Abstract
DNA sequences that can form unusual secondary structures are implicated in regulating gene expression and causing genomic instability. H-palindromes are an important class of such DNA sequences that can form an intramolecular triplex structure, H-DNA. Within an H-palindrome, the H-DNA and canonical B-DNA are in a dynamic equilibrium that shifts toward H-DNA with increased negative supercoiling. The interplay between H- and B-DNA and the fact that the process of transcription affects supercoiling makes it difficult to elucidate the effects of H-DNA upon transcription. We constructed a stable structural analog of H-DNA that cannot flip into B-DNA, and studied the effects of this structure on transcription by T7 RNA polymerase in vitro. We found multiple transcription blockage sites adjacent to and within sequences engaged in this triplex structure. Triplex-mediated transcription blockage varied significantly with changes in ambient conditions: it was exacerbated in the presence of Mn2+ or by increased concentrations of K+ and Li+. Analysis of the detailed pattern of the blockage suggests that RNA polymerase is sterically hindered by H-DNA and has difficulties in unwinding triplex DNA. The implications of these findings for the biological roles of triple-stranded DNA structures are discussed.
INTRODUCTION
In addition to the canonical double helix described by Watson and Crick over 60 years ago (1), DNA can adopt a number of non-canonical conformations (reviewed in (2)). These unusual DNA structures appear to play important roles in major DNA transactions (reviewed in (3)). Of particular interest are the effects of these structures on transcription, as this can influence gene expression and trigger a number of regulatory responses (reviewed in (4)).
Among the DNA structures that affect transcription are DNA triplexes, formed when the Watson–Crick DNA duplex accumulates a third strand via Hoogsteen-type hydrogen bonding. Because only purines are capable of participating in both Watson–Crick and Hoogsteen-type hydrogen bonding, triplex formation requires sequences in which one strand contains a sufficiently long stretch of purines (while the complementary strand contains a long stretch of pyrimidines). These are usually referred to as homopurine-homopyrimidine sequences. There are two major types of triplexes, Hoogsteen and reverse Hoogsteen, in which the third strand interacts with the purine strand within the duplex via Hoogsteen or reverse Hoogsteen base pairing, respectively. The first type usually contains protonated cytosines in the homopyrimidine third strand, which causes it to depend upon acidic pH for stabilization, while the second type usually has a homopurine third strand and is very stable under physiological pH and ionic conditions. Because of that, the reverse Hoogsteen triplex is a more likely candidate for biologically relevant roles (for detailed review of various types of triplexes and their properties see (5)).
One way to form a triplex at a homopurine-homopyrimidine DNA region is to add an appropriate DNA (or its analog) oligonucleotide to serve as the third strand of the triplex. These intermolecular triplexes are widely recognized as potential tools for artificial gene regulation, mutagenesis and other genetic manipulations (reviewed in (6,7)). Specifically, they have been shown to arrest transcription elongation such that the blockage occurs as RNA polymerase (RNAP) attempts to enter the triplex-forming region (8,9). This observation is easy to explain, given that binding of the third strand additionally stabilizes the Watson–Crick duplex, making it difficult for RNAP to unwind the three-stranded region.
H-DNA is an intramolecular DNA triplex, which does not require an ‘external’ third strand, (reviewed in (5)). It is formed by two adjacent homopurine-homopyrimidine DNA stretches that are mirror images of one another (H-palindrome), where one stretch serves as an acceptor and another as a donor of the third strand (Figure 1A). During H-DNA formation, the donor stretch dissociates into two single strands. One strand winds back down the major groove of the acceptor stretch forming the triplex via Hoogsteen-type base pairing, while the other remains single-stranded. Both Hoogsteen and reverse Hoogsteen versions of H-DNA have been experimentally detected, and several nomenclatures to distinguish these two isoforms of H-DNA were suggested (reviewed in (5)). Here we have only studied the reverse-Hoogsteen structure (see below), referring to it as ‘H-DNA’. The net energetic balance of base pairing (i.e. formation of the reverse-Hoogsteen base pairs within the acceptor region versus disruption of the Watson–Crick base pairing within the donor region together with the base pairing disruptions and distortions in the center and at the borders of the H-palindrome) is in favor of B-DNA. Therefore, in the absence of additional factors facilitating H-DNA formation, the equilibrium strongly shifts toward B-DNA, rendering the B-to-H transition virtually undetectable under these conditions. One factor that strongly shifts the equilibrium in favor of H-DNA is negative supercoiling. This is because the formation of H-DNA is topologically equivalent to unwinding the entire H-palindrome and this unwinding partially ‘absorbs’ negative superhelical tension (reviewed in (5)). Therefore, the presence of negative supercoiling favors B-H transitions even in solution conditions that otherwise favor B-DNA.
Figure 1.
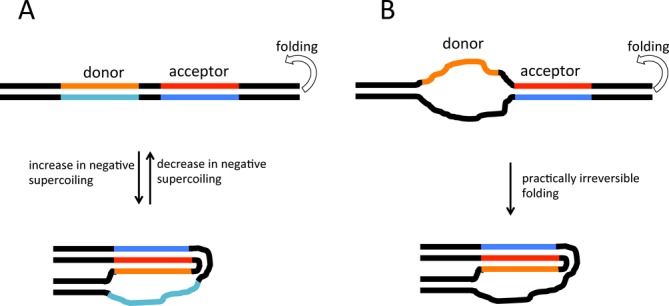
H-DNA and its stable analog. (A) H-DNA formation by completely complementary double-stranded sequences. Homopurine strand in the acceptor part of the H-palindrome, which would become a central strand of H-DNA triplex, is shown in red; its complementary homopyrimidine strand is shown in blue; homopurine strand from the donor part of H-palindrome, which would be donated to a triplex is shown in orange; its Watson–Crick complement is shown in turquoise; flanking sequences are in black. The donor and acceptor parts of the H-palindrome could be separated by several base pairs of arbitrary sequence. In the process of H-DNA formation, the donor part of an H-palindrome unwinds and donates one of its strands to the triplex. Usually, the energy of triplex formation is not sufficient to compensate the melting of duplex regions required for triplex formation. Consequently, transition to H-DNA requires additional energetic input from negative supercoiling. (B) Stable H-DNA analog. Strands in the donor part of the H-palindrome (as well as in the middle, and a few nucleotides at the flank) are not complementary, eliminating the energetically unfavorable step of duplex melting from the triplex formation process. Consequently, H-DNA is exceptionally stable and its formation is practically irreversible.
