Abstract
c-MYC is an important oncogene that is considered as an effective target for anticancer therapy. Regulation of this gene's transcription is one avenue for c-MYC-targeting drug design. Direct binding to a transcription factor and generating the intervention of a transcriptional programme appears to be an effective way to modulate gene transcription. NM23-H2 is a transcription factor for c-MYC and is proven to be related to the secondary structures in the promoter. Here, we first screened our small-molecule library for NM23-H2 binders and then sifted through the inhibitors that could target and interfere with the interaction process between NM23-H2 and the guanine-rich promoter sequence of c-MYC. As a result, a quinazolone derivative, SYSU-ID-01, showed a significant interference effect towards NM23-H2 binding to the guanine-rich promoter DNA sequence. Further analyses of the compound–protein interaction and the protein–DNA interaction provided insight into the mode of action for SYSU-ID-01. Cellular evaluation results showed that SYSU-ID-01 could abrogate NM23-H2 binding to the c-MYC promoter, resulting in downregulation of c-MYC transcription and dramatically suppressed HeLa cell growth. These findings provide a new way of c-MYC transcriptional control through interfering with NM23-H2 binding to guanine-rich promoter sequences by small molecules.
INTRODUCTION
The human c-MYC gene or its product is a key regulator of cellular proliferation and cell growth. The aberrant overexpression of c-MYC is associated with a variety of malignant tumours and amplification of c-MYC is among the most common genetic alterations observed in cancer genomes (1). Anti-c-MYC therapies could involve multiple routine approaches, including inhibition or modulation of c-MYC gene transcription and/or translation, prevention of c-MYC-Max heterodimer formation, inhibition of c-MYC or Max in DNA binding and inhibition of key c-MYC target gene products (2). A few reports on direct inhibitors of c-MYC could be found (3), while various transcription inhibitors have been reported, because c-MYC is a classical G-quadruplex-relating gene (4). Although several G-quadruplex ligands exhibit good selectivity for quadruplex versus duplex DNA, it is hard to find a real selective ligand only for the c-MYC gene in vivo, because G4-motif-mediated regulation is a general mode of transcriptional control (5). This makes us start to think about an alternative strategy for modulating c-MYC transcription.
Transcription factors are proteins that play crucial roles in gene regulation, and deregulation of transcription factor networks is a major pathogenic event (6). In general, mutations in upstream regulators and aberrant gene amplification may destabilize the proper function of the transcription factor network and drive disease (7,8). Small-molecule intervention is an attractive approach to intervene directly with transcription factors (9). NM23-H2 has been identified as a transcriptional factor of the oncogene c-MYC, which is responsible for the transcriptional activation of the proto-oncogene c-MYC (10–12). The overexpression of NM23-H2 was observed in a wide range of cancers, such as chronic myeloid leukaemia (13), hepatocellular carcinoma (14,15), breast cancer (16) and oral squamous cell carcinoma (17), making it a promising anticancer drug target.
Some studies have shown that NM23-H2 could specifically recognize and bind to purine-rich sequence domains, including the nuclease hypersensitive element III1 (NHE III1) of the c-MYC promoter (18–20). In addition, more detailed studies revealed that, unlike other classical transcription activators, NM23-H2 might be involved in the alteration or removal of unusual DNA conformations in the promoter through the breaking and rejoining of DNA strands instead of directly stimulating transcription by binding to the sequence of CCCTCCCCA (termed the CT element) (18,21). These phenomena suggested that the DNA-binding activity of NM23-H2 was likely to be the foundation of NM23-H2 function as a transcription factor (18,22), and the NM23-H2/purine-rich sequence interaction and corresponding transcriptional regulation may be important processes for NM23-H2 to act as a biological regulator. Thus, interfering with NM23-H2 binding to a guanine-rich sequence within the promoter of targeting genes by a small molecule may be a novel way of gene transcriptional control. Some G-quadruplex stabilizers have shown abilities to prevent NM23-H2 binding to the target gene c-MYC (23), however, there was few reports on small-molecule ligands that may interfere with the DNA–protein interaction by directly interacting with NM23-H2 protein only or binding to both DNA and protein, and thus control the level of gene transcription.
First, we constructed a screening and evaluation platform, which included the expression and purification of NM23-H2, and the establishment of analytical methods to probe protein–DNA interactions. Then, we proceeded to screen our compound library (built by the School of Pharmaceutical Science, Sun Yat-sen University) containing 146 natural products and related derivatives with diverse structures. Among them, SYSU-ID-01, a quinazolone derivative, was identified as a potent NM23-H2 binder and inhibitor for the protein–DNA interaction. In vitro evaluation revealed that SYSU-ID-01 showed good binding affinity for NM23-H2. Studies on the interaction of the compound and/or DNA with the wild-type and seven mutants of the NM23-H2 protein showed possible binding sites for SYSU-ID-01 on the protein. Further studies indicated that SYSU-ID-01 was capable of abrogating the binding of NM23-H2 with the NHE III1 region of c-MYC, resulting in downregulation of c-MYC transcription and translation. Moreover, SYSU-ID-01 exhibited significant inhibitory effects on HeLa cells similar to those obtained by RNA interference (RNAi) of NM23-H2. Additionally, the results of DNA microarray analysis and a reverse transcription-polymerase chain reaction (RT-PCR) assay indicated that SYSU-ID-01 was actually targeting NM23-H2 intracellularly. These findings illustrated that transcriptional regulatory activity that was derived from the NM23-H2/guanine-rich sequence binding could be controlled by a small molecule.
MATERIALS AND METHODS
Cell culture and treatments, plasmid construction, NM23-H2 expression and purification
Detailed information is provided in the Supplementary Methods section.
Fluorescence resonance energy transfer assays
Fluorescence resonance energy transfer (FRET) assay was carried out on a real-time PCR apparatus as follows. The fluorescent labelled oligonucleotide Pu18 (Supplementary Table S1) [5′-FAM- Pu18-TAMRA-3′] was used in the assays, 6-carboxyfluorescein (FAM) as donor fluorophore and 6-carboxytetramethylrhodamine (TAMRA) as acceptor fluorophore. The oligonucleotide was diluted to 400 nM in Tris-HCl buffer (10 mM, pH 7.4) containing 60 mM KCl, and then annealed by heating to 95°C for 5 min followed by cooling to room temperature. Samples were composed of 10 μl of the annealed Pu18 and 10 μl of the compound solutions (at 2 μM) and incubated at 4°C overnight. Measurements were made in LightCycler capillaries with excitation at 470 nm and detection at 530 nm. Analysis of the data was carried out using Origin 8 (OriginLab Corp.).
Surface plasmon resonance
Surface plasmon resonance (SPR) measurements were performed on a ProteOn XPR36 Protein Interaction Array system (Bio-Rad Laboratories, Hercules, CA, USA) using a GLH chip. In a typical experiment, wild-type and mutant NM23-H2 proteins were immobilized (∼12 000 RU) in flow cells, with one flow cell used as a blank. For the screening experiment, the ligands were diluted with running buffer (12 mM HEPES, 4 mM Tris, 1 mM EDTA, 1.5 mM MgCl2 and 0.005% Tween-20) to 10 μM. The SYSU-ID-01 solution was diluted to different concentrations with running buffer. The ligand was injected at a flow rate of 25 μl/min for 180 s during the association phase, which was followed by a 300 s disassociation phase at 25°C. The GLH chip was regenerated with a short injection of 1 M NaCl between consecutive measurements. The final graphs were obtained by subtracting blank sensorgrams from the NM23-H2 protein sensorgrams. Kinetic and equilibrium analyses were performed using the ProteOn manager software. The theoretical max RU of compounds were evaluated via the equation as Rmax = Rprotein rprotein/ligand (MWprotin/MWligand) (Rprotein is the amount of protein immobilized on the chip, which is 12000RU in our study; rprotein/ligand is the ratio of protein–ligand interaction, which is 1:1 in our study; MWprotein is the molecular weight of NM23-H2, which is 60 000 g/mol in our study; MWligand is the molecular weight of compounds). The kinetic SPR sensorgrams were fitting with a 1:1 Langmuir binding mode, KD values were achieved by the Analysis module in ProteOn XPR36 software. The equation of association process is d[AL]/dt = ka[A][L] -kd[AL], whereas in the dissociation process, [A] = 0, d[AL] /dt = -kd[AL]. ([AL] is the concentration of complex, t is time, [A] and [L] are the concentration of analyte and the substance, respectively. ka and kd are the association and dissociation rate coefficients, respectively). KD was calculated from ka and kd, KD = kd/ka.
