Abstract
Polyploidization is an important speciation mechanism in the barley genus Hordeum. To analyze evolutionary changes after allopolyploidization, knowledge of parental relationships is essential. One chloroplast and 12 nuclear single-copy loci were amplified by polymerase chain reaction (PCR) in all Hordeum plus six out-group species. Amplicons from each of 96 individuals were pooled, sheared, labeled with individual-specific barcodes and sequenced in a single run on a 454 platform. Reference sequences were obtained by cloning and Sanger sequencing of all loci for nine supplementary individuals. The 454 reads were assembled into contigs representing the 13 loci and, for polyploids, also homoeologues. Phylogenetic analyses were conducted for all loci separately and for a concatenated data matrix of all loci. For diploid taxa, a Bayesian concordance analysis and a coalescent-based dated species tree was inferred from all gene trees. Chloroplast matK was used to determine the maternal parent in allopolyploid taxa. The relative performance of different multilocus analyses in the presence of incomplete lineage sorting and hybridization was also assessed. The resulting multilocus phylogeny reveals for the first time species phylogeny and progenitor-derivative relationships of all di- and polyploid Hordeum taxa within a single analysis. Our study proves that it is possible to obtain a multilocus species-level phylogeny for di- and polyploid taxa by combining PCR with next-generation sequencing, without cloning and without creating a heavy load of sequence data.
Keywords: Evolution, Hordeum, in silico cloning, multispecies coalescent, nuclear single-copy genes, phylogeny, polyploidy, systematics
Hordeum L., belonging to the economically important grass tribe Triticeae, consists of about 33 species including cultivated barley, H. vulgare. The species are disjunctly distributed in arid and temperate areas of the Northern Hemisphere, South America, and South Africa (Bothmer et al. 1995; Blattner 2006; Blattner et al. 2010). With nearly half of the species being polyploids (tetra- and hexaploids), including allo- and autopolyploids, the genus Hordeum is a good model to study speciation through polyploidization. However, even after 50 years of research in species relationships, the phylogeny of the genus is still not completely resolved (for a review, see Blattner 2009). Recently, a consensus seems to have emerged for the relationships among diploid Hordeum taxa from the analysis of multilocus data sets (Blattner 2009; Petersen et al. 2011), although analyses including multiple individuals per species are still rare. For the polyploids, most of the phylogenetic studies published to date have been aimed at resolving the relationships of only a few polyploid taxa (Salomon and Bothmer 1998; Petersen and Seberg 2004; Taketa et al. 2005, 2009; Blattner 2006; Jakob et al. 2007; Jakob and Blattner 2010; Tanno et al. 2010) or used only single DNA marker regions to analyze all polyploid taxa (Blattner 2004; Brassac et al. 2012). Thus, a comprehensive data set including all species with multiple individuals and multiple loci, which is necessary to arrive at a good hypothesis of relationships, is still lacking.
Studies on polyploid taxa are generally impeded by the complex evolution of these organisms, involving recurrent formation (Brochmann et al. 1992; Soltis and Soltis 1999), gene loss or retention (Blattner 2004; Kotseruba et al. 2010; Buggs et al. 2012), and homoeologous recombination (Doyle et al. 2008; Brassac et al. 2012; Weiss-Schneeweiss et al. 2013). Chloroplast DNA is usually maternally inherited in angiosperms (Reboud and Zeyl 1994) and therefore not able to detect hybridization events. They can, however, be used to identify the direction of hybrid speciation in polyploids, that is, to determine maternal parents (Doebley et al. 1992; Nishikawa et al. 2002). Also the often employed nuclear ribosomal DNA region might result in effectively uniparental inheritance due to its peculiar mode of evolution with homogenization of tandem repeats (Álvarez and Wendel 2003; Blattner 2004) or the loss of entire rDNA clusters (Kotseruba et al. 2010). As an alternative approach, low- or single-copy nuclear loci have been proposed as a source of phylogenetic information and for improving resolution and robustness in comparison to plastid and ribosomal DNA (Sang 2002; Small et al. 2004), particularly if polyploid taxa are studied (Sang et al. 2004). Low-copy nuclear markers also have some disadvantages; there are no universal PCR primers available that are applicable in all plant groups, and additional costs and lab work arise due to the necessity of cloning PCR amplicons prior to sequencing (Triplett et al. 2012). Single-molecule PCR (Marcussen et al. 2012) or using homoeolog-specific primers (Petersen and Seberg 2004) are alternatives to cloning when working with polyploids, but require intensive lab work and/or a priori knowledge of the allele diversity at the analyzed loci.