A priori, one would expect H-DNA to affect transcription similarly to intermolecular triplexes: RNA polymerase pausing upstream of the H-palindrome when it bumps into the triplex. Surprisingly, the actual blockage pattern is just the opposite: blockage occurs downstream of an H-palindrome (10,11). To explain this paradox, it was hypothesized that H-DNA is formed behind the transcribing RNA polymerase owing to an increased level of negative supercoiling upstream of the enzyme (12). As a result, RNA polymerase becomes sterically sequestered at the downstream flank of the H-palindrome (10,11). Since disruption of mirror symmetry in the H-palindrome destabilizes H-DNA, this model predicts that it would reduce transcription blockage as well. This was indeed observed for the short, imperfect H-palindrome from the c-myc gene promoter (13).
Surprisingly, for longer non-interrupted homopurine-homopyrimidine sequences, disruption of mirror symmetry does not affect the blockage signal (14). It has been proposed that the principal cause for transcription blockage in these cases is the formation of R-loops behind elongating RNA polymerase—a process that favors homopurine runs in the coding strand, but that does not require mirror symmetry (14). Thus, the contribution of H-DNA to transcriptional blockage could vary depending on the composition of a homopurine-homopyrimidine sequence. Note that the R-loop and H-DNA models of transcription blockage are not mutually exclusive: an H-DNA-like structure could serve as an intermediate in the process of R-loop formation (15). The fact that G-rich, homopurine-homopyrimidine sequences during transcription can also form quadruplexes ((16), reviewed in (17)), which are known to inhibit transcription (18,19), additionally complicates these interpretations. Given that H-DNA was detected in vivo and has been proposed to regulate transcription (reviewed in (20); also see Discussion), it is of prime interest to understand its interference with the transcription machinery. To clarify the effects of H-DNA on transcription, it is useful to study a model DNA template containing this structure in a ‘frozen’ state. The formation of B-DNA in the donor region is the main H-DNA destabilizing factor at an H-palindrome. To preclude this, we designed a stable analog of H-DNA in which DNA strands comprising the donor region and the center of the H-palindrome are rendered non-complementary (Figure 1B) and cannot form a duplex to compete with triplex formation. Triplex formation in this case does not require negative supercoiling, and the transcribing RNA polymerase encounters a preformed stable triplex. This experimental system allows us to directly determine whether the presence of H-DNA in the transcriptional template blocks RNA polymerase and to learn the mechanisms of triplex-mediated transcription blockage. We found that RNA polymerase is sterically hindered by H-DNA and has severe problems in unwinding the triple helical part of this structure.
MATERIALS AND METHODS
Preparation of DNA substrates
Supplementary Figure S1 illustrates the general strategy for the preparation of DNA substrates, similar to that described previously (21,22). Briefly, a double-stranded DNA fragment containing the T7 promoter was obtained by EcoRI and BsgI restriction digest of the pG4 plasmid (14) followed by its ligation to various triplex-and duplex-containing templates, which were obtained by annealing of synthetic oligonucleotides (see Supplementary Materials for details).
In vitro T7 transcription assays
T7 RNA polymerase transcription reaction and its product analysis were performed as previously described (e.g. (21,22)). Briefly, DNA substrates were transcribed in the presence of non-radioactive NTPs mixed with radioactive CTP, and radioactive transcripts were then analyzed on sequencing gel. The longest transcript (run-off product, RO) results from unperturbed transcription, while transcription blockage produces various truncated transcripts (see Supplementary Materials). Transcription was performed under the standard conditions: 6 mM MgCl2, 8.3 mM NaCl, 37.8 mM Tris HCl (pH 7.9), 1.7 mM Spermidine, 4.2 mM DTT, or in special buffers. Mn-buffer contained 1 mM MgCl2 and 2 mM MnCl2 instead of 6 mM MgCl2; K-buffer contained 80 mM KCl instead of 8.3 mM NaCl, and Li-buffer contained 80 mM LiCl instead of 8.3 mM NaCl.
Restriction protection assay
The restriction protection assay was performed as described in (14). It exploits the presence of a BsgI cleavage site inside the triplex-forming sequence, since formation of a triplex inhibits BsgI digest (Supplementary Figure S2 and Supplementary Materials).
DNA melting experiments
DNA melting experiments were performed as previously described (23) (Supplementary Figure S5 and Supplementary Materials).
RESULTS
T7 transcription is arrested by triplex-forming sequences
We have designed an artificial triplex-forming H-palindrome sequence, in which the two DNA strands of the donor half are non-complementary and cannot form a duplex. This ensures that this intramolecular triplex is thermodynamically stable, even in the absence of negative supercoiling (Figure 1). As a basic triplex-forming motif, we have chosen a sequence comprised of CGG and TAT triads (Figure 2A). These sequences have been previously studied and shown to form a very stable triplex that blocks DNA replication in vitro ((23) and references therein). These templates were obtained by annealing of specific synthetic oligonucleotides in different combinations, followed by their ligation to a DNA fragment containing the promoter for T7 RNA polymerase (Supplementary Figure S1).
Figure 2.