Saturation transfer difference nuclear magnetic resonance experiments
Saturation transfer difference nuclear magnetic resonance (STD-NMR) experiments were performed at 298 K using a Bruker Avance 500 NMR spectrometer equipped with a 1.7 mm TCI MicroCyroProbe. The water solvent signal was suppressed using the 3–9–19 WATERGATE gradient spin echo. The number of scans and dummy scans in the STD experiments were 256 and 32, respectively. All spectra were acquired using a spectral width of 10 000 Hz (20 ppm), an acquisition time of 2 s and a relaxation delay of 3 s. Selective saturation of protein resonances was achieved at −1 ppm. The NMR sample containing 200 μM SYSU-ID-01 and 12 μM NM23-H2 was prepared in phosphate buffer [20 mM K2HPO4 and 100 mM KCl (pH 7.0) in D2O]. Control experiments using the free ligand (SYSU-ID-01 only) and a control protein (NM23-H2) were performed under the same experimental conditions and no STD signal was observed.
Molecular modelling
The crystal structure of NM23-H2 complexed with dinucleotide d(AG) (code ID:3BBB) was obtained from the Protein Data Bank. Docking studies were performed using the AUTODOCK 4.0 programme. Docking calculations were carried out using the Lamarckian genetic algorithm. Molecular dynamics (MD) simulations were performed using the sander module of the AMBER 10.0 programme suite with ff03 force field. Periodic boundary conditions and the particle mesh Ewald algorithm were used. The complexes were solvated in an octahedral box of TIP3P water molecules with solvent layers that were 10 Å thick. Chlorine counterions were added to neutralize the complexes. The hydrogen bonds were constrained using SHAKE. The solvated structures were subjected to initial minimization to equilibrate the solvent and counter cations. The system was then heated from 0 to 300 K in a 100 ps simulation followed by a 100 ps simulation to equilibrate the density of the system. Then, a constant-pressure MD simulation was performed for 40 ns in an NPT ensemble at 1 atm and 300 K. The Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) method was used to calculate the binding free energy. All of the water molecules and counterions were stripped off. A set of 200 snapshots from the last MD trajectories was collected to calculate the binding free energies.
Filter-binding assay
Filter-binding assays were performed as follows. In brief, a nylon membrane was placed directly below the nitrocellulose membrane to trap any DNA not retained on the nitrocellulose. The nitrocellulose membrane was treated with 0.5 M KOH for 10 min at 4°C and washed with 0.5×TB prior to use. Single- or double-stranded biotin-labelled c-MYC Pu27 (200 fmol) (Supplementary Table S1) was incubated with increasing concentrations of SYSU-ID-01 in 12 μl of binding buffer (20 mM HEPES, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.01 mM EDTA, 20% glycerol, pH 7.4) (11) at 25°C for 30 min and then further incubated for 1 h at 25°C after the addition of 2 μg NM23-H2 protein. All samples were applied to the membrane under vacuum and washed 10 times with 0.5×TE buffer. The cross-linking reaction was carried out under UV irradiation at 265 nm for 10 min. Detection of biotin-labelled DNA was carried out using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific) according to the protocol supplied by the manufacturer. In the part of Disruption of the Protein–DNA Interaction by SYSU-ID-01, the grey levels of dots were measured by Quantity One. We set the grey level of the second dot (sample without compound) as 100%, and the later dots were compared to the second one to obtain the relative level of the protein–DNA complex. The IC50 value was evaluated via a Hill model in Origin 8, the Hill equation is R (d) = λ+ vdn (kn + dn) [R (d) is the response under ligand treatment, λ is the background effect, v is the maximum response, k is half of the maximum dose, n is Hill coefficient, respectively]. In the part of Binding Mode of SYSU-ID-01 with NM23-H2, the grey levels of dots were measured by Quantity One. We set the grey level of the protein–DNA complex of wild-type NM23-H2 as 100%, and the mutant proteins were compared to the wild-type to obtain the relative level of the protein–DNA complex. The column chart was drawn in Origin 8.
Electrophoretic mobility shift assay
The 5′-FAM-labelled c-MYC Pu27 (Supplementary Table S1) was diluted in 10 μl binding buffer (20 mM HEPES, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.01 mM EDTA, 20% glycerol, pH 7.4) at a concentration of 5 μM, and incubated with NM23-H2 (protein/oligomer strand ratio was 1:1) and SYSU-ID-01 at concentrations of 0 μM, 1.25 μM, 2.5 μM, 5 μM and 10 μM at 25°C for 1 h. The binding buffer was 10 mM HEPES (pH 7.4), composed of 100 mM KCl and 2 mM MgCl2. The samples were run on 8% native polyacrylamide gels at 4°C and 10 V·cm−1 in 0.5×TB buffer (Tris/borate) followed by detection using a gel imaging system with a UV light. The grey levels of protein–DNA complex at the upper panel of the gel were measured by Quantity One. We set the grey level of the complex in the second lane (without compounds) as 100%, and the later lanes were compared to the second lane to obtain the relative level of the protein–DNA complex. The IC50 value was evaluated via a Hill model in Origin 8.
Two microgram NM23-H2 were diluted in 40 μl binding buffer (20 mM HEPES, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.01 mM EDTA, 20% glycerol, pH 7.4) and incubated with compounds at a concentration of 10 μM at 25°C for 1 h. The c-MYC Pu27 was added to the system at a concentration of 0.75 μM and further incubated at 25°C for 1 h. The binding buffer was 20 mM HEPES (pH 7.4) containing 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.01 mM EDTA and 20% glycerol. The samples were run on 8% native polyacrylamide gels at 4°C and 10 V·cm−1 in 0.5×TB buffer followed by silver staining and photography.
ELISA assays
The streptaWell High Bind plates (Roche) were used in the ELISA assays. The 5′-biotin-labelled c-MYC Pu18 (Supplementary Table S1) was diluted in 200 μl Tris-HCl buffer (10 mM, pH 7.4) containing 100 mM KCl at a concentration of 100 nM, and then annealed by heating to 95°C for 5 min followed by cooling to room temperature. To prepare the protein samples, the concentration of NM23-H2 protein was initially 4000 nM and half-diluted nine times in blocking buffer, after adding DMSO or 20 μM compounds, the samples were incubated at 25°C for 1 h. Wells were rehydrated with 200 μl PBS, then incubated with the DNA samples at 4°C overnight; washed three times by ELISA buffer (50 mM KH2PO4, 100 mM KCl, pH 7.4); blocked in 200 μl of blocking buffer (3% BSA in ELISA buffer) for 1.5 h at RT; washed three times by ELISA buffer; incubated with protein samples at 37°C for 3 h; washed three times by washing buffer (ELISA buffer containing 0.1% Tween-20) and incubated with primary antibody (SantaCruz: sc-100400) at 4°C overnight; washed three times by washing buffer; incubated with 100 μl secondary HRP-conjugated antibody (CST: 7076s) for 2 h at 37°C and washed three times with washing buffer. Following addition of 100 μl of TMB Chromogen Solution (Life Technologies), plates were incubated in the dark for 5 min and the reaction was stopped by addition of 50 μl 1M H2SO4. Absorbance was read at 450 nm. The fitting curve was obtained using Graphpad Prism 5 via Hill fitting, and the KD is the numeric equivalent of the concentration of NM23-H2 when the absorbance is half of the plateau absorbance (Amax) on the fitting curve.
Microscale thermophoresis experiments
Labelling procedure
The protein concentration was diluted to 20 μM using the labelling buffer (50 mM HEPES with 150 mM NaCl, pH 8.0). The NT-647-NHS dye was dissolved in 100% DMSO at a concentration of ∼650 μM and mixed with the protein in a 1:1 ratio followed by incubation for 30 min at room temperature in the dark. Unreacted ‘free’ dye was removed by a gel filtration column (Sephadex G25, GE Healthcare). The purity was monitored by measuring the ratio of protein to dye (spectroscopically by measuring absorption at 280 nm for the protein and 650 nm for the dye) after the clean-up procedure.
Measuring procedure
The concentration of NT-647 labelled NM23-H2 was kept constant as 200 nM, while the concentrations of the compounds were initially 200 μM and half-diluted 15 times in binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.05% Tween-20). A similar process was performed for the detection of the NM23-H2 and FAM-Pu27 interaction with constant concentrations of FAM-Pu27 at 250 nM. After an incubation of 30 min, the samples were loaded into microscale thermophoresis (MST)-grade glass capillaries. The intensities of the LED and laser were set at 40%. The MST analyses were performed using a Monolith NT.115 and the fitting curve was obtained using NT Analysis 1.4.23 via Hill fitting, and the KD is the numeric equivalent of the concentration of NM23-H2 or SYSU-ID-01 when the response is half of the plateau response (Rmax) on the fitting curve.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed using a Magna ChIPTM kit (Millipore) according to the manufacturer's protocol. In brief, HeLa cells were fixed with 1% formaldehyde for 10 min and then lysed after 12 h of treatment. Chromatin was sheared to an average size of 0.2–0.5 kb using a SCIENTZ-II D sonicator (SCIENTZ, China). Then, 10% of the lysate was removed for use as an input, and 3 μg of antibody against NM23-H2 (SantaCruz: sc-100400) was used for ChIP. Normal rabbit IgG (SantaCruz: sc-3888) was used as the negative control. ChIP was performed overnight at 4°C, and immune complexes were collected using the protein A magnetic beads provided in the kit. After extensive washing, the DNA was extracted from immunoprecipitated chromatin and amplified by PCR using c-MYC-ChIP-A and c-MYC-ChIP-S primers (Supplementary Table S1). The amplified products were separated on a 1.5% agarose gel, and the gel was photographed on a Gel Doc 2000 Imager System.
