, we performed loss of function experiments using two different specific shRNAs against Myc, Nanog and Oct3/4 messengers. Since we recently demonstrated that Myc is an important component for the stem cell pluripotency and synergistically acts together its family member MycN in ESCs (34), we performed a double knockdown of Myc/Mycn. The silencing efficiency was evaluated by RT-qPCR (Supplementary Figure S1C–E). In the silenced ESCs, we observed a significant decrease of the Tet1.1 expression, while no changes were observed in the Tet1.2 mRNA level (Figure 1E). Taken together these results show that the Tet1.1 isoform is highly expressed in ESCs and its transcriptional activation is controlled by ESC-specific factors. In differentiated cells and in adult tissues is only expressed the Tet1.2 isoform, which does not appear to be regulated by the pluripotency-transcriptional network.
Figure 1.
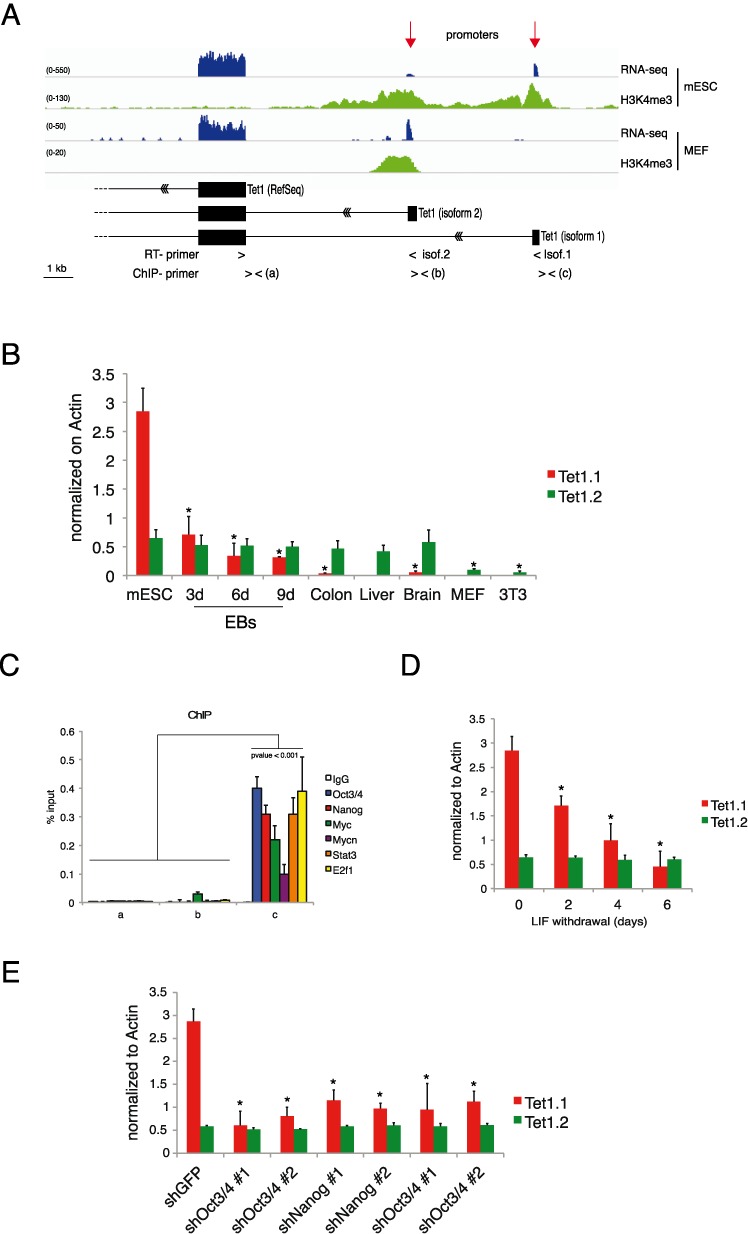
Regulation of Tet1 in mouse ES cells. (A) Genomic view of RNA-seq and H3K4me3 in MEFs and mouse ESCs together with reconstructed Tet1 gene structure. (B) Isoform-specific RT-qPCR of Tet1 mRNA in the indicated samples. (C) qPCR of ChIP analysis for the indicated transcriptional factors on Tet1.1 promoter in mESCs. (D) Isoform-specific RT-qPCR of Tet1 mRNA in ESCs cultured without LIF for the indicated times. (E) Isoform-specific RT-qPCR of Tet1 mRNA in control and cMyc/Mycn, Nanog and Oct3/4 knockdowns ESCs. Error bars represent the standard deviation of three independent experiments. P-value was calculated by using t-test. (* = P-value < 0.01).