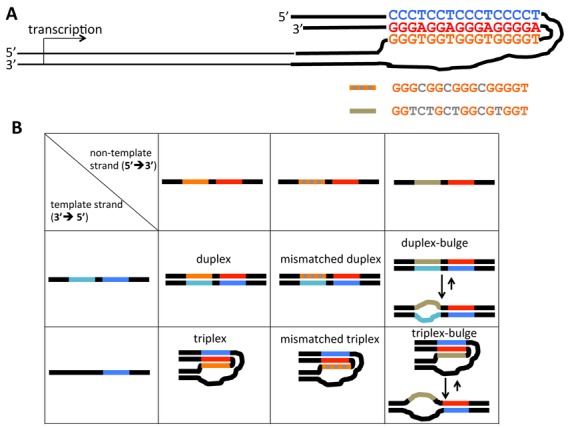
DNA substrates used in transcription experiments. (A) General scheme of a transcription template containing a perfect YR*R triplex. It consists of the promoter-containing fragment (thin black lines) ligated to a triplex-forming construct pre-assembled from synthetic DNA oligonucleotides (the color-coding is the same as in Figure 1). Modifications of the third strand within the triplex are shown in gray underneath. A sequence with three T-to-C point substitutions is called ‘mismatched’, a sequence with seven point substitutions, which efficiently prevents duplex or triplex formation, is referred to as ‘bulged’. (B) Various triplex- and duplex-forming constructs studied. T-to-C substitutions in mismatched substrates are shown as gray squares; highly mismatched sequences in ‘bulge’ substrates are shown in yellow-gray.
Figure 2B shows the expected structures for various experimental and control templates used in our transcription experiments. When both strands are completely complementary, the substrate is predicted to form a perfect Watson–Crick duplex (first column, first row) in the absence of DNA supercoiling. When the homopyrimidine run in the donor part of the sequence is replaced by a random sequence (first column, second row), a stable (‘perfect’) triplex is predicted to form.
We also used templates, in which three thymines were replaced by cytosines in the donor region, resulting in a mismatched duplex and triplex (second column, first and second rows, respectively), as well as templates with four guanine-to-thymine substitutions along with the three T-to-C substitutions (third column; exact sequence is in the scheme at the top). The latter templates (designated as ‘duplex’ and ‘triplex’ bulges) contained seven mismatches in a sixteen bp-long sequence and are not expected to form stable duplexes or triplexes within the mismatched area. Formation of triplexes in these transcription templates was monitored by protection against BsgI digestion (Supplementary Figure S2). As expected, the perfect triplex, but not the triplex bulge, protects the acceptor duplex against BsgI digestion. In the case of the mismatched triplex, a partial protection from BsgI digestion was observed, suggesting that the mismatches do not abolish the triplex formation completely, likely owing to the fact that they do not perturb the stabilizing GGC triads.
Because oligonucleotides within these constructs were not phosphorylated, an unsealed nick remains in the non-template strand between the promoter fragment and the structure-forming sequence. We have previously shown that a nick in this position does not affect the results of our transcription assay (21). Typical results of in vitro transcription for these templates are shown in Figure 3A. The most intense signal, between 320 and 330 nucleotides, corresponds to the full length ‘run off’ RNA product (designated ‘RO’), which forms when the polymerase reaches the end of the template. The second most intense signal, at ∼240 nucleotides, corresponds to the ‘run off’ from those promoter fragments that did not ligate to the structure-forming oligonucleotide construct (see Supplementary Figure S1). Bands between these two signals correspond to truncated transcription products resulting from transcription blockage, and are referred to as ‘blockage signals’.
Figure 3.
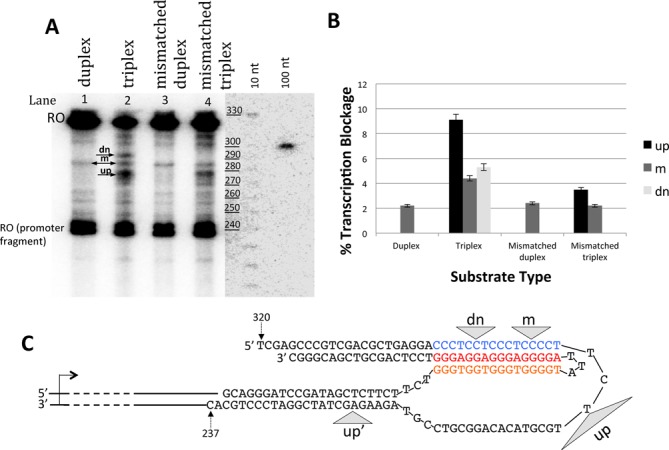
T7 RNAP transcription is arrested by triplexes. (A) Gel analysis of transcription products at standard conditions. The most pronounced blockage bands (designated as ‘up’, ‘m’ and ‘dn’ are marked by arrows. DNA size standards (10 and 100 nt ladders) are shown at right. (B) Quantification of the results from panel (A). Each construct was transcribed at least twice, and the columns correspond to average blockage intensity for different constructs at different positions along the template. Error bars show standard deviations. (C) Mapping of the blockage sites in H-DNA structure obtained by interpolation from the reference size ladder. Blockage sites are shown by gray triangles. Signals ‘up’, ‘m’ and ‘dn’ are seen at all conditions (e.g. Figure 3A), while the additional signal ‘up’ appears only in the presence of Mn2+ ions (see Figure 4). Triangle width roughly corresponds to the resolution of the mapping (about 5 nt).
In the perfect duplex template (Figure 3A, lane 1), there are low intensity blockage signals, slightly above background. The most prominent of these blockages is the ‘m-signal’ at the middle of the synthetic sequence. We believe that these signals simply correspond to sequences for which the probability of transcription termination is above average. In the case of the triplex-bearing template (Figure 3A, lane 2), at least two significantly stronger blockage signals appear: one upstream (up), and another downstream (dn) from the m-signal. Furthermore, blockage at the m-signal position becomes more pronounced relative to the run off product (Figure 3B). For triplex-forming templates overall, truncated transcripts constitute 19% of total transcripts, with roughly half of those corresponding to the upstream signal (see Methods for calculation). Figure 3C shows the results of mapping of the most prominent blockage positions. We found that the up-signal corresponds to the junction between the single-stranded part of the template strand and the triplex, while m- and dn-signals are within the triplex-forming region. In addition, the triplex seems to exacerbate minor blockage signals over the entire oligonucleotide construct as compared to the duplex, including areas outside the region directly involved in H-DNA formation. This could be caused by extended RNA-DNA hybrid formation and by collisions between stalled and elongating RNA polymerases (see Discussion).