Hoechst 33342/PI staining and confocal imaging
HeLa cells or primary cultured mouse mesangial cells (2.0 × 105) were plated on 3 cm Petri dishes. After 72 h of ligand treatment, the cells were washed with cold PBS and incubated with Hoechst 33342 solution (Sigma, 5 μg/ml) for 10 min at 37°C in the dark. The Hoechst 33342 solution was removed, and the cells were washed with cold PBS for three times. Then the PI solution (Sigma, 5 μg/ml) was added and incubated at room temperature for 10 min in the dark. After washing with cold PBS for three times, the stained cells were imaged by a Zeiss LSM 710 laser scanning confocal microscope.
FITC Annexin V/PI apoptosis detection
FITC Annexin V/PI apoptosis detection was performed using the FITC Annexin V/PI Apoptosis Detection Kit (KeyGEN). Briefly, after 72 h treatment of SYSU-ID-01 or transfection of siRNA, HeLa cells were digested and resuspended in 500 μl binding buffer. FITC Annexin V (5 μl) and 5 μl PI were added, and the cells were disturbed by gently vortexing the samples prior to incubation at RT (25°C) for 15 min in the dark. Emitted fluorescence was quantitated by Epics Elite flow cytometry (Beckman-Coulter).
Protein extracts and western blots
After 72 h treatment of SYSU-ID-01 or transfection of siRNA, cells harvested from each well of the culture plates were counted, and western blots were performed as described in the Supplementary Methods section.
Silencing NM23-H2 using siRNA
A commercially available synthetic siRNA duplex (SantaCruz: sc 40774) against NM23-H2 was used to knock down NM23-H2 in HeLa cells. An siRNA duplex (SantaCruz: sc 37007) with a scrambled sequence was used as a negative control. Transfection of siRNA duplexes was accomplished using Lipo2000 (Invitrogen) based on the manufacturer's protocol. Cells were harvested for various experiments 72 h after transfection with the siRNA.
Total RNA isolation, microarray hybridization and analysis
Total cellular RNA was isolated from NM23-H2 knockdown clones (by RNAi) and untreated control cells using the TRIzol Reagent (Life Technologies) according to the manufacturer's instructions. The integrity of the total RNA was analysed on 1% agarose gel and quantitated using UV light. For microarray analyses, the standard procedure for preparing the hybridization of total RNA was followed as recommended by Affymetrix's standard protocol (available online, www.Affymetrix.com). An Ambion® WT Expression Kit (Ambion), GeneChip WT Terminal Labelling Kit, and Controls Kit (Affymetrix) were used. Labelled and fragment cRNA (complementary RNA) was further hybridized to probes on an Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix) using a GeneChip. Hybridization, washing and staining were performed according to the manufacturer's instructions (Affymetrix). Expression levels of mRNA were evaluated using the GeneChip Scanner 3000 7G (Affymetrix). The relative change in abundance between the untreated control cells and the experimental sample (NM23-H2 knockdown cells) was evaluated for each transcript. The major pathway of the differential genes was identified according to KEGG, Biocarta and Reactome. Fisher's exact test and the χ2 test were also used to select the major pathway. The threshold of significance was defined by the P-value and false discovery rate. The enrichment Re was calculated using the equation described above.
Reverse transcription-polymerase chain reaction
HeLa cells were treated with SYSU-ID-01 or NM23-H2 siRNA, or left untreated, as indicated in the individual figures and legends. Total RNA was purified and analysed by RT-PCR as described in the Supplementary Methods section.
RESULTS
Survey of small molecules targeting NM23-H2
In order to screen compound that may suppress the DNA–protein interaction by interacting with NM23-H2 protein only or binding to both protein and DNA, we started our work with a small survey through an in-house small-molecule library with 146 compounds which was constructed during our studies on the discovery of G-quadruplex ligands (24–32). As shown in Supplementary Figure S1, some of compounds in this library were strong c-MYC G-quadruplex DNA stabilizing agents and some of them had a weak effect on G-quadruplex stability.
ProteOn XPR36 instrument (BioRad) is a good SPR analyser for affinity determination of binders and for the simultaneous analysis of up to six ligands. Thus, SPR was used here with wild-type NM23-H2 immobilized to a GLH chip, and activity of the immobilized protein was determined by its interaction with Pu27 (results were shown in Supplementary Figure S2a and b). The molecules screened here are alkaloid natural products and their analogues, including berberine derivatives (No. 1–38), quindoline derivatives (No. 39–77), bis(quinolinylmethylene)cycloalkanone derivatives (No. 78–86), quinazolone derivatives (No. 87–114) and rutaecarpine derivatives (No. 115–146) (the general structures of these compounds were shown in Figure 1B).
Figure 1.
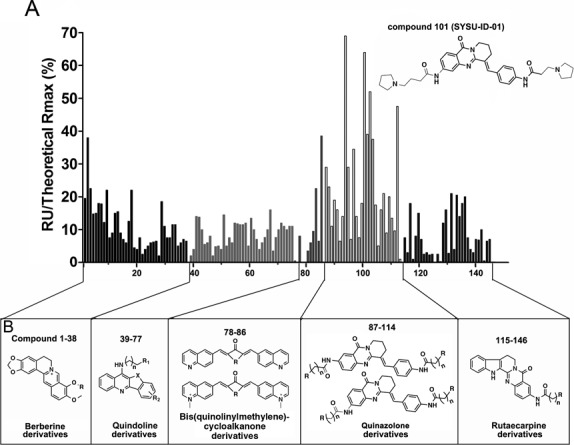
SPR screening results. NM23-H2 proteins were immobilized in flow cells. For the screening experiment, the ligands were diluted with running buffer to 10 μM. The ligand was flowed for 180 s during the association phase, and followed by a 300 s disassociation phase. This figure showed the histogram of relative SPR response (% RU theoretical max) for all screened compounds that flowed through immobilized NM23-H2; the top performer was SYSU-ID-01 (compound ID as 101). The structures and numbers of screened compounds were listed in (B).
The SPR data were shown in Figure 1A with a y-axis of the percentage of the resonance units (RU) from compound treatment compared with the theoretical max RU, and this index reflects the level of binding affinity of the compounds to the immobilized NM23-H2. We made a statistical evaluation of all the SPR data (Supplementary Figure S3). The average RU% of all the compounds was 12, the minimum RU% was 0 and the maximum RU% was 69. Most of the relative RU values (75%) were ∼15. Among all the groups, the group of quinazolone derivatives (compound 87–114) exhibited extraordinary binding activities to NM23-H2, with an average RU% of 23, and a quarter of the compounds in this group possessed relative RU values of ∼33. The classical G-quadruplex stabilizers developed by our group, the berberine and quindoline derivatives, possessed stronger stabilizing activities to quadruplex but lower binding affinity to the NM23-H2 protein; the rutaecarpine derivatives showed weak abilities on both stabilizing of G-quadruplex DNA and binding to NM23-H2 protein. These results implied that the binding to NM23-H2 was not relative to whether the compound was a G-quadruplex ligand or not.
Our aim was to find a way to regulate c-MYC expression, and binding to NM23-H2 just gave us a hint for further regulation of its targeting gene c-MYC. Thus, to further confirm the effects of the compounds in the SPR screening, most of the compounds (No. 1–77, 87–135, 139–146) were further evaluated for their ability to disrupt the binding process between NM23-H2 and the c-MYC G-rich sequence Pu27 by the electrophoretic mobility shift assay (EMSA) method (Supplementary Figure S4). The result showed that part of the quinazolone derivatives exhibited strong disruption activities on the NM23-H2–DNA complex, some of the berberine derivatives and quindoline derivatives, and few of the rutaecarpine derivatives also showed meaningful disruption activities on the complexes. Comparing the results from EMSA (Supplementary Figure S4) with FRET (Supplementary Figure S1) or SPR (Figure 1) assays, it was found that most of compounds with great stabilizing potentials to G-quadruplex (such as 3, 7, 9, 74, 93 and 106) showed good disruption abilities on complex. However, compounds with remarkable binding effects on protein, such as 113, might not have good disruption activity on NM23-H2–DNA complex. Some of compounds (90 and 141) with unconspicuous activities on both G-quadruplex stabilization and protein binding also seem to possess notable disruption activities on complex.