Tet1 expression is downmodulated in proliferating cells
The above experiments show that Tet1.2 is expressed at similar levels during development and in adult tissues, but it is downmodulated in proliferating MEFs and NIH-3T3 cells (Figure 1B) suggesting that cell proliferation inhibits its expression in differentiated tissues. Because Tet1.2 downmodulation could be an effect limited to the in vitro culture conditions we explored whether the downregulation of Tet1.2 occurs also in proliferating cells in vivo. To this end we analyzed the expression of Tet1.2 in mouse hepatocytes during liver regeneration after partial hepatectomy. We observed a peak of Pcna+ cells (proliferating hepatocytes) at 48 h after resection. Hepatocytes proliferation was accompanied by a reduced expression of Tet1.2 together with a significant reduction of 5hmC staining (Figure 2A, B and Supplementary Figure S2) indicating that Tet1.2 is downmodulated in adult tissues when cells proliferate. Accordingly, we observed in primary MEFs a drop in the level of 5hmC together with reduced Tet1.2 mRNA after few passages (Figure 2C and D). Moreover, the inhibition of cell proliferation, either by cell confluence or by mitomycin treatment, inhibited both the Tet1.2 and 5hmC downregulation (Figure 2E). Interestingly, the block of cell proliferation did rescue the Tet1.2 expression and the 5hmC level even at later passages (Figure 2F).
Figure 2.
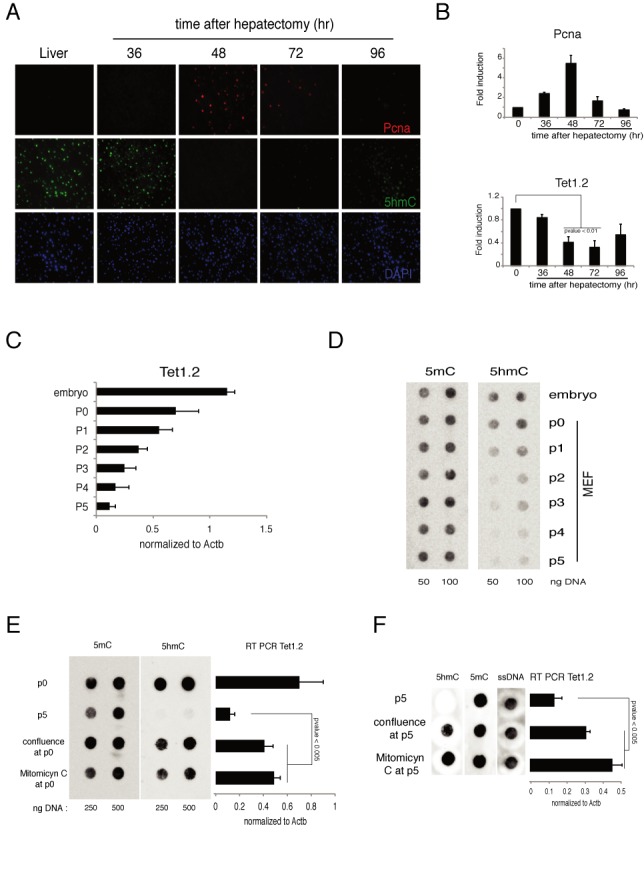
Regulation of Tet1 in mouse proliferating cells. (A) Immunofluorescence analysis of Pcna and 5hmC in liver after hepatectomyzed the indicated time. DAPI is used for nuclei staining. (B) RT-qPCR of Tet1 and Pcna mRNA in liver after hepatectomy. (C) RT-qPCR of Tet1 mRNA in mouse embryo or in MEFs after several passages. (D) Dot-blot analysis of 5hmC and 5mC in mouse embryo or in MEFs after several passages. (E) Left panel: dot-blot analysis of 5hmC and 5mC in MEFs just extracted (p0) or maintained proliferating (p5) or blocked (by confluence or Mitomicyn C) from p0. Right panel: RT-qPCR of Tet1 mRNA in MEF in the same conditions. (F) Same experiment as in (E), but the blocking of proliferation was induced at p5 and maintained for other 5 days. Error bars represent the standard deviation of three independent experiments. P-value was calculated by using t-test.