Another disadvantage of nuclear loci is their potential discordance with each other. Incongruences between gene trees, and eventually with the species tree, have been long recognized (Pamilo and Nei 1988), as have the processes from which they may arise (reviewed in Maddison 1997; Edwards 2009). A simple method to integrate phylogenetic information of multiple loci consists in concatenating the different sequences to obtain an average phylogeny. However, it has been shown that this method can lead to overconfidence in incorrect species trees (Kubatko and Degnan 2007; Edwards 2009). New methods accounting for the stochastic history of genes have since been developed, among them are Bayesian concordance analysis (BCA) and the multispecies coalescent (MSC) method (Degnan and Rosenberg 2009; Heled and Drummond 2010).
Here we present an analysis that is based on 12 nuclear loci, distributed on six of the seven barley chromosomes, and one chloroplast region. Eight of the nuclear loci were newly explored for phylogenetics and are derived from rice genes (Ishikawa et al. 2009), while four genes had previously been used (Petersen et al. 2011; Brassac et al. 2012). Initially, we started out with a higher number of genes and then removed loci that did not easily amplify, were not single copy, or comprised motifs that turned out to be problematic to sequence. All species were included with one to four individuals per species to additionally sample part of the intraspecific diversity. Taking advantage of next-generation sequencing (NGS) technologies including barcoding (Meyer et al. 2008b) and in silico cloning of multiplex-sequenced DNA fragments, we present here an extension of the method described in Griffin et al. (2011). In contrast to Griffin et al. (2011), our approach allows sequencing PCR amplicons of a large size (the longest sequence had a length of 3500 base pairs [bp]) not specifically designed to fit the read length of the employed NGS platform. Furthermore, we propose a method to disentangle reads from different genomes in a contig. Phylogenetic analyses were conducted on single loci and concatenated data from all loci. A dated species tree was inferred from the MSC from multiple gene trees. Due to the lack of fossils for the tribe Triticeae, calibration points were taken from Marcussen et al. (2014) who used four Pooideae fossils and a chloroplast data set for 92 representatives of Pooideae to determine the age of the Brachypodium stem node that was further used to infer the crown-group age of the Triticeae based on 275 gene phylogenies. We also inferred incongruences between gene trees. These approaches allowed us to explore the allelic diversity simultaneously at numerous loci in multiple individuals to retrieve the phylogeny of the genus Hordeum and infer relationships between diploid and polyploid taxa and cytotypes. Also, the efficiency of multilocus phylogenetic methods in the presence of incomplete lineage sorting (ILS) and hybridization could be tested.
Materials and Methods
Plant Materials
We included 105 individuals representing all 33 species and most subspecies of the genus plus 1–2 individuals from each of five diploid Triticeae species outside Hordeum plus Bromus as out-groups (Table 1). For genome nomenclature in Hordeum, we follow Blattner (2009), with the H genome occurring in H. vulgare and H. bulbosum, the Xu genome in H. murinum, the Xa genome in H. marinum and H. gussoneanum while all the remaining species have the I genome. Included individuals (Supplementary Table S1 available on Dryad at http://dx.doi.org/10.5061/dryad.fn2nt were chosen to reflect the intraspecific diversity observed in TOPO6 (Brassac et al. 2012). Herbarium vouchers of the analyzed materials were deposited in the herbaria of the IPK Gatersleben (GAT) or the Museum of Natural History, Buenos Aires (BA).
Table 1.