As we know, EMSA is a semiquantitative method to investigate protein–DNA interaction, while aggregation might occur under some conditions (just like ligand interference and allosteric property of DNA/protein). In order to investigate the interference with NM23-H2–DNA complex by the compounds more accurately, and to exclude the possibility that the compounds might induce aggregation of DNA–protein complex and result in disappearance of the complex signal, five prominent compounds (58, 74, 90, 101 and 141) in EMSA were further tested by ELISA assay (Supplementary Figure S5), a quantitative assay that was carried out on a solid phase. NM23-H2 was treated with 20 μM compound and then interacted with c-MYC G-quadruplex that immobilized previously, detection was achieved with primary antibody, secondary antibody and TMB (3, 3′, 5, 5′-Tetramethylbenzidine). The results revealed that NM23-H2 possessed high binding affinity for c-MYC G-quadruplexes (KD was 0.53 ± 0.018 μM), while 20 μM of the compounds could suppress the protein–DNA combination, and led to a weakened protein–DNA interaction which could be deduced from the change of KD values in the presence of compound 58, 74, 90, 101 and 141. Among them, 58 and 74, the top two compounds with protein–DNA interaction inhibitory activities, possessed strong stabilization activities on c-MYC G-quadruplex (ΔTm value was 16°C and 22°C, respectively) but weak protein binding abilities. While compound 141 and 90, which exhibited moderate inhibitory activities on complex, showed a weaker performance both on binding to NM23-H2 protein and stabilizing of c-MYC G-quadruplex DNA. Finally, compound 101, a quinazolone derivative, named as SYSU-ID-01, whose structure was shown in Figure 1, was selected for further studies due to its stronger interfering effect on the NM23-H2–DNA complex and binding affinity to the NM23-H2 protein but weaker stabilization activity on c-MYC G-quadruplex (ΔTm value was 9°C).
SYSU-ID-01 specifically binds to NM23-H2
Several additional biophysical assays were performed to confirm whether SYSU-ID-01 could specific bind to NM23-H2. First, a thorough SPR experiment was used to quantitatively determine the kinetic binding constants of SYSU-ID-01 to NM23-H2 wild-type protein. After evaluating the SPR data of SYSU-ID-01 binding to immobilized NM23-H2 at a range of concentrations between 0 and 8 μM, Figure 2A showed the plots obtained from the 1:1 Langmuir binding analysis using the kinetic fitted SPR sensorgrams, and the KD value was 5.29 μM. This was a relatively high binding affinity between a small molecule and a hexamer protein (33). Because SPR experiments need immobilized NM23-H2 on a chip and the hexamer situation is easy to affect by this immobilized status, some methods measured in an absolute solution environment should be applied to give more convincing binding data. MST is such a sensitive method for quantitative analysis of molecular interactions in solution at the microliter scale (34), and we further applied this method to investigate the binding affinity of SYSU-ID-01 to NM23-H2. The equilibrium dissociation constant (KD) from MST detection was 9.8 ± 0.039 μM (Figure 2B), which was of the same order but slightly different from that in the SPR assay.
Figure 2.
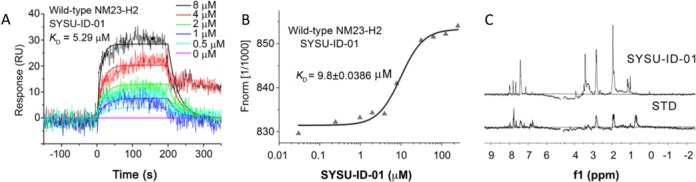
Binding affinity of SYSU-ID-01 to NM23-H2. (A) The plots of SYSU-ID-01 binding to immobilized wild-type NM23-H2 in SPR assay. The SYSU-ID-01 solution was diluted to the concentrations of 0, 1 μM, 2 μM, 4 μM and 8 μM with running buffer, and was analysed with an association phase of 180 s and a dissociation phase of 300 s. The KD value was obtained by fitting with 1:1 Langmuir binding mode. (B) The MST results of the binding of fluorescently labelled wild-type NM23-H2 to SYSU-ID-01. The concentration of NT-647 labelled NM23-H2 was 200 nM, and the SYSU-ID-01 was titrated from 6 nM to 200 μM in binding buffer. After an incubation of 30 min, the samples were loaded into MST-grade glass capillaries and tested. The data were evaluated using NT Analysis 1.4.23 via Hill model. (C) 1H STD NMR spectrum of NM23-H2 solution together with SYSU-ID-01 (bottom) and normal 1H NMR spectrum of SYSU-ID-01 (top).
Furthermore, STD 1H-NMR spectroscopy was used to provide more evidence for the binding of SYSU-ID-01 to NM23-H2. The NMR-STD experiment is a well-established method for investigation of ligand-protein interactions, in which binding epitopes may be mapped by comparing signals of the ligand with and without saturation of the protein (35). The 1H NMR spectrum of SYSU-ID-01 and the STD NMR experiment for SYSU-ID-01 in complex with NM23-H2 were shown in Figure 2C. Comparing the spectrum of SYSU-ID-01 with the STD spectrum, varying signals were visible between 6.7 and 8.0 ppm and between 2.0 and 3.0 ppm, which revealed that NM23-H2 was interacted with SYSU-ID-01, the aromatic moiety and amide side chain of SYSU-ID-01 may participate in the interaction.
Disruption of the protein–DNA interaction by SYSU-ID-01
The DNA-binding activity of NM23-H2 is the foundation for NM23-H2 to function as a transcription factor (18,22,36), and the interaction between NM23-H2 and the putative G-quadruplex sequence (PQS) in the NHE III1 region of c-MYC directly relates to its regulatory effects on the c-MYC gene (23). SYSU-ID-01 showed interfering activity on the NM23-H2–DNA complex at 10 μM in the screening phase (Supplementary Figure S4), and a thorough EMSA was performed to further evaluate SYSU-ID-01 in the dose-response. The results illustrated that SYSU-ID-01 could significantly inhibit the formation of an NM23-H2/DNA complex (Figure 3A and B) in a dose-dependent manner; the corresponding IC50 value was 5.9 μM. To better understand the binding between NM23-H2 and Pu27 DNA, we further applied a filter-binding assay to evaluate SYSU-ID-01's effects (37). In this assay, the amount of DNA is very low (16.7 nM in our study), while the amount of protein is relatively high (2.5 μM in our study), which is very similar to the situation in cells where every sequence has only one copy in the whole genome, while there are much more than one copy of the expression proteins. As shown in Figure 3C and D, SYSU-ID-01 blocked NM23-H2's binding to Pu27 in a dose-dependent manner, and the corresponding IC50 value was 3.7 μM. Both data suggested that SYSU-ID-01 could interfere with the binding between NM23-H2 and PQS in c-MYC. In addition, the interfering effects of this compound depended on the binding to the NM23-H2 protein, because the IC50 values maintained a similar ratio with the concentration of NM23-H2 (5.9 μM versus 5 μM protein, and 3.7 μM versus 2.5 μM protein), despite the huge difference in DNA concentration (5 μM and 16.7 nM).
Figure 3.
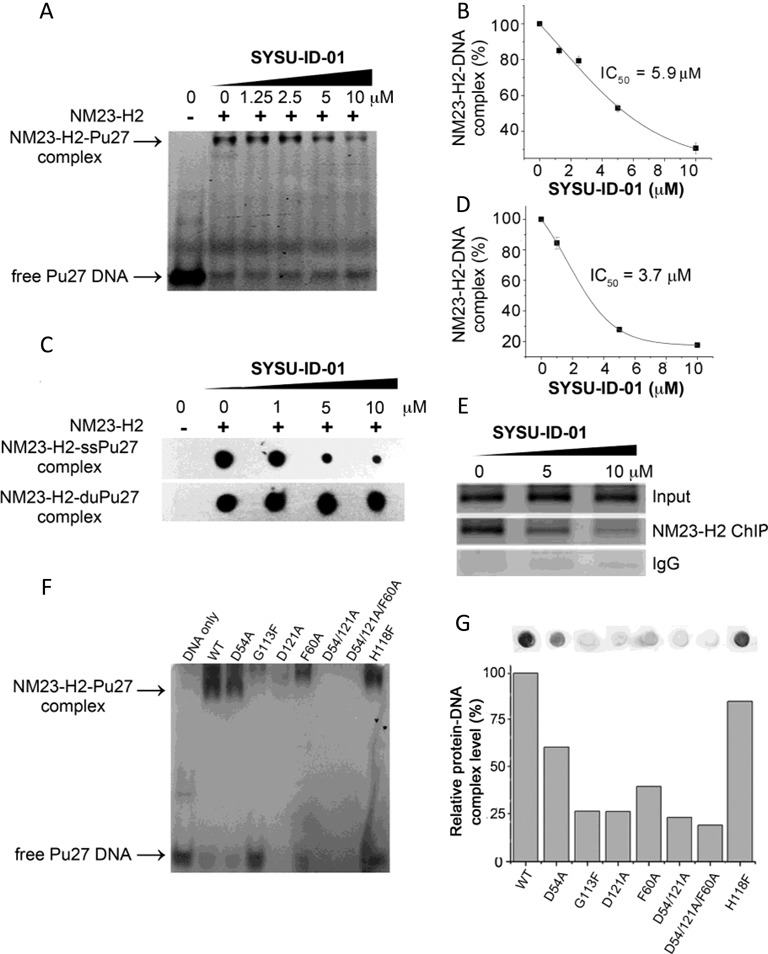
Disruption of the protein–DNA interaction by SYSU-ID-01. (A) Electrophoretic mobility shift of 5′-FAM-labelled Pu27 from c-MYC promoter in presence or absence of NM23-H2 and/or SYSU-ID-01 in 8% native PAGE. Top panel: NM23-H2–DNA complex, bottom panel: free Pu27 DNA. (B) Dose-dependent inhibition curve for NM23-H2 binding to Pu27 in the presence of increasing concentrations of SYSU-ID-01 obtained from a gel mobility shift assay. The data were derived from three experiments and were shown as the means ± S.E.M. (C) Filter-binding assay results reflected the effect of SYSU-ID-01 on the combination of NM23-H2 with the c-MYC G-rich sequence Pu27 (SS-Pu27) and double-stranded c-MYC Pu27 (DS-Pu27). (D) Dose-dependent inhibition curve for NM23-H2 binding to SS-Pu27 in the presence of increasing concentrations of SYSU-ID-01 obtained from a filter-binding assay. The data were derived from three experiments and were shown as the means ± S.E.M. (E) ChIP assays to evaluate the disruption of NM23-H2–DNA interaction by SYSU-ID-01 in the HeLa cell line. Normal rabbit IgG was used as the negative control for mock immunoprecipitation. Immunoprecipitated DNA samples were PCR-amplified to show NM23-H2 occupancy of the c-MYC promoter, the amplified products were separated on a 1.5% agarose gel. (F) Gel mobility shift results of wild-type and mutant NM23-H2 binding to Pu27. (G) Filter-binding assay results of wild-type and mutant NM23-H2 binding to Pu27 in the graph and quantitative columns. (A) and (F) had 60 pmol Pu27 in each sample, while the DNA amount was 100 fmol in (C) and (G) samples. SYSU-ID-01 treatments were for 1 h at 25°C before detection.