To check whether a destabilization of the triplex would reduce transcription blockage, we replaced thymines in the third strand with cytosines, which cannot form Hoogsteen base pairs with adenines. As expected, transcription blockage at this mismatched triplex was significantly weaker (Figure 3A and B, lane 4 versus lane 2). These substitutions, however, did not abolish transcription blockage completely, consistent with only partially impaired triplex formation, as evident from the BsgI protection assay in Supplementary Figure S2. Furthermore, transcription reactions performed under the same ambient conditions as the restriction protection assay resulted in transcription pattern similar to that observed in our standard conditions (Supplementary Figures S3 and S4).
As expected, mismatched duplex constructs produced virtually the same transcription pattern as the respective normal duplex (Figure 3A and B, lane 3 versus lane 1).
Transcription blockage is sensitive to bivalent cations in the reaction
Bivalent cations are known to differentially affect the stability of pyrimidine-purine-purine DNA triplexes (24,25). Specifically, these triplexes are more stable in the presence of manganese than magnesium (24). To confirm our hypothesis that the strength of transcriptional blockage is correlated with the stability of triplexes, we conducted a T7 RNA polymerase transcription reaction in the presence of 2mM Mn2+.
As expected, the intensity of triplex-mediated transcriptional blockage increased significantly in the presence of Mn2+(Figure 3A, lane 2 versus Figure 4A, lane 2). In these conditions, truncated transcripts accounted for nearly 90% of total RNA products (see quantitation below Figure 4A) as compared to ∼20% in the presence of Mg2+ (see quantitation in Figure 3B). Furthermore, a new arrest band (designated as ‘up’), which was not observed in standard transcription conditions, appeared approximately 5 bases upstream of the H-DNA boundary (see Figure 3C). This truncated transcript corresponded to 33.5% of the total RNA product. This new blockage signal may have arisen from the collision of an oncoming RNA polymerase with the stalled one at the triplex edge.
Figure 4.
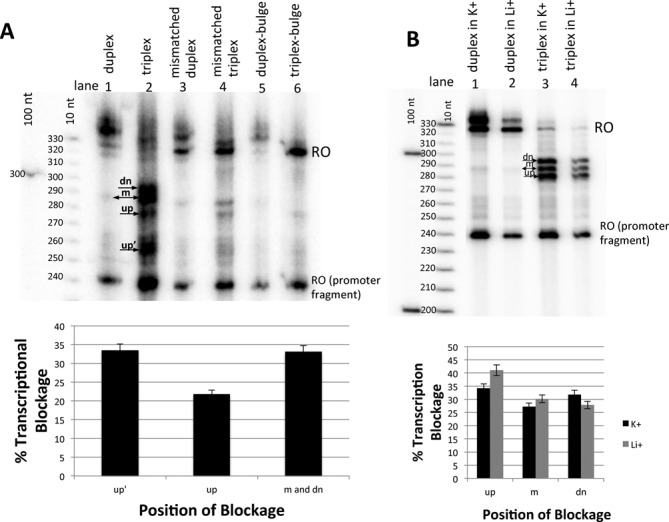
The intensity and pattern of transcription blockages changes with the type of cation in solution. (A) Upper panel. In vitro transcription by T7 RNA polymerase conducted in the presence of 2 mM Mn2+. Note the new product corresponding to a transcript truncated 5 bases upstream of the H-DNA edge (designated ‘up’) in addition to the previously observed ‘up’, ‘m’ and ‘dn’ blockage positions (see the scheme in Figure 3C). Due to poor resolution of the gel, the ‘m’ and ‘dn’ positions form a single blockage band. Full-length products appear at their usual 320 nt location, but also in positions above 330 nt. Lower panel. Quantification of the results in the upper panel reveals that arrest at up’-position constitutes 33.5% of the all RNA transcripts; up-transcripts and (m- + dn-) transcripts account for 21.7 and 33.1% respectively. The full-length product makes only 11.6% of the total product. (B) Upper panel. Triplex-mediated transcription arrests observed at 80 mM K+ or at 80 mM Li+. Lower panel. Quantification of these data reveal that intensities of blockages in quadruplex-favoring (K+) conditions are similar to intensities in conditions (Li+) that do not favor quadruplexes.
In concordance with our results under standard conditions, the mismatched triplex with T-to-C substitutions causes less transcription blockage than the perfect triplex in the presence of Mn2+ (Figure 4A, lane 4 versus lane 2). In fact, the difference between these two constructs is even more pronounced in manganese transcription buffers. The sequence with seven mismatches in the third strand (Figure 4A, ‘triplex bulge’, lane 6) does not give rise to any strong blockage signals (Figure 4A, lane 1). This is consistent with the idea that the triplex is completely destabilized by these mismatches. A more detailed analysis shows that the bulge in the substrate exacerbates weak, diffusive blockage signals in the downstream duplex region, which we explain by the R-loop formation (see Supplementary Figure S3 and its legends).
Note that in the presence of manganese, the run-off product is heterogeneous, i.e. comprised of several bands. This heterogeneity has been occasionally observed for T7 RNA polymerase (26). We suggest that it is caused by sub-standard conditions (the presence of Mn2+) in our transcription reaction.
Transcription blockage is exacerbated by monovalent metal cations.
When a DNA strand contains guanine runs, another non-B DNA structure, called a G- quadruplex, can form (reviewed in (27)). In our transcription reaction, G-quadruplexes could form in either the non-template DNA strand or in the nascent RNA transcript, both of which might contribute to transcription arrest (reviewed in (4)). To evaluate the role of G-quadruplexes in our system, we capitalized upon their dependence on monovalent cations. G-quadruplexes are more readily formed in the presence of potassium (K+) than in the presence of lithium (Li+) ions (28). If G-quadruplexes contribute to transcription blockage in our system, we would expect to observe more truncated transcripts in the presence of K+ than Li+ ions.
We conducted T7 transcription reactions on triplex forming DNA templates in two different buffers, containing either 80 mM K+ or 80 mM Li+ ions. In both conditions, we observed the same pattern of triplex-mediated blockage comprising the three well-pronounced up-, m-, and dn-signals (Figure 4B, lanes 3 and 4). The intensities of blockage signals relative to run-off products were similar in the presence of potassium or lithium (Figure 4B, lower panel). These data suggest that transcription blockage observed in our experiments is not caused by G-quadruplexes.