Tet1 regulates the cell proliferation in primary cells
The above experiments show a strict correlation between cell proliferation and downregulation of Tet1.2/5hmC levels. It has been demonstrated that Tet1 expression can reduce breast cancer malignancy and metastasis (29–31) and we recently demonstrated that Tet1 downmodulation is necessary for colon cancer growth (32).
To study the physiological function of Tet1.2 we performed Tet1.2 silencing in freshly isolated MEFs (Figure 3A and Supplementary Figure S3A). Tet1 knockdown in MEFs at p0 induced a faster decay of 5hmC, which was accompanied by an increased cellular growth rate and a higher number of cells in S phase (Figure 3B–E) suggesting a functional role of Tet1.2 in controlling cell proliferation. In agreement with this observation, the ectopic expression of TET1 in these cells reduced the growth rate maintaining high the levels of 5hmC even at late passages (Figure 3F, G and Supplementary Figure S3B and C). This function is dependent on the TET1 enzymatic activity because the expression of a catalytically dead mutant did not affect fibroblasts cell growth (Supplementary Figure S4). These data underline the importance of the understanding of the regulation of Tet1.2 in adult cells because it is very strictly connected to the proliferative activity of the cells.
Figure 3.
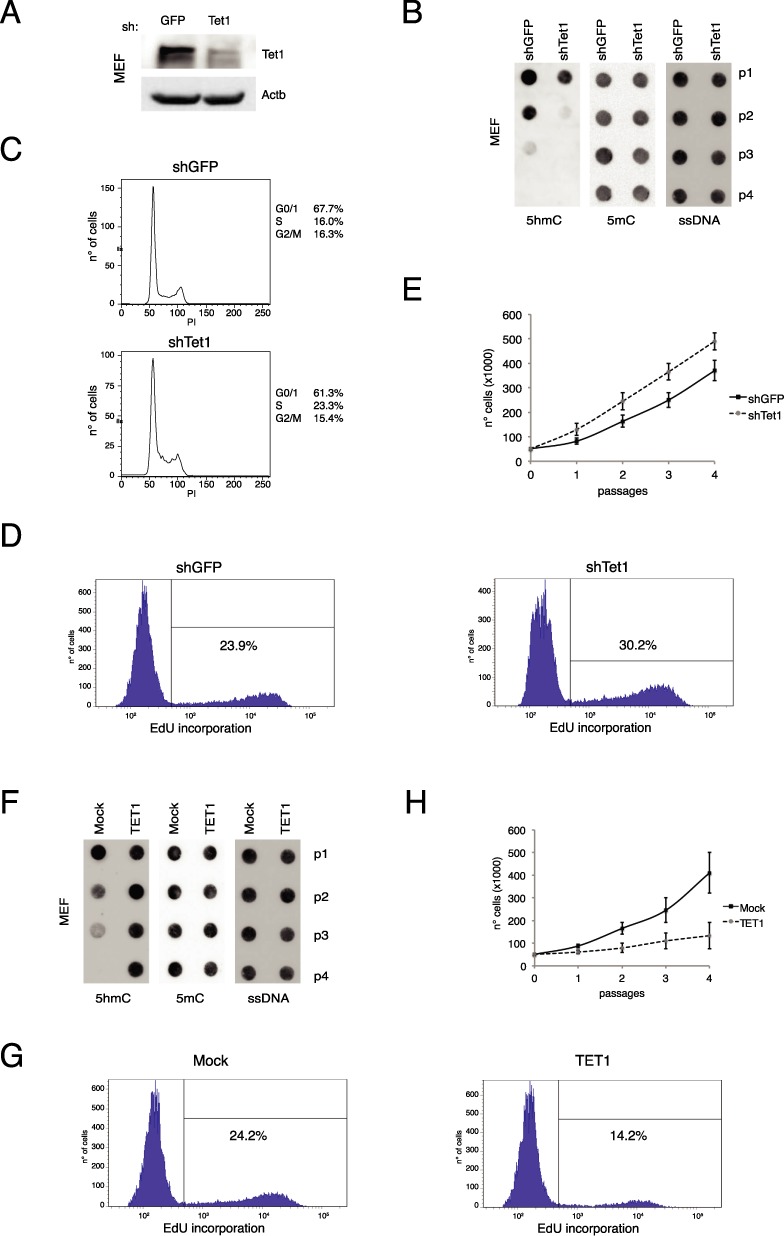
Tet1 regulates proliferation in primary cells. (A) Western blot analysis of Tet1 in control or Tet1 silenced MEF, after 24 h from silencing. Actb was used as loading control. (B) Dot-blot analysis of 5hmC and 5mC in Tet1 silenced MEFs. ssDNA was used as loading control. (C and D) FACS cell cycle and EdU incorporation analysis (at p2) in Tet1 silenced MEFs. (E) Cell growth assay in Tet1 silenced MEFs. (F) Dot-blot analysis of 5hmC and 5mC in control or Tet1 expressing MEFs at several passages. ssDNA was used as loading control. (G) FACS EdU incorporation analysis (at p2) in control or Tet1 expressing MEFs. Error bars represent the standard deviation of three independent experiments. (H) Cell growth assay in control or Tet1 expressing MEFs at several passages.