NM23-H2 binds to both double-stranded and single-stranded DNA according to previous reports (23,38–39). To determine the binding position of SYSU-ID-01 on the NM23-H2 protein, a filter-binding assay was performed. As shown in Figure 3C, SYSU-ID-01 could block the binding of NM23-H2 to a single G-rich strand (SS-Pu27) in a dose-dependent manner, while having no effect on NM23-H2's ability to bind double-stranded DNA (DS-Pu27).
To determine whether compound SYSU-ID-01, an blocker of NM23-H2–DNA in vitro, could inhibit transcription factor binding to the c-MYC promoter in HeLa cervical cancer cells, we further applied ChIP assays (Figure 3E). The chromatin of HeLa cells treated with 5 and 10 μM of SYSU-ID-01 were isolated and sonicated. First, the cytotoxicity of SYSU-ID-01 in tumour cells was tested using the HeLa cell line by the MTT assay. To be noted, the concentrations used here and in the following cellular experiments were subcytotoxic concentrations according to the MTT cell toxicity assays (data shown in Supplementary Figure S6). Amplification of the immunoprecipitated samples covering the PQS in the promoter of c-MYC observed a decrease of NM23-H2-bound DNA with increasing SYSU-ID-01 in a dose-dependent manner, revealing that SYSU-ID-01 could disrupt the NM23-H2–DNA interaction in HeLa cells.
SYSU-ID-01 regulates a gene's expression via intervention on NM23-H2
The observation that SYSU-ID-01 interferes with NM23-H2 occupancy on DNA led us to initially investigate its effect on the gene target c-MYC. Routine RT-PCR and western blot analyses were applied to determine the transcription and expression levels of this gene. As shown in Figure 4, SYSU-ID-01 could repress the transcription of c-MYC in a dose-dependent manner and slightly repress the expression of c-MYC at a concentration of 10 μM.
Figure 4.
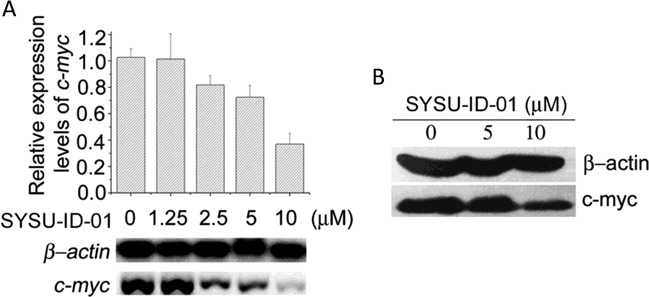
SYSU-ID-01 downregulated c-MYC transcription (A) and expression (B) after 72 h treatment. (A) The semiquantitative RT-PCR detecting the transcription level of c-MYC gene (bottom). The grey levels of the bands were measured by Quantity One and summarized as column chart by Origin 8. (B) Western blot showing the level of c-MYC protein under the treatment of increased concentrations of SYSU-ID-01 in HeLa cells, beta action was used as a loading control.
A critical property of an NM23-H2 ligand would be the downregulation of genes downstream of NM23-H2-occupied promoters. Here we need to determine the genes with NM23-H2 occupancy in HeLa cells and SYSU-ID-01's specific effect on these genes’ transcription. Thus, we started this part with the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Invitrogen, China) comparing the cells knocking down NM23-H2 using siRNA with the cells untreated. The expression profiles of HeLa cells transfected with NM23-H2 siRNA were compared with those of the untreated cells to establish the interfering effect of siRNA (Supplementary Figure S7). A total of 1665 known and hypothetical genes were differentially expressed by ≥2-fold on average (611 were upregulated, 1054 were downregulated) after NM23-H2 was knocked down (Supplementary Table S2). Pathway analysis revealed that these genes were involved in many tumour-associated pathways in cancer cells (Supplementary Figure S8), especially in transcriptional misregulation in cancers.
RT-PCR was then applied to analyse the effect of SYSU-ID-01 on 14 NM23-H2 downstream genes obviously changed after siRNA treatment from the gene-chip data, including MEF2D, FOSL1, MMP28, FNBP4, ANKH, MAMDC2, CDKN1A, DOCK1, BIRC3, PTH2R, MDM2, CCL20, HRK and CLTA. The semiquantitative RT-PCR results exhibited that, although the expression of the NM23-H2-related genes did not change in the presence of SYSU-ID-01, the mRNA levels of all 14 NM23-H2-related genes after SYSU-ID-01 treatment showed good correlations with the data from the knockdown assay (Figure 5, and the origin full gels were shown in Supplementary Figure S9). These results were also verified by quantitative RT-PCR (the qRT-PCR results were shown in Supplementary Table S3 and the primers used in qRT-PCR were shown in Supplementary Table S1) and the data showed similar outcomes. That SYSU-ID-01 treatment specifically regulates these NM23-H2-related genes provided strong evidence that the inhibitory effect of SYSU-ID-01 on NM23-H2 binding translates in cells.
Figure 5.
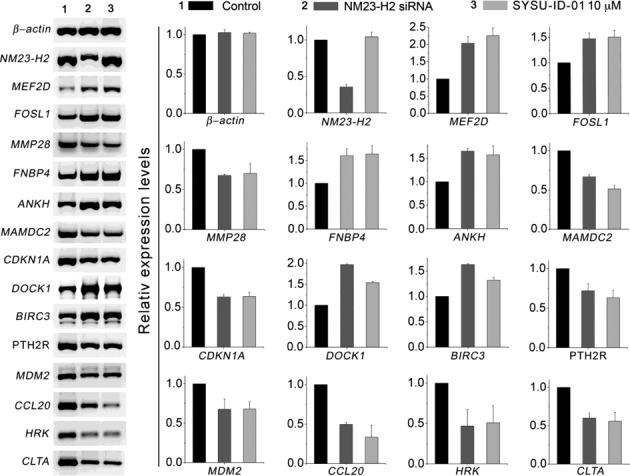
The effects of SYSU-ID-01 on 14 NM23-H2-associated genes. The mRNA levels of 14 genes in untreated HeLa cells (left), 10 μM SYSU-ID-01-treated HeLa cells (right), and NM23-H2 knockdown HeLa cells (middle). The detected genes included 14 NM23-H2-associated genes, which showed significant changes in transcription levels during the knockdown of the NM23-H2 genes. Total RNA was purified and then reverse transcribed and analysed by RT-PCR for the indicated genes. Expression data for each gene were derived by quantitatively calculating the optical density of the left electrophoresis gels and graphing the results (original full gels were shown in Supplementary Figure S9). The y-axis indicates the relative expression data for each gene normalized to the untreated samples (n = 3; error bars, S.E.M.). Data from untreated samples and samples treated for 72 h with 10 μM SYSU-ID-01 or NM23-H2 siRNA were shown for each gene.