Though we did not observe significant differences between the effects of Li+ and K+ ions on transcription blockage, both caused strong increase in transcription blockage compared to standard conditions (Figure 4B, lanes 3, 4 versus Figure 3A, lane 2), in which the concentration of monovalent metal cations was about 10-fold lower. This effect could be due to non-specific stabilization of DNA triplexes by increased salt concentration. Alternatively, it may be the result of destabilization of the transcription complex by increased salt concentration ((29) and references therein), which could make RNA polymerase more ‘sensitive’ to obstacles in general.
To check whether our triplex is additionally stabilized by increased salt concentration, we performed DNA melting experiments using self-folding oligonucleotides described in (23) that form triplexes of the same base composition as our transcription substrates (Supplementary Figure S5). It was previously shown that these triplexes melt in one step, i.e. from a triplex to three separate strands (23). Consequently, the presence of a triplex manifests itself as an increase in the melting temperature compared to that of the corresponding duplex DNA. Melting curves in Supplementary Figure S5 confirm triplex formation under our standard transcription conditions. In fact, the triplexes were so stable in these conditions that the midpoints of their melting curves (Tms) were technically impossible to reach. We did observe, however, an increase in absorbance with increasing temperature, indicative of partial triplex dissociation. The rates of these absorbance increases were similar in both standard and high salt conditions, implying that our triplex structures are not additionally stabilized by increased salt concentration. That might seem surprising, given that the increase in salt concentration is expected to stabilize nucleic acid complexes by screening of the repulsion between negatively charged phosphates. However, in our case, triplexes are pre-formed in the presence of bivalent (magnesium) cations, which are much more potent DNA binders than monovalent cations. Therefore, the presence of additional monovalent ions does not produce the apparent increase in triplex stability. Also note that in certain cases, triplex formation could be suppressed by monovalent cations. For example, usual H-DNA formation requires higher negative supercoiling at higher concentration of sodium salt (30), and intermolecular pyrimidine-purine-purine triplex formation is inhibited by potassium (31–33). Both these effects are likely to be due to stronger stabilization of the structure competing with the triplex formation: B-DNA in the donor half of the H-palindrome in the first case, and G-quadruplex in the triplex-forming oligonucleotide in the second case (33). In our case, competition with B-DNA is excluded by design of the system (Figure 1) and quadruplex formation in the Hoogsteen strand is avoided by pre-forming the triplex in the absence of monovalent cations.
Therefore, high salt conditions do not have an apparent effect upon the triplex in our system; thus, we believe that the increase in transcription blockage observed in high salt conditions is likely due to destabilization of the transcription complex (29), rendering it particularly prone to blockage by local DNA structures.
Orientation of a DNA triplex changes the pattern of transcription blockage
In our initial transcription experiments, elongating T7 RNA polymerase transcribes the strand that does not participate in the formation of reversed Hoogsteen base pairs. To further investigate the mechanism of transcriptional arrest in DNA triplexes, we created a ‘reverse triplex’ template. In this template, the orientation of the promoter was reversed such that RNA polymerase transcribes a central strand of the triplex, which is involved in both Watson–Crick and Hoogsteen base pairing (Figure 5A). Since this would require T7 RNA polymerase to unwind the DNA triplex as a whole, this configuration was expected to result in a stronger triplex-mediated blockage of transcription. To ensure maximum stability of the reverse triplex, transcription assays with this template were performed in the presence of Mn2+.
Figure 5.
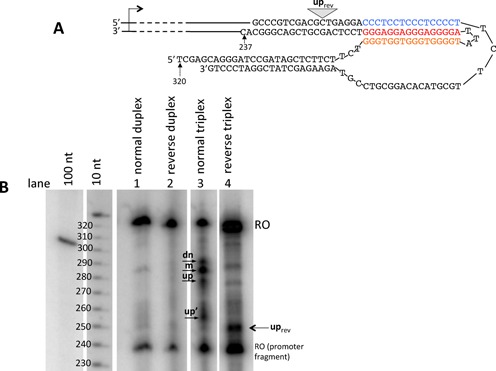
Pattern of transcriptional blockages changes upon changing the orientation of triplex. (A) Schematic representation of the ‘reverse triplex’ showing triplex-mediated transcription blockage sites. (B) Gel analysis of transcription products. Transcription of the reversed triplex construct was conducted in the presence of Mn2+ ions. The ‘reverse’ triplex exhibits one strong truncated transcript at 250 bp, a position corresponding to a few bases upstream of the triplex/duplex junction.
Figure 5B (lane 4) shows a well-pronounced blockage (uprev) that occurs at a position adjacent to the edge of three-stranded region in the reverse triplex (Figure 5A). The remaining blockage signals appear to be superficial for this construct. These data suggest that when RNA polymerase bumps into the reverse triplex, it dismantles it via an ‘all-or-none’ mechanism. This is in contrast to the more subtle process of dismantling the opposite orientation triplex (Figure 5B, lane 3), in which RNA polymerase can progress inside the triplex zone more easily. Surprisingly, the blockage caused by the reverse triplex was unexpectedly small: about 12% of the total run-off product, as compared to the 30% blockage observed for the m-signal alone in the original orientation (see Discussion).
Removal of steric strain in a triplex simplifies the pattern of transcription blockage
In addition to the difficult-to-unwind triplex, H-DNA may also inhibit transcription by sterically sequestering RNAP in sharply bent elements of the structure (see Discussion). This type of steric sequestration was invoked to explain transcription blockage at the end of homopurine-homopyrimidine runs (10,11). To test if steric blocks contribute to triplex-mediated transcription blockage, we designed a structure called a ‘cut triplex’, which contained a break at the duplex-to-triplex junction in the non-template strand of our original triplex structure. This break relieved the steric strain associated with the bulky H-DNA structure. Upon transcription of the cut triplex, the most promoter-proximal up-band disappears (Figure 6B, lane 3 versus lane 2). This suggests that this band could be due to a steric obstacle to transcription (see Discussion).
Figure 6.