Although Tet1 and Tet2 knockout mice are viable, depletion of both the genes causes epigenetic abnormalities during mouse development, suggesting some compensatory role of these two proteins (49–51) Li:2011cn, (51).
Tet1 is transcriptionally and epigenetically regulated by Polycomb complex
The mRNA level of Tet1.2 during cell proliferation can be mediated either by transcriptional or post-transcriptional mechanisms. To elucidate this point, we performed an RT-qPCR analysis on the heterologous nuclear RNA (hnRNA) of Tet1.2. In actively proliferating MEFs, we observed a decline of the Tet1.2 hnRNA demonstrating that the downregulation of the Tet1.2 is a transcriptional event (Figure 4A). In adult tissues, Tet1.2 is a ‘bivalent’ promoter since it shows the repressive H3K27me3 histone modification together with the H3K4me3 mark associated with active transcription (Figure 4B). We performed ChIP analysis for these two epigenetics marks in freshly isolated MEFs and during the first five passages. Although the Polycomb components Suz12 and Ezh2 are slightly downregulated following MEFs culturing (Supplementary Figure S5A), we observed a specific increase of H3K27me3 and reduction of H3K4me3 on the Tet1.2 promoter (Figure 4C) indicating a progressive repression of the promoter. ChIP analysis of Ezh2, the enzymatic subunit of the PRC2 complex responsible for the H3K27 trimethylation, showed a progressive engagement to the Tet1.2 promoter along with MEF passages (Figure 4D) suggesting that Ezh2 recruitment contributes to switch off the transcription driven by the Tet1.2 promoter in proliferating cells. To confirm this hypothesis, we performed the silencing of Suz12, another component of the PRC2 core complex, which has been demonstrated to be required for the correct enzymatic activity of the complex (52,53). Knockdown of Suz12 in MEFs (at P2) resulted in the upregulation of Tet1.2 transcript and protein (Figure 4E and F), with a correspondent decrease of H3K27me3 signal and Ezh2 binding on the promoter (Figure 4G). Importantly, the upregulation of Tet1.2 in Suz12 silenced MEFs correlated with an increase of total 5hmC signal (Figure 4H). The use of GSK343, an Ezh2 inhibitor (54,55) confirmed the results obtained by performing knockdown of Suz12 (Supplementary Figure S5B and D). These results demonstrated a strict connection between PRC2 and Tet1, revealing a link between H3K27me3 histone modification and DNA methylation, two of the major repressive epigenetic features. PRC2 complex and H3K27me3 histone mark are involved and, sometimes, necessary in a notable number of human cancers (56–59). Our results could explain one of the mechanism by which Polycomb proteins can control cell proliferation and chromatin structure during cancer development. For this reason we extended our work also to human system and we checked whether the same mechanisms of Tet1.2 regulation are maintained also in human cells.
Figure 4.