SYSU-ID-01 induces cell apoptosis
The results above prompted us to investigate the anti-tumour activity of SYSU-ID-01 and its relationship with NM23-H2. Four concentrations (2.5 μM, 5 μM, 10 μM and 20 μM) of SYSU-ID-01 were chosen for real-time cellular analysis (RTCA) to detect the effects of this molecule on long-term proliferation of HeLa cells. The RTCA result showed that the arrest of the proliferation of HeLa cells appeared after 50 h treatment with SYSU-ID-01 at all four concentrations (Supplementary Figure S10a). Whereas for Primary cultured mouse mesangial cells, in which the proliferation does not depend on c-MYC expression (Supplementary Figure S10b), instead of proliferation arrest, SYSU-ID-01 can promote cellular proliferation at all four concentrations. Moreover, Hoechst 33342 and propidium iodide (PI) double-staining results showed that Hoechst-positive and PI-positive clones obviously increased after SYSU-ID-01 treatment (Figure 6A), implying cell apoptosis and subsequent cell death. We further stained primary cultured mouse mesangial cells with Hoechst 33342 and PI. As shown in Supplementary Figure S11, the primary cultured cells were stained by Hoechst 33342 in light blue but could not be stained by PI in both SYSU-ID-01 treated and untreated samples, and the cell morphology was not changed obviously, which revealed that cell apoptosis does not occur in primary cultured cells under SYSU-ID-01 treatment. The SYSU-ID-01-treated HeLa cells were also analysed by flow cytometry after double-staining by Annexin V and PI. As shown in Figure 6B, SYSU-ID-01 could induce early and late apoptosis after SYSU-ID-01 treatment, showing FITC-positive and PI-negative, and FITC-positive and PI-positive, respectively. In addition, a dose-dependent increase of the apoptosis marker, cleaved caspase-3, was also detected in HeLa cells after SYSU-ID-01 treatment (Figure 6C), which confirmed the apoptosis-inducing capacity of SYSU-ID-01. Thus, the proliferation arrest and apoptosis of HeLa cells by SYSU-ID-01 might be produced by regulating c-MYC transcription and expression. Besides, we found that SYSU-ID-01 was incapable to induce further proliferation inhibition after knocking down of NM23-H2 in Hela cells by using siRNA (Supplementary Figures S6 and S10c). Considering of the function of NM23-H2 in c-MYC regulation, it was hypothesized that the proliferation arrest and apoptosis induced by SYSU-ID-01 in Hela cells might be the effect of suppressing NM23-H2 function. According to the previous reports, c-MYC is very important for DNA damage-induced apoptosis (40); our finding that SYSU-ID-01 repressed c-MYC transcription and induced cell apoptosis gave us a hint that a NM23-H2 binder could be an alternative strategy for c-MYC targeting therapy.
Figure 6.
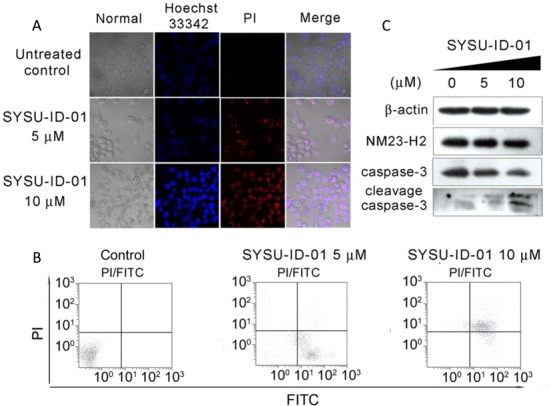
The effects of SYSU-ID-01 on HeLa cells. The cells were either untreated or treated with 5 μM or 10 μM SYSU-ID-01. (A) The confocal images of HeLa cells. After treated with SYSU-ID-01 for 72 h, all cells were stained with Hoechst 33342 (blue) and PI (red), and imaged by a Zeiss LSM 710 laser scanning confocal microscope. The Hoechst 33342 signal was detected at 408 nm and the PI signal was detected at 453 nm, the samples were also photographed in normal visible light. (B) Apoptosis was measured by flow cytometry. Cells were stained with Annexin V-FITC and PI and analysed via flow cytometry. The horizontal and vertical axes represent labelling with Annexin V-FITC and PI, respectively. (C) Western blot was used to detect the expression level of cleaved caspase-3 protein, which is a marker of apoptosis response, under SYSU-ID-01 treatment. The expression level of NM23-H2 protein was also detected, beta actin was used as a loading control.
Binding mode of SYSU-ID-01 with NM23-H2
SYSU-ID-01 showed good activity on the binding to NM23-H2, intervention on the NM23-H2–DNA complex, repression on c-MYC transcription and induction of tumour cell apoptosis, implying it could be used as a leading compound for our study. Thus, we needed more binding information for further drug development. Molecular modelling was firstly carried out to predict the possible binding mode and binding pocket of SYSU-ID-01 with the NM23-H2 protein. The NM23-H2 protein structures were obtained from the reported X-ray structure (PDB ID: 3BBB) (23). Four binding modes with ligands in different orientation were clustered form 200 docking runs by the AUTODOCK 4.0 programme and validated by MD simulation (Supplementary Figure S12). All of the models were quite stable during the dynamics runs (Supplementary Figure S13). The estimated free energies in MM-PBSA calculations for each model were strong (Supplementary Table S4). The modelling results showed that the SYSU-ID-01 molecule was well suited for the narrow, slightly curved pocket that the dinucleotide possessed, as was previously reported (23), implying that SYSU-ID-01 could bind to NM23-H2 by replacing single-stranded DNA.
Figure 7A showed the average minimized structure for the most stable binding mode 2, as indicated by the maximum free energy of −27.50 kcal/mol (Supplementary Table S4), and Figure 7B showed the crystal structure of the complex of NM23-H2 and DNA fragment (dAG). The amino acids Asp54, Asp121, His118, Gly113 and Phe60 in this protein pocket might be the crucial sites interacting with the compound SYSU-ID-01.
Figure 7.
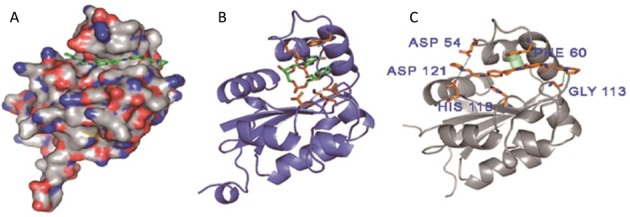
(A) Binding mode 2 of SYSU-ID-01 bound to one subunit of NM23-H2. (B) Binding mode of dinucleotide binding to NM23-H2. (C) Binding mode 2 of SYSU-ID-01 bound to NM23-H2, indicating hydrogen bonding to ASP54, HIS118 and ASP121, and π–π stacking with PHE60.
To further validate the binding modes, five single mutants (D54A, G113F, H118F, D121A, F60A) and two multiple mutants (D54A/D121A and D54A/D121A/F60A) were constructed and verified (construction process could be found in the supplementary methods, and Supplementary Tables S5 and S6) basing on the binding mode 2 (Figure 7A and Supplementary Table S4). Both MST (Figure 8A) and SPR (Figure 8B) experiments showed that SYSU-ID-01 could barely bind to the D54A/D121A/F60A three-point mutant. In addition, MST data showed that SYSU-ID-01 could still bind to the D54A mutant with a reduced binding affinity (with a KD value of 55.6 ± 7.6 μM compared to 9.8 ± 0.039 μM for the wild-type NM23-H2), while barely binding to the other single mutants (G113F, H118F, D121A, F60A) or the two-point mutant D54A/D121A. All these data indicated that Asp121, His118, Gly113 and Phe60 are indispensable for the binding between SYSU-ID-01 and NM23-H2, just like the prediction from molecular modelling. On the other hand, what are the roles of these sites on NM23-H2–DNA binding? We further applied EMSA, filter-binding assays and MST experiments for elucidation. EMSA and filter-binding assays, shown in Figure 3F and G, demonstrated that the binding affinities of the NM23-H2 mutants to Pu27 decreased significantly, and the G113F, D121A, D54/121A and D54/121A/F60A mutants did not have the capability to bind c-MYC G-rich DNA again. The MST experiments gave us more quantitative evidence to support the results from the EMSA and filter-binding assays. As shown in Supplementary Figure S14, the KD value for the wild-type NM23-H2 was 0.14 ± 0.010 μM, while the binding affinity was reduced to 7.8 ± 0.63 μM for the D54A mutant, and 0.7 ± 0.029 μM for the H118F mutant. For G113F, D121A, F60A, D54/121A and D54/121A/F60A mutants, a quite weak binding ability or not even the ability to obtain a binding curve to Pu27 was observed. The results indicated that three amino acid residues, Phe60, Gly113, and Asp121, were more crucial to the protein–DNA interaction than Asp54 and His118. Thus, SYSU-ID-01 and Pu27 DNA possess the same binding sites with NM23-H2 in Asp121, Gly113 and Phe60, implying that SYSU-ID-01 bind to NM23-H2 by replacing the single-stranded DNA.
Figure 8.