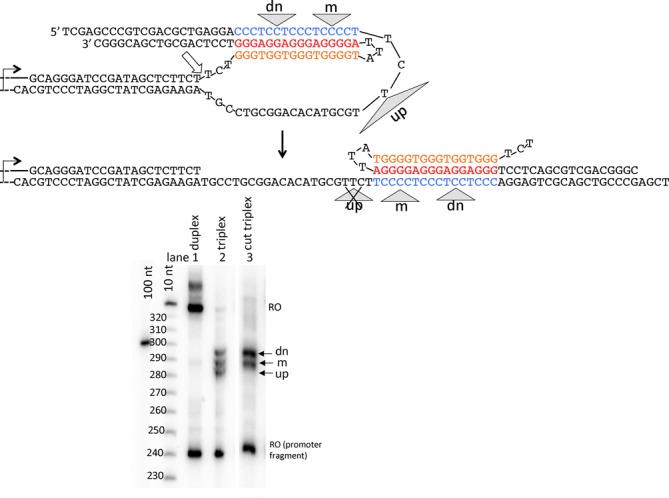
Removal of steric strain from intramolecular triplex modifies the blockage pattern. (A) The ‘cut triplex’ was designed from the standard triplex structure by introducing a break at the duplex/triplex junction (indicated by a white block arrow) to release the steric tension. (B) Transcription of cut and standard triplexes, as well as duplex controls were performed at K+ conditions. While a regular triplex (lane 2) shows blockage signals at up, m and dn positions, in the cut triplex (lane 3) the up-signal disappears.
DISCUSSION
To elucidate the effect of H-DNA on transcription, we created synthetic analogs that fold into an intramolecular triplex configuration regardless of supercoiling and studied transcription by T7 RNA polymerase through these structures. Depending on specific design, RNA polymerase could first encounter either a donor, or an acceptor part of the H-palindrome (designated (d→a), or (a→d), respectively) with either a pyrimidine (y), or a purine (r) strand as the transcriptional template. In total, one can imagine four different transcriptional substrates (Figure 7); of which we studied two—(d→a)-y and (a→d)-r in this work. Note that the (d→a)-y configuration is similar to the one responsible for ‘suicidal replication’ (34) as well as to H-DNA that might be induced by transcription (10,12,14). It also mimics the orientation of many natural H-palindromes, including that found in the c-myc promoter (e.g. (13) and references therein). Furthermore, H-DNA formation in this orientation could compete with the formation of R-loops and/or G-quadruplexes in the non-template strand during transcription.
Figure 7.
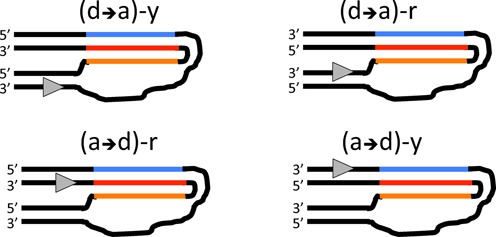
Possible orientations of H-DNA relative to direction of transcription. RNA polymerase (shown as gray triangle on the template strand pointing in the direction of elongation) can either proceed from a donor part of the H-palindrome toward its acceptor part (d→a), or from an acceptor half to the donor (a→d) using either homopyrimidine (y), or homopurine (r) strands as templates. This results in four possible combinations of transcription substrates. Only substrates from the first column were studied in the current work.
The simplest pattern of transcription blockage was observed for the (a→d)-r construct (Figure 8A): a well-pronounced blockage signal at the duplex-to-triplex junction. This pattern is likely due to RNA polymerase ‘bumping’ into the triplex and being unable to unwind it (scheme in dashed-bordered box below). In this orientation, the template strand for transcription is involved in both Watson–Crick and reverse Hoogsteen base pairing, requiring RNA polymerase to disrupt the entire triplex in order to pass through. This presents a much higher energetic barrier for the RNAP as compared to a construct in which the template strand participates in Watson–Crick base pairing only ((d→a)-y). That being said, the two displaced strands do not pair in the (a→d)-r structure resulting in a larger entropic barrier for triplex reformation than in the (d→a)-y structure (Figure 8B), where the displaced strands could remain base paired via reverse Hoogsteen interactions (35,36). Thus, when the template strand participates in both Watson–Crick and reverse Hoogsteen base pairing, it is more difficult to unwind the triplex, but it is also more difficult to reverse the unwinding once it is initiated. That could explain efficient RNAP passage through the (a→d)-r structure, once the triplex is unwound.
Figure 8.
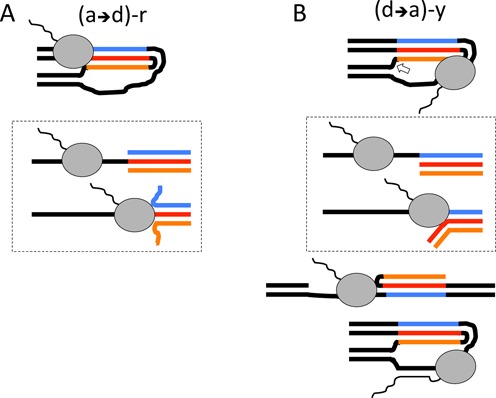
Possible mechanisms of transcription blockage by stable H-DNA analog in various orientations. (A) The simplest mechanism of blockage is expected for (a→d)-r configuration, in which RNA polymerase (RNAP) (gray oval) is bumping into the triplex and is unable to unwind it. In this case, the template strand (red) is involved in Watson–Crick and Hoogsteen interactions and RNA polymerase has to disrupt the triplex as a whole (see the scheme in the dashed line-border box below). At the same time, two displaced strands are not pairing with each other resulting in the high entropic barrier for triplex re-formation. (B) The mechanism of blockage in the case of (d→a)-y configuration is more complex: on one hand, the energetic barrier for the triplex dismantling is lower, since only Watson–Crick interactions have to be disrupted to make the template available; on the other hand Hoogsteen interactions at the displaced strands could persist, decreasing the entropic barrier for triplex reformation (see the scheme in the dashed line-border box below). Altogether, it is easier for an elongating RNAP to unwind the triplex in (d→a)-y than in (a→d)-r construct (in Figure 8, A), but the probability that it would be ‘pushed back’ by triplex re-formation is greater in (d→a)-y construct. In the (d→a)-y configuration, RNAP could be sequestered by sterical constraints even before it started to unwind the triplex. This sequestration is gone if the H-DNA-like structure is cut at its triplex-duplex junction (bottom). Finally, hybridization between the nascent RNA and the template strand (bottom) could also contribute to the blockage in the (d→a)-y configuration.