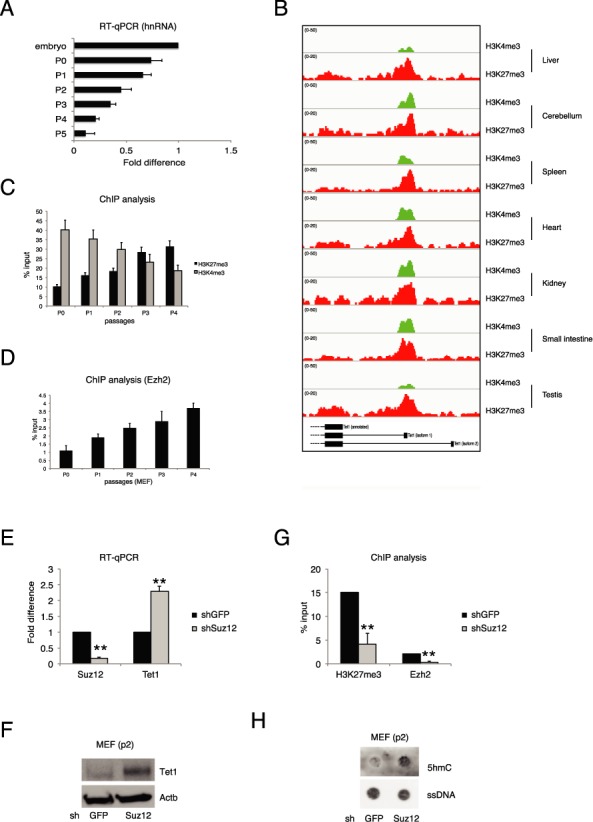
Transcriptional regulation of Tet1.2 in proliferating cells. (A) RT-qPCR of Tet1 hnRNA in MEFs at the indicated passages. (B) Genomic view of the Tet1 promoters for the indicated ChIP-seq analysis in several mouse adult tissues. (C) qPCR of ChIP analysis for H3K4me3 and H3K27me3 in MEFs at the indicated passages. (D) qPCR of ChIP analysis for Ezh2 in MEFs at the indicated passages. (E) RT-qPCR of Suz12 and Tet1 mRNAs in control and Suz12 knockdown MEFs. (F) Western blot analysis of Tet1b in control or Suz12 silenced MEFs. Actb was used as loading control. (G) qPCR of ChIP analysis for H3K27me3 and Ezh2 in MEFs in the indicated knockdown. (H) Dot-blot analysis of 5hmC and 5mC in control or Suz12 silenced MEFs. ssDNA was used as loading control. Error bars represent the standard deviation of three independent experiments. P-value was calculated by using t-test. ** P-value < 0.001.
TET1 is regulated by ESC-specific factors and Polycomb complex also in human cells
To verify whether a similar regulation takes place in human cells we performed RT-qPCR of TET1 mRNA in human tissues, primary cells in culture, including human ESCs (hESCs), hEBs, human umbilical endothelial cells (HUVECs), human microvascular endothelial cells (HMECs), colon and several colon cell lines (Figure 5A). Similarly to the mouse gene, the human TET1 was highly expressed in hESCs and was downregulated during hEBs differentiation. TET1 mRNA was low in human adult tissues such as colon and umbilical cord and it was nearly absent in proliferating primary and transformed cell lines. These results showed an expression pattern similar to what we observed in mouse cells and tissues, suggesting analogous mechanisms of gene regulation. We therefore analyzed RNA-seq and ChIP-seq of histone modifications datasets from hESCs (Figure 5B) (40). These analyses did not identify alternative TET1 isoforms but revealed the presence of an active intragenic enhancer, marked by H3K4me1 and H3K27ac, localized at about 40 kb downstream of the TET1 TSS (Figure 5B). Interestingly, the region sequence surrounding the center of the enhancer contained OCT3/4 and MYC binding motifs (Supplementary Figure S6A). By ChIP analysis we identified a significant binding of OCT3/4, MYC and NANOG proteins on this enhancer (Figure 5C). To validate the functional role of these transcriptional factors on the TET1 transcription level we performed loss of function experiments by using two different shRNAs for OCT3/4, MYC or NANOG (Supplementary Figure S6B–D). Silencing of each of these three factors reduced the level of TET1 mRNA in hESCs (Figure 5D). These results demonstrate that pluripotency-associated transcriptional factors regulate the expression of the TET1 gene in hESCs.
Figure 5.