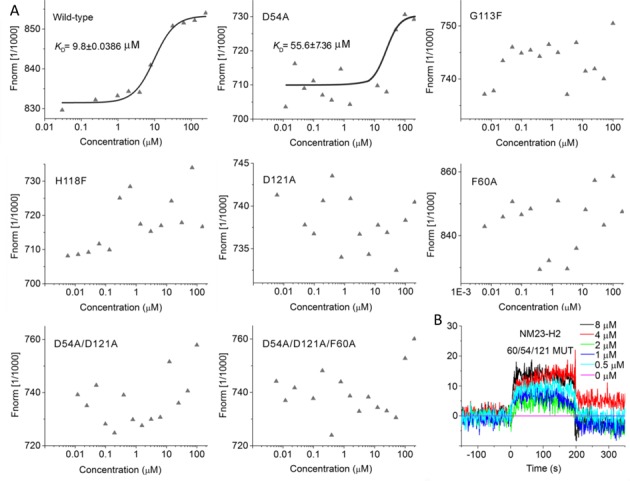
Binding affinity of wild-type and mutant NM23-H2 to SYSU-ID-01. (A) The MST results provided us with a quantitative impression of the interaction of wild-type and mutant NM23-H2 to SYSU-ID-01. Only wild-type NM23-H2 gives a typical binding curve and leads to a KD = 9.8 ± 0.0386 μM, while the data points for mutant NM23-H2 proteins were chaotic, which demonstrated the weaker binding affinities. The SYSU-ID-01 was titrated from 6 nM to 200 μM, the concentration of NT-647 labelled NM23-H2 was kept constant at 200 nM. (B) SPR sensorgrams for the binding of SYSU-ID-01 to immobilized D54A/D121A/F60A mutant NM23-H2 in SPR assays, the ligand concentrations in the flow solutions were 0–8 μM. The sensorgrams of SYSU-ID-01 binding to D54A/D121A/F60A mutant NM23-H2 were comparatively low and chaotic, which revealed the weaker combining capacity.
DISCUSSION
Efforts to disturb transcriptional pathways have focused on protein–protein interactions or particular recognition on DNA, such as protein dimerization antagonist Stabilized Alpha-Helices of BCL-2 domains (SAHBs) that binds to multidomain BCL-2 member pockets to activate apoptosis (41). Ligands directly binding to transcription factors are hard to find, because transcription factors lack substrate binding pockets and are often characterized by hydrophobic surfaces with few druggable regions (42). However, some recent publications report novel small molecules from high throughput screening that directly bind to transcription factors, such as FDI-6 that binds to FOXM1 (9) and BRD32048 that is an ETV1 perturbagen (43). We also started our work with small-scale screening to identify small molecule binders of the transcription factor NM23-H2 using an SPR assay. Quinazolone derivatives showed good binding affinities to NM23-H2, most likely arising from the quinazolone pharmacophore (44). The compounds possess high binding affinities that further interfere with the NM23-H2–DNA complex, indicating that the screening was effective. As we indicated previously, the screening was performed on our in-house G-quadruplex ligand library, although some of compounds among them were not good G-quadruplex ligands, we actually hardly exclude the contribution of compounds stabilizing quadruplexes from the final output that interferes with the NM23-H2–DNA complex. It is worth noting that some quindoline derivatives (such as compound 74) that have been identified as strong quadruplex stabilizers (25) possess weak binding affinity to NM23-H2, while still powerfully disrupting NM23-H2–DNA in our study, which indicate that G-quadruplex ligands may also be a good candidate for the chemical intervention of NM23-H2 transcriptional programme on c-MYC and need a further study in the future. In addition, we did not find a good suppressor of the DNA–protein interaction by strongly binding both DNA and protein in the library. It is true that we focused on the direct binder of NM23-H2 here, and thus we selected the quinazolone derivative SYSU-ID-01 as a potential candidate for this study.
Thus, the first thing we needed to confirm with more evidence was whether this compound could directly bind to the NM23-H2 protein. Data from SPR, MST and STD-NMR all supported that SYSU-ID-01 bound directly to NM23-H2. Further assays indicated that SYSU-ID-01 not only bound to NM23-H2 but also replaced the single-stranded DNA binding to the protein, because the compound may share the same binding pocket as single-stranded DNA in NM23-H2 and interact with the same amino acid residues, including Asp121, Gly113 and Phe60. Molecular modelling and site-directed mutagenesis data revealed that the shape of this compound was slender, which ensured that it could fit within the narrow DNA binding pocket of NM23-H2. Moreover, the fused aromatic ring system facilitated π–π stacking with Phe60, a critical residue within the active pocket. The introduction of amide side chains with hydrogen bond donors or acceptors enhanced the interaction of the side chain with polar amino acid residues. Nevertheless, we have to admit that the most convincing evidence for the binding should be crystallographic data and we are still working on it, because the crystal structure of a ternary complex containing an active hexamer protein is very hard to obtain.
Our aim is to determine an active transcription factor binder with obvious effects on c-MYC transcription and thus to provide an alternative strategy for c-MYC targeting therapy. Therefore, we further verified the effect of SYSU-ID-01 on c-MYC transcription, expression and subsequent cellular events. Data showed that SYSU-ID-01 could regulate the transcription of c-MYC and other NM23-H2-associated genes, resulting in the induction of HeLa cell apoptosis and death, and finally, the inhibition of tumour cell proliferation. These findings support a possible mechanism whereby a small molecule binds directly to NM23-H2 and then prevents the interaction of NM23-H2 with several gene promoters. The more direct evidence of NM23-H2 as the target of compound in cell needs further efforts by using biotin-labelled compound to pull down the protein.
Because NM23-H2 exhibits selective binding to purine-rich DNA sequences (23), which prefer to form non-B DNA structures that play roles in gene regulation, and the functional non-B DNA structures are always under the control of proteins through specific interactions, it is suggested that NM23-H2 is likely to act as a regulatory factor dependent on the structure of its binding elements. Specifically, the protein-binding elements in the genes’ promoters adopt specific secondary structures (such as a G-quadruplex) when no protein binding occurs and act as repressing or activating regulators towards gene transcription. Once NM23-H2 recognizes and binds to one of these elements, the secondary structure changes to another structure (such as a single strand) that cannot function as a repressor or activator. Based on the common mechanism demonstrated here, we believe that searching for small molecules that can block the interaction of the NM23-H2 protein with DNA structures is likely to be an effective method for the development of NM23-H2-targeting regulators, and this strategy also gives a new perspective on the development of agents that regulate special DNA structures.
Supplementary Material
Acknowledgments
We thank Qing-song Liu, Na Zhang and Ao-li Wang from the High Magnetic Field Laboratory, Chinese Academy of Sciences for technical assistance with Saturation Transfer Difference (STD) NMR experiments, as well as Shankar Balasubramanian and Kaiping Deng for help with manuscript preparation. We also thank Liang Zhou (Quantum Design China & NanoTemper Technologies GmbH) for supporting the Microscale Thermophoresis (MST) assay.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [81330077, 21172272 and 91213302 to Z.-S.; 21372263 to T.-M.]; the Fundamental Research Funds for the Central Universities [11ykzd04 to T.-M.]; the Zhujiang Nova Programme [2011J2200075 to T.-M.]; the Foundation for Distinguished Young Talents in Higher Education of Guangdong [Yq2013002 to T.-M.]; the Academic Scholarship for Distinguished Doctoral Candidates [by the Ministry of Education to J.L.]; the Guangdong Provincial Key Laboratory of Construction Foundation (2011A060901014). Funding for open access charge: Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prochownik E.V. c-Myc as a therapeutic target in cancer. Expert Rev. Anticancer Ther. 2004;4:289–302. doi: 10.1586/14737140.4.2.289. [DOI] [PubMed] [Google Scholar]
- 3.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y., Tan J., Ou T., Huang Z., Gu L. DNA G-quadruplex binders: a patent review. Expert Opin. Ther. Patents. 2013;23:1495–1509. doi: 10.1517/13543776.2013.833187. [DOI] [PubMed] [Google Scholar]