The pattern of transcription blockage is more complex in the (d→a)-y construct (Figure 8B). RNAP could be sterically sequestered at the bent parts of this H-DNA structure (10,11) even before it faces the triplex per se. Furthermore, the nascent RNA could re-hybridize with the single-stranded part of the template strand, forming an extended RNA-DNA hybrid (R-loop), which could destabilize the transcription complex (37). Transcription through this construct generates three blockage signals (Figure 3A): the most upstream one is at the junction between the single- and three-stranded DNA, while the two other signals are inside the triplex-forming region. The upstream and downstream bands are only evident in triplex-forming constructs; the middle band is seen in all constructs, but is much more intense in triplex-forming constructs. We conclude that these blockages are either caused by or exacerbated by H-DNA. To test whether the blockages are due to a triplex per se or to a sterical impediment to transcription by bent elements of H-DNA, we produced a construct called ‘cut triplex’, in which the connection between the flanking duplex and the third strand of H-DNA is severed (white block arrow in Figure 8). In this construct, steric constraints are removed while the overall H-DNA structure is upheld. Transcription through this structure did not produce the most upstream blockage signal, while the middle and the downstream signals remained intact (Figure 6B). Thus, the upstream signal appears to be due to steric constraints, while the other two signals are caused by the triplex per se. Supporting this conclusion, transcription blockage is additionally enhanced in the presence of manganese ions, which are known to be potent stabilizers of DNA triplexes (24).
Judging from the results for the triplex-bulge and duplex-bulge constructs, the presence of long single-stranded regions does not cause any strong blockages, indicating that single-stranded DNA present in our triplex-forming substrates is not a major cause for transcription blockage. However, in the case of a bulge substrate we did observe some increase of weak, diffusive blockage signals localized in the downstream duplex region (Supplementary Figure S3). Previously, we had suggested that the exacerbation of these weak downstream blockages could be due to the R-loop formation (37). R-loops can easily be formed during transcription of our triplex-forming templates, as they contain single-stranded DNA regions, which might enhance triplex-mediated blockage.
In addition, transcription blockage does not appear to be sensitive to the type of monovalent metal cation (potassium versus lithium) arguing against the role of G-quadruplexes.
Rather unexpectedly, transcription blockage was exacerbated by increased concentrations of monovalent metal cations regardless of their type. Our DNA melting experiment shows that triplex stability is not increased by the increased concentrations of monovalent cations. We believe that monovalent cation-induced blockage is likely due to a destabilization of the transcription complex (29) making it more sensitive to inherent obstacles in DNA templates. The latter observation could be of high biological relevance as the intracellular concentration of monovalent metal ions (∼150 mM) is comparable to that used in our experiments.
Sequences that can form intramolecular triplexes are unusually frequent in genomic DNA. Bioinformatics analyses of the human genome has revealed that long homopurine-homopyrimidine tracts occur once per 50 000 bps (38). Moreover, mirror repeats are overrepresented among these homopurine-homopyrimidine sequences (39). Since H-DNA is the only known structure that requires its sequence motifs to be both homopurine-homopyrimidine and mirror symmetrical, the overrepresentation of mirror repeats among homopurine-homopyrimidine sequences implies some biological function of H-DNA. Clusters of H-palindromes are found in the pseudoautosomal region of the sex chromosomes, which are essential for meiotic segregation and recombination, as well as in genes involved in cell communications in the brain (40). H-palindromes are also common elements of eukaryotic promoters, including c-myc, and have been implicated in the expression of many disease-linked genes ((41) and references therein). Finally, formation of H-DNA has been implicated in transcription blockage by expanded (GAA)n repeats responsible for the human genetic disease Friedreich's ataxia (11).
Transcription blockage mediated by H-DNA-like structures may lead to various biological consequences, including transcription-related genome instability (reviewed in (4)). Of a particular interest is the model of ‘gratuitous’ transcription-coupled repair (gratuitous TCR) ((42), reviewed in (43)), proposing that these structures may be recognized as lesions by the TCR machinery. This could trigger futile cycles of excision and repair replication, eventually causing mutagenesis. Collisions between blocked transcription complexes and DNA replication machinery may further contribute to genomic instability. We can also think of positive biological effects coming from triplex-mediated transcription arrest. A recent report suggests that DNA lesions that do not block transcription could still be repaired by TCR, when a stalled RNA polymerase is present in the vicinity (44). Thus, it is possible that unusual DNA structures arresting transcription could sensitize the TCR machinery to nearby DNA lesions that do not block transcription per se.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Alexander Neil for his helpful editorial comments.
FUNDING
NIH [CA07712 to P.C.H., GM105473 to S.M.M.]; RFBR-DFG [14-04-91343 to A.M.O.]. Funding for open access charge: NIH from the National Cancer Institute to the laboratory of PCH at Stanford University [RO1 CA077712].
Conflict of interest statement. None declared.