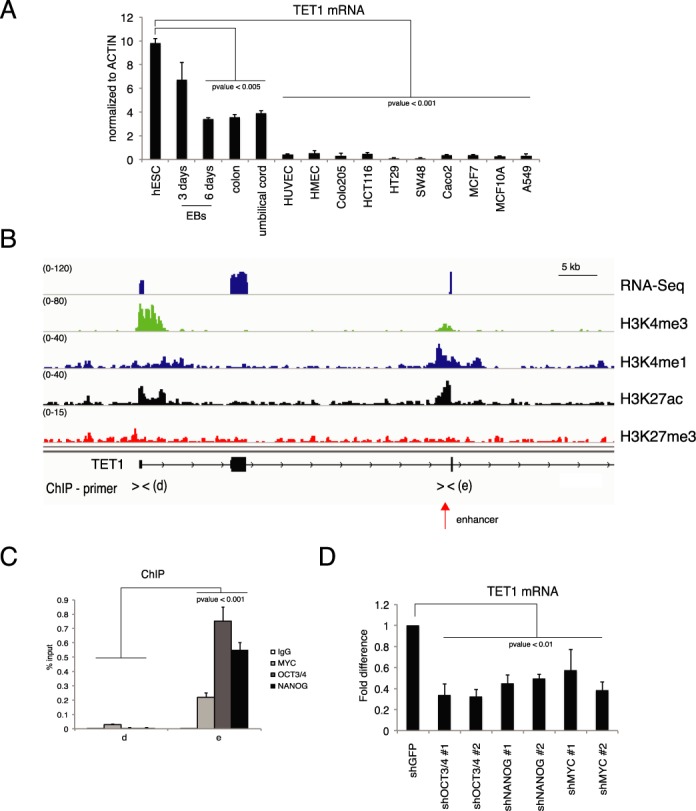
Regulation of TET1 in human embryonic stem cells. (A) RT-qPCR of TET1 mRNA in the indicated samples. (B) Genomic view of RNA-seq and of the indicated ChIP-seq in human ESCs. (C) qPCR of ChIP analysis of the indicated transcriptional factors on the TET1 promoter in human ESCs. (D) RT-qPCR of TET1 mRNA in hESCs silenced for the indicated transcriptional factors. Error bars represent the standard deviation of three independent experiments. P-value was calculated by using t-test.
Next we analyzed the expression of TET1 in differentiated primary cells. To this end we isolated HUVECs and we measured the TET1 mRNA level during the cell culturing. We observed that, similar to MEFs, in HUVECs TET1 was downregulated by cell passages (Figure 6A). Accordingly, we also observed a progressive reduction of 5hmC (Figure 6B). Analogously to the mouse model, the reduced levels of TET1 hnRNA confirmed that TET1 downregulation was triggered by transcriptional mechanisms (Supplementary Figure S6E). We then analyzed the chromatin status of TET1 promoter in a subset of primary cells and we confirmed the presence of the repressive H3K27me3 signal (Figure 6C). Moreover, we performed ChIP analysis on the TET1 promoter in proliferating HUVECs and we detected a progressive reduction of H3K4me3 mark coupled to an increase of H3K27me3 mark and EZH2 binding (Figure 6D and E). These results demonstrate that, although the gene structures are different, both in human and mouse ESCs TET1 is highly expressed in stem cells where it is regulated by stemness-specific factors, while in differentiated cells TET1 is downregulated by cell growth via the PRC2 complex through the deposition of repressive histone mark H3K27me3 on its promoter.
Figure 6.
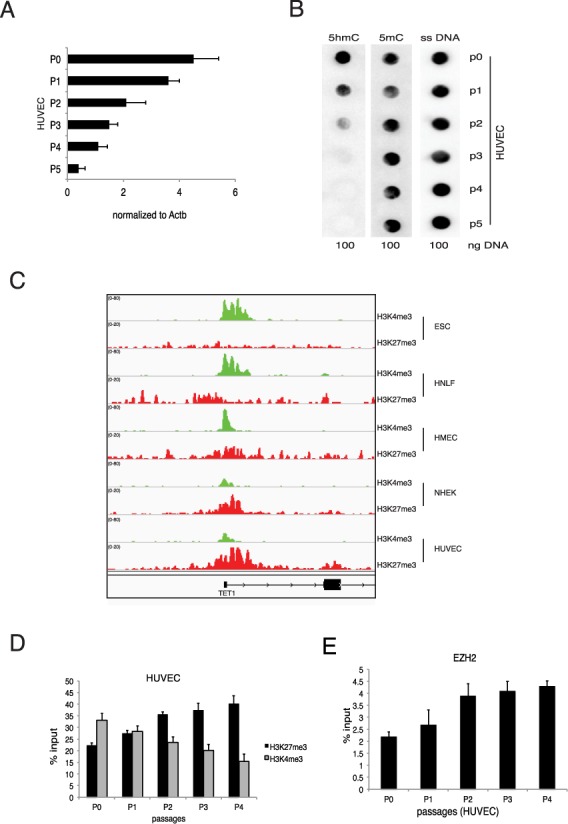
Regulation of TET1 in human adult cells. (A) RT-qPCR of TET1 mRNA in HUVECs at the indicated passages (B) Dot-blot analysis of 5hmC and 5mC level in HUVECs at the indicated passages. ssDNA was used as a loading control. (C) Genomic view of the TET1 promoter for the indicated ChIP-seq analysis in hESCs and several human differentiated cells. (D) qPCR of ChIP analysis for H3K4me3 and H3K27me3 in HUVECs at the indicated passages. (E) qPCR of ChIP analysis for H3K4me3 and H3K27me3 in HUVECs at the indicated passages. Error bars represent the standard deviation of three independent experiments.