- 5.Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 6.Libermann T.A., Zerbini L.F. Targeting transcription factors for cancer gene therapy. Curr. Gene Ther. 2006;6:17–33. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- 7.Darnell J.E. Jr. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 8.Koehler A.N. A complex task: Direct modulation of transcription factors with small molecules. Curr. Opin. Chem. Biol. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gormally M.V., Dexheimer T.S., Marsico G., Sanders D.A., Lowe C., Matak-Vinkovic D., Michael S., Jadhav A., Rai G., Maloney D.J., et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat. Commun. 2014;5:5165–5175. doi: 10.1038/ncomms6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postel E.H., Mango S.E., Flint S.J. A nuclease-hypersensitive element of the human c-myc promoter interacts with a transcription initiation factor. Mol. Cell. Biol. 1989;9:5123–5133. doi: 10.1128/mcb.9.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postel E.H., Berberich S.J., Flint S.J., Ferrone C.A. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 12.Berberich S.J., Postel E.H. PuF/NM23-H2/NDPK-B transactivates a human c-myc promoter-CAT gene via a functional nuclease hypersensitive element. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 13.Tschiedel S., Bach E., Jilo A., Wang S.Y., Lange T., Al-Ali H.K., Vucinic V., Niederwieser D., Cross M. Bcr-Abl dependent post-transcriptional activation of NME2 expression is a specific and common feature of chronic myeloid leukemia. Leuk Lymphoma. 2012;53:1569–1576. doi: 10.3109/10428194.2012.656631. [DOI] [PubMed] [Google Scholar]
- 14.Lee M.J., Xu D.Y., Li H., Yu G.R., Leem S.H., Chu I.S., Kim I.H., Kim D.G. Pro-oncogenic potential of NM23-H2 in hepatocellular carcinoma. Exp. Mol. Med. 2012;44:214–224. doi: 10.3858/emm.2012.44.3.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi A., Urano T., Goi T., Takeuchi K., Niimoto S., Nakagawara G., Furukawa K., Shiku H. Expression of human nm23-H1 and nm23-H2 proteins in hepatocellular carcinoma. Cancer. 1994;73:2280–2284. doi: 10.1002/1097-0142(19940501)73:9<2280::aid-cncr2820730908>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Yokdang N., Buxton N.D., Buxton I.L. Measurement of human breast tumor cell-secreted shNDPK-B in a murine breast cancer model suggests its role in metastatic progression. Proc. West Pharmacol. Soc. 2009;52:88–91. [PMC free article] [PubMed] [Google Scholar]
- 17.Herak Bosnar M., Bago R., Konjevoda P., Pavelic J. Gene expression profiling of Nm23-H2 overexpressing CAL 27 cells using DNA microarray. Neoplasma. 2008;55:447–454. [PubMed] [Google Scholar]
- 18.Postel E.H., Berberich S.J., Rooney J.W., Kaetzel D.M. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J. Bioenerg. Biomembr. 2000;32:277–284. doi: 10.1023/a:1005541114029. [DOI] [PubMed] [Google Scholar]
- 19.Pinon V.P., Millot G., Munier A., Vassy J., Linares-Cruz G., Capeau J., Calvo F., Lacombe M.L. Cytoskeletal association of the A and B nucleoside diphosphate kinases of interphasic but not mitotic human carcinoma cell lines: specific nuclear localization of the B subunit. Exp. Cell Res. 1999;246:355–367. doi: 10.1006/excr.1998.4318. [DOI] [PubMed] [Google Scholar]
- 20.Kraeft S.K., Traincart F., Mesnildrey S., Bourdais J., Veron M., Chen L.B. Nuclear localization of nucleoside diphosphate kinase type B (nm23-H2) in cultured cells. Exp. Cell Res. 1996;227:63–69. doi: 10.1006/excr.1996.0250. [DOI] [PubMed] [Google Scholar]
- 21.Michelotti E.F., Sanford S., Freije J.M., MacDonald N.J., Steeg P.S., Levens D. Nm23/PuF does not directly stimulate transcription through the CT element in vivo. J. Biol. Chem. 1997;272:22526–22530. doi: 10.1074/jbc.272.36.22526. [DOI] [PubMed] [Google Scholar]
- 22.Postel E.H., Ferrone C.A. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J. Biol. Chem. 1994;269:8627–8630. [PubMed] [Google Scholar]
- 23.Dexheimer T.S., Carey S.S., Zuohe S., Gokhale V.M., Hu X., Murata L.B., Maes E.M., Weichsel A., Sun D., Meuillet E.J., et al. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III(1) Mol. Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J.L., Lu Y.J., Ou T.M., Zhou J.M., Huang Z.S., Zhu X.F., Du C.J., Bu X.Z., Ma L., Gu L.Q., et al. Synthesis and evaluation of quindoline derivatives as G-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J. Med. Chem. 2005;48:7315–7321. doi: 10.1021/jm050041b. [DOI] [PubMed] [Google Scholar]
- 25.Ou T.M., Lu Y.J., Zhang C., Huang Z.S., Wang X.D., Tan J.H., Chen Y., Ma D.L., Wong K.Y., Tang J.C., et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y.J., Ou T.M., Tan J.H., Hou J.Q., Shao W.Y., Peng D., Sun N., Wang X.D., Wu W.B., Bu X.Z., et al. 5-N-methylated quindoline derivatives as telomeric g-quadruplex stabilizing ligands: effects of 5-N positive charge on quadruplex binding affinity and cell proliferation. J. Med. Chem. 2008;51:6381–6392. doi: 10.1021/jm800497p. [DOI] [PubMed] [Google Scholar]
- 27.Hou J.Q., Tan J.H., Wang X.X., Chen S.B., Huang S.Y., Yan J.W., Chen S.H., Ou T.M., Luo H.B., Li D., et al. Impact of planarity of unfused aromatic molecules on G-quadruplex binding: learning from isaindigotone derivatives. Org. Biomol. Chem. 2011;9:6422–6436. doi: 10.1039/c1ob05884c. [DOI] [PubMed] [Google Scholar]
- 28.Tan J.H., Ou T.M., Hou J.Q., Lu Y.J., Huang S.L., Luo H.B., Wu J.Y., Huang Z.S., Wong K.Y., Gu L.Q. Isaindigotone derivatives: a new class of highly selective ligands for telomeric G-quadruplex DNA. J. Med. Chem. 2009;52:2825–2835. doi: 10.1021/jm801600m. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W.J., Ou T.M., Lu Y.J., Huang Y.Y., Wu W.B., Huang Z.S., Zhou J.L., Wong K.Y., Gu L.Q. 9-Substituted berberine derivatives as G-quadruplex stabilizing ligands in telomeric DNA. Bioorg. Med. Chem. 2007;15:5493–5501. doi: 10.1016/j.bmc.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 30.Ma Y., Ou T.M., Tan J.H., Hou J.Q., Huang S.L., Gu L.Q., Huang Z.S. Synthesis and evaluation of 9-O-substituted berberine derivatives containing aza-aromatic terminal group as highly selective telomeric G-quadruplex stabilizing ligands. Bioorg. Med. Chem. Lett. 2009;19:3414–3417. doi: 10.1016/j.bmcl.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 31.Ma Y., Ou T.M., Tan J.H., Hou J.Q., Huang S.L., Gu L.Q., Huang Z.S. Quinolino-benzo-[5, 6]-dihydroisoquindolium compounds derived from berberine: a new class of highly selective ligands for G-quadruplex DNA in c-myc oncogene. Eur. J. Med. Chem. 2011;46:1906–1913. doi: 10.1016/j.ejmech.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Peng D., Tan J.H., Chen S.B., Ou T.M., Gu L.Q., Huang Z.S. Bisaryldiketene derivatives: a new class of selective ligands for c-myc G-quadruplex DNA. Bioorg. Med. Chem. 2010;18:8235–8242. doi: 10.1016/j.bmc.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Webb P.A., Perisic O., Mendola C.E., Backer J.M., Williams R.L. The crystal structure of a human nucleoside diphosphate kinase, NM23-H2. J. Mol. Biol. 1995;251:574–587. doi: 10.1006/jmbi.1995.0457. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W., Duhr S., Baaske P., Laue E. Microscale thermophoresis for the assessment of nuclear protein-binding affinities. Methods Mol. Biol. 2014;1094:269–276. doi: 10.1007/978-1-62703-706-8_21. [DOI] [PubMed] [Google Scholar]
- 35.Mayer M., Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 36.Postel E.H. NM23/Nucleoside diphosphate kinase as a transcriptional activator of c-myc. Curr. Top Microbiol. Immunol. 1996;213:233–252. doi: 10.1007/978-3-642-61109-4_11. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez V., Guo K., Hurley L., Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postel E.H., Weiss V.H., Beneken J., Kirtane A. Mutational analysis of NM23-H2/NDP kinase identifies the structural domains critical to recognition of a c-myc regulatory element. Proc. Natl Acad. Sci. U.S.A. 1996;93:6892–6897. doi: 10.1073/pnas.93.14.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raveh S., Vinh J., Rossier J., Agou F., Veron M. Peptidic determinants and structural model of human NDP kinase B (Nm23-H2) bound to single-stranded DNA. Biochemistry. 2001;40:5882–5893. doi: 10.1021/bi001085j. [DOI] [PubMed] [Google Scholar]
- 40.Phesse T.J., Myant K.B., Cole A.M., Ridgway R.A., Pearson H., Muncan V., van den Brink G.R., Vousden K.H., Sears R., Vassilev L.T., et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21:956–966. doi: 10.1038/cdd.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walensky L.D., Kung A.L., Escher I., Malia T.J., Barbuto S., Wright R.D., Wagner G., Verdine G.L., Korsmeyer S.J. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdine G.L., Walensky L.D. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 43.Pop M.S., Stransky N., Garvie C.W., Theurillat J.P., Hartman E.C., Lewis T.A., Zhong C., Culyba E.K., Lin F., Daniels D.S., et al. A small molecule that binds and inhibits the ETV1 transcription factor oncoprotein. Mol. Cancer Ther. 2014;13:1492–1502. doi: 10.1158/1535-7163.MCT-13-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava V., Sinha D., Tiwari A.K., Sharma H., Bala Sharma R., Singh V.K., Mishra A.K. Quantitative structure-activity relationship analysis of 4(3H)-quinazolone derivatives as tyrosine kinase inhibitors by multiple linear regression. Cancer Biother. Radiopharm. 2010;25:559–562. doi: 10.1089/cbr.2010.0791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