REFERENCES
- 1.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin S.M. Discovery of alternative DNA structures: a heroic decade (1979–1989) Front. Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 3.Wang G., Vasquez K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair (Amst.) 2014;19:143–151. doi: 10.1016/j.dnarep.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belotserkovskii B.P., Mirkin S.M., Hanawalt P.C. DNA sequences that interfere with transcription: implications for genome function and stability. Chem. Rev. 2013;113:8620–8637. doi: 10.1021/cr400078y. [DOI] [PubMed] [Google Scholar]
- 5.Mirkin S.M., Frank-Kamenetskii M.D. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 6.Knauert M.P., Glazer P.M. Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet. 2001;10:2243–2251. doi: 10.1093/hmg/10.20.2243. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee A., Vasquez K.M. Triplex technology in studies of DNA damage, DNA repair, and mutagenesis. Biochimie. 2011;93:1197–1208. doi: 10.1016/j.biochi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rando R.F., DePaolis L., Durland R.H., Jayaraman K., Kessler D.J., Hogan M.E. Inhibition of T7 and T3 RNA polymerase directed transcription elongation in vitro. Nucleic Acids Res. 1994;22:678–685. doi: 10.1093/nar/22.4.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannangeli C., Perrouault L., Escude C., Gryaznov S., Helene C. Efficient inhibition of transcription elongation in vitro by oligonucleotide phosphoramidates targeted to proviral HIV DNA. J. Mol. Biol. 1996;261:386–398. doi: 10.1006/jmbi.1996.0471. [DOI] [PubMed] [Google Scholar]
- 10.Grabczyk E., Fishman M.C. A long purine-pyrimidine homopolymer acts as a transcriptional diode. J. Biol. Chem. 1995;270:1791–1797. doi: 10.1074/jbc.270.4.1791. [DOI] [PubMed] [Google Scholar]
- 11.Grabczyk E., Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reaban M.E., Lebowitz J., Griffin J.A. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- 13.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 14.Belotserkovskii B.P., Liu R., Tornaletti S., Krasilnikova M.M., Mirkin S.M., Hanawalt P.C. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabczyk E., Mancuso M., Sammarco M.C. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broxson C., Beckett J., Tornaletti S. Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry. 2011;50:4162–4172. doi: 10.1021/bi2002136. [DOI] [PubMed] [Google Scholar]
- 19.Tornaletti S., Park-Snyder S., Hanawalt P.C. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buske F.A., Mattick J.S., Bailey T.L. Potential in vivo roles of nucleic acid triple-helices. RNA Biol. 2011;8:427–439. doi: 10.4161/rna.8.3.14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas-Rios V., Belotserkovskii B.P., Hanawalt P.C. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neil A.J., Belotserkovskii B.P., Hanawalt P.C. Transcription blockage by bulky end termini at single-strand breaks in the DNA template: differential effects of 5′ and 3′ adducts. Biochemistry. 2012;51:8964–8970. doi: 10.1021/bi301240y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krasilnikov A.S., Panyutin I.G., Samadashwily G.M., Cox R., Lazurkin Y.S., Mirkin S.M. Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 1997;25:1339–1346. doi: 10.1093/nar/25.7.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkov V.A., Voloshin O.N., Soyfer V.N., Frank-Kamenetskii M.D. Cation and sequence effects on stability of intermolecular pyrimidine-purine-purine triplex. Nucleic Acids Res. 1993;21:585–591. doi: 10.1093/nar/21.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potaman V.N., Soyfer V.N. Divalent metal cations upon coordination to the N7 of purines differentially stabilize the PyPuPu DNA triplex due to unequal Hoogsteen-type hydrogen bond enhancement. J. Biomol. Struct. Dyn. 1994;11:1035–1040. doi: 10.1080/07391102.1994.10508050. [DOI] [PubMed] [Google Scholar]
- 26.Nacheva G.A., Berzal-Herranz A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003;270:1458–1465. doi: 10.1046/j.1432-1033.2003.03510.x. [DOI] [PubMed] [Google Scholar]
- 27.Huppert J.L. Structure, location and interactions of G-quadruplexes. FEBS J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 28.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 29.Sastry S.S., Hearst J.E. Studies on the interaction of T7 RNA polymerase with a DNA template containing a site-specifically placed psoralen cross-link. I. Characterization of elongation complexes. J. Mol. Biol. 1991;221:1091–1110. [PubMed] [Google Scholar]
- 30.Lyamichev V.I., Mirkin S.M., Frank-Kamenetskii M.D. Structure of (dG)n.(dC)n under superhelical stress and acid pH. J. Biomol. Struct. Dyn. 1987;5:275–282. doi: 10.1080/07391102.1987.10506393. [DOI] [PubMed] [Google Scholar]
- 31.Cheng A.J., Van Dyke M.W. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993;21:5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milligan J.F., Krawczyk S.H., Wadwani S., Matteucci M.D. An anti-parallel triple helix motif with oligodeoxynucleotides containing 2′-deoxyguanosine and 7-deaza-2′-deoxyxanthosine. Nucleic Acids Res. 1993;21:327–333. doi: 10.1093/nar/21.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivas W.M., Maher L.J., 3rd Competitive triplex/quadruplex equilibria involving guanine-rich oligonucleotides. Biochemistry. 1995;34:278–284. doi: 10.1021/bi00001a034. [DOI] [PubMed] [Google Scholar]
- 34.Samadashwily G.M., Dayn A., Mirkin S.M. Suicidal nucleotide sequences for DNA polymerization. EMBO J. 1993;12:4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panyutin I.G., Kovalsky O.I., Budowsky E.I. A transition from a twice-folded structure to a hairpin in oligo(dG) in the presence of magnesium ions. J. Biomol. Struct. Dyn. 1991;8:967–975. doi: 10.1080/07391102.1991.10507860. [DOI] [PubMed] [Google Scholar]
- 36.Cheng A.J., Wang J.C., Van Dyke M.W. Self-association of G-rich oligodeoxyribonucleotides under conditions promoting purine-motif triplex formation. Antisense Nucleic Acid Drug Dev. 1998;8:215–225. doi: 10.1089/oli.1.1998.8.215. [DOI] [PubMed] [Google Scholar]
- 37.Belotserkovskii B.P., Neil A.J., Saleh S.S., Shin J.H., Mirkin S.M., Hanawalt P.C. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013;41:1817–1828. doi: 10.1093/nar/gks1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroth G.P., Ho P.S. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox R., Mirkin S.M. Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5237–5242. doi: 10.1073/pnas.94.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bacolla A., Collins J.R., Gold B., Chuzhanova N., Yi M., Stephens R.M., Stefanov S., Olsh A., Jakupciak J.P., Dean M., et al. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang G., Vasquez K.M. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanawalt P.C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 43.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 44.Haines N.M., Kim Y.I., Smith A.J., Savery N.J. Stalled transcription complexes promote DNA repair at a distance. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4037–4042. doi: 10.1073/pnas.1322350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