Our experiments showed a regulatory circuit between Tet1 expression and cell proliferation in adult cells by which Tet1 is both controlled and a controller of cell growth while in ESCs the high levels of Tet1 do not alter the growth rate. As stem cells are hypomethylated, it is likely that the different phenotypic outputs depend on the DNA methylation pattern of the cells where Tet1 is expressed.
It was previously observed a reduction of TET1 and 5hmC in cancers (60–62) with the resulting hypermethylation and switch off of tumor suppressor genes (29,31,32). Our results unveiled the interplay between the epigenetic modifications mediated by Polycomb and TET1. These results will improve the understanding of how this epigenetic modification crosstalk contributes to cancer development. After revising this manuscript, it has been published a work that independently demonstrated the presence of the two Tet1 isoforms by 5'-RACE (63).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC) [IG 2011/11982]. Funding for open access charge: AIRC [ IG 2011/11982].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Robertson K.D. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 3.Jones P.A. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 4.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H.G., Oliviero S. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 6.Bagci H., Fisher A.G. DNA demethylation in pluripotency and reprogramming: the role of tet proteins and cell division. Cell Stem Cell. 2013;13:265–269. doi: 10.1016/j.stem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Tan L., Shi Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz L.A., Kota S.K., Feil R. Genome-wide DNA demethylation in mammals. Genome Biol. 2010;11:110. doi: 10.1186/gb-2010-11-3-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohr F., Döhner K., Buske C., Rawat V.P.S. TET Genes: new players in DNA demethylation and important determinants for stemness. Exp. Hematol. 2011;39:272–281. doi: 10.1016/j.exphem.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Guo J.U., Su Y., Zhong C., Ming G.-L., Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimmino L., Abdel-Wahab O., Levine R.L., Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A.C., Rappsilber J., Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent J.J., Huang Y., Chen P.-Y., Feng S., Calvopiña J.H., Nee K., Lee S.A., Le T., Yoon A.J., Faull K., et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neri F., Incarnato D., Krepelova A., Rapelli S., Pagnani A., Zecchina R., Parlato C., Oliviero S. Genome-wide analysis identifies a functionalassociation of Tet1 and Polycomb repressivecomplex 2 in mouse embryonic stem cells. Genome Biol. 2013;14:R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 21.Pastor W.A., Pape U.J., Huang Y., Henderson H.R., Lister R., Ko M., McLoughlin E.M., Brudno Y., Mahapatra S., Kapranov P., et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeifer G.P., Kadam S., Jin S.-G. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenet. Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A., et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccolo F.M., Bagci H., Brown K.E., Landeira D., Soza-Ried J., Feytout A., Mooijman D., Hajkova P., Leitch H.G., Tada T., et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farthing C.R., Ficz G., Ng R.K., Chan C.-F., Andrews S., Dean W., Hemberger M., Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y., et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Hu X., Zhang L., Mao S.-Q., Li Z., Chen J., Zhang R.-R., Wu H.-P., Gao J., Guo F., Liu W., et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu C.-H., Peng K.-L., Kang M.-L., Chen Y.-R., Yang Y.-C., Tsai C.-H., Chu C.-S., Jeng Y.-M., Chen Y.-T., Lin F.-M., et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 30.Sun M., Song C.-X., Huang H., Frankenberger C.A., Sankarasharma D., Gomes S., Chen P., Chen J., Chada K.K., He C., et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song S.J., Poliseno L., Song M.S., Ala U., Webster K., Ng C., Beringer G., Brikbak N.J., Yuan X., Cantley L.C., et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neri F., Dettori D., Incarnato D., Krepelova A., Rapelli S., Maldotti M., Parlato C., Paliogiannis P., Oliviero S. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene. 2014 doi: 10.1038/onc.2014.356. doi:10.1038/onc.2014.356. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6:278–284. doi: 10.1016/j.celrep.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neri F., Zippo A., Krepelova A., Cherubini A., Rocchigiani M., Oliviero S. Myc regulates the transcription of the prc2 gene to control the expression of developmental genes in embryonic stem cells. Mol. Cell. Biol. 2012;32:840–851. doi: 10.1128/MCB.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galvagni F., Cantini M., Oliviero S. The utrophin gene is transcriptionally up-regulated in regenerating muscle. J. Biol. Chem. 2002;277:19106–19113. doi: 10.1074/jbc.M109642200. [DOI] [PubMed] [Google Scholar]
- 36.Galvagni F., Lestingi M., Cartocci E., Oliviero S. Serum response factor and protein-mediated DNA bending contribute to transcription of the dystrophin muscle-specific promoter. Mol. Cell. Biol. 1997;17:1731–1743. doi: 10.1128/mcb.17.3.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Incarnato D., Neri F., Diamanti D., Oliviero S. MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets. Nucleic Acids Res. 2013;41:8421–8433. doi: 10.1093/nar/gkt629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krepelova A., Neri F., Maldotti M., Rapelli S., Oliviero S. Myc and max genome-wide binding sites analysis links the myc regulatory network with the polycomb and the core pluripotency networks in mouse embryonic stem cells. PLoS One. 2014;9:e88933. doi: 10.1371/journal.pone.0088933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Incarnato D., Krepelova A., Neri F. High-throughput single nucleotide variant discovery in E14 mouse embryonic stem cells provides a new reference genome assembly. Genomics. 2014;3:6–7. doi: 10.1016/j.ygeno.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 40.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 42.Loh Y.-H., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 43.Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 44.Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith K.N., Singh A.M., Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartwright P. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 47.Niwa H., Burdon T., Chambers I., Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kidder B.L., Yang J., Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dawlaty M.M., Ganz K., Powell B.E., Hu Y.-C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.-W., Page D.C., et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z., Cai X., Cai C.L., Wang J., Zhang W., Petersen B.E., Yang F.C., Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawlaty M.M., Breiling A., Le T., Raddatz G., Barrasa M.I., Cheng A.W., Gao Q., Powell B.E., Li Z., Xu M., et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neri F., Incarnato D., Krepelova A., Rapelli S., Pagnani A., Zecchina R., Parlato C., Oliviero S. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013;14:R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verma S.K., Tian X., LaFrance L.V., Duquenne C., Suarez D.P., Newlander K.A., Romeril S.P., Burgess J.L., Grant S.W., Brackley J.A., et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med. Chem. Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amatangelo M.D., Garipov A., Li H., Conejo-Garcia J.R., Speicher D.W., Zhang R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle. 2013;12:2113–2119. doi: 10.4161/cc.25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laugesen A., Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Bender S., Tang Y., Lindroth A.M., Hovestadt V., Jones D.T.W., Kool M., Zapatka M., Northcott P.A., Sturm D., Wang W., et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 58.Neff T., Sinha A.U., Kluk M.J., Zhu N., Khattab M.H., Stein L., Xie H., Orkin S.H., Armstrong S.A. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Y.-W., Chen H.-M., Fang J.-Y. Gene silencing by the Polycomb group proteins and associations with cancer. Cancer Invest. 2011;29:187–195. doi: 10.3109/07357907.2010.512605. [DOI] [PubMed] [Google Scholar]
- 60.Jin S.G., Jiang Y., Qiu R., Rauch T.A., Wang Y., Schackert G., Krex D., Lu Q., Pfeifer G.P. 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with idh1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haffner M.C., Chaux A., Meeker A.K., Esopi D.M., Gerber J., Pellakuru L.G., Toubaji A., Argani P., Iacobuzio-Donahue C., Nelson W.G., et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W., Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids. 2011;2011:1–5. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sohni A., Bartoccetti M., Kouerly R., Spans L., Velde J.V., De Troyer L., Pulakanti K., Claessens F., Rao S., Koh K.P. Dynamic Switching of Active Promoter and Enhancer Domains Regulates Tet1 and Tet2 Expression during Cell State Transitions between Pluripotency and Differentiation. Mol. Cell. Biol. 2015;35:1026–1042. doi: 10.1128/MCB.01172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