Taxa included in the study
| Taxon | Ploidy level (N)a | Haploid genome | Distribution area |
|---|---|---|---|
| Hordeum subgenus Hordeum | |||
| Section Hordeum | |||
| H. vulgare L. | |||
| subsp. spontaneum (C.Koch.) Thell. | 2x (2) | H | SW Asia |
| H. bulbosum L. | 2x (1), 4x (3) | H, HH | Mediterranean to C Asia |
| Section Trichostachys Dum. | |||
| H. murinum L. | |||
| subsp. glaucum (Steud.) Tzvel. | 2x (2) | Xu | Mediterranean to C Asia |
| subsp. murinum | 4x (2) | XuXu | NW Europe to Caucasus |
| subsp. leporinum (Link) Arc. | 4x (2), 6x (1) | XuXu, XuXuXu | Mediterranean to C Asia |
| Hordeum subgenus Hordeastrum (Doell) Rouy | |||
| Section Marina (Nevski) Jaaska | |||
| H. gussoneanum Parl. | 2x (2), 4x (2) | Xa, XaXa | Mediterranean to C Asia |
| H. marinum Huds. | 2x (2) | Xa | Mediterranean |
| Section Stenostachys Nevski | |||
| Series Sibirica Nevski | |||
| H. bogdanii Will. | 2x (3) | I | C Asia |
| H. brevisubulatum (Trin.) Linkb | 2x (5), 4x (4), 6x (3) | I,II, III | C Asia |
| H. roshevitzii Bowden | 2x (2) | I | C Asia |
| Series Critesion (Raf.) Blattner | |||
| H. californicum Covas & Stebb. | 2x (3) | I | SW California |
| H. chilense Roem. & Schult. | 2x (2) | I | Chile and W Argentina |
| H. comosum Presl | 2x (3) | I | S Argentina |
| H. cordobenseBothmer et al. | 2x (2) | I | C Argentina |
| H. erectifolium Bothmer et al. | 2x (1) | I | C Argentina |
| H. euclaston Steud. | 2x (3) | I | C Argentina, Uruguay |
| H. flexuosum Steud. | 2x (1) | I | E+C Argentina |
| H. intercendens Nevski | 2x (3) | I | SW California, NW Mexico |
| H. muticum Presl | 2x (2) | I | C to N Andes |
| H. patagonicum (Haum.) Covasb | 2x (3) | I | S Argentina |
| H. pubiflorum Hook.f.b | 2x (2) | I | S Argentina |
| H. pusillum Nutt. | 2x (2) | I | C+E USA |
| H. stenostachys Godr. | 2x (2) | I | C Argentina |
| H. depressum (Scribn. & Sm.) Rydb. | 4x (2) | II | W USA |
| Interserial allopolyploids of series Critesion and Sibirica | |||
| H. brachyantherum Nevski | 4x (2) | II | W North America, Kamchatka, Newfoundland |
| H. fuegianum Bothmer et al. | 4x (2) | II | S Argentina, S Chile |
| H. guatemalense Bothmer et al. | 4x (1) | II | Guatemala, S Mexico |
| H. jubatum L. | 4x (2) | II | NE Asia, NW+W North America, C Argentina |
| H. tetraploidum Covas | 4x (4) | II | C Argentina |
| H. arizonicum Covas | 6x (3) | III | SW USA |
| H. lechleri (Steud.) Schenk | 6x (3) | III | C+S Argentina |
| H. parodii Covas | 6x (3) | III | C Argentina |
| H. procerum Nevski | 6x (2) | III | S Argentina |
| Section Nodosa (Nevski) Blattner | |||
| H. brachyantherum Nevski | 6x (1) | IIXa | C California |
| H. capense Thunb. | 4x (2) | IXa | S Africa |
| H. secalinum Schreb. | 4x (2) | IXa | Mediterranean to W Europe |
| Out-group species | |||
| Eremopyrum triticeum (Gaertn.) Nevski | 2x (1) | FXe | |
| Dasypyrum villosum (L.) Candargy | 2x (2) | V | |
| Taeniatherum caput-medusae (L.) Nevski | 2x (1) | Ta | |
| Triticum monococcum L. | 2x (1) | Am | |
| Secale vavilovii Grossh. | 2x (1) | R | |
| Bromus tectorum L. | 2x (1) | ||
| Brachypodium distachyon (L.) P.Beauv. ex J.Presl. & C.Presl. | 2x (1) | ||
aNumber of individuals included per species or cytotype.
bSpecies with subspecies not further detailed here.
Molecular Methods
Genome size and ploidy level of the Hordeum individuals were initially verified by flow cytometry according to Jakob et al. (2004). Genomic DNA was extracted from approximately 10 mg of silica gel-dried leaves with the DNeasy Plant Mini Kit (Qiagen) according to the protocol of the manufacturer. DNA quality and concentrations were checked on 1% agarose gels.
To arrive at PCR primers amplifying putative single-copy regions in Hordeum, the PCR-based Landmark Unique Gene (PLUG) system (Ishikawa et al. 2007) was used. It consists of mapped single-copy rice genes, which were used to detect conserved regions in wheat expressed sequence tag (EST) sequences, and map orthologous loci to the three genomes of bread wheat (Ishikawa et al. 2009). The PLUG database (http://plug.dna.affrc.go.jp/, last accessed June 19, 2015) was screened for rice landmark loci with unique mapped orthologs in wheat and barley. PCR primers different from the suggested PLUG primers were designed in conserved exons with the intention to amplify regions of ∼2000 bp and spanning one or several introns from all seven Hordeum chromosomes. The target regions thus resulted in larger amplicons in comparison to the PLUG markers, which have an average length of 951 bp (Ishikawa et al. 2009). Initially, 24 loci were PCR-screened with diploid representatives of the four genomes occurring in Hordeum. PCR products were checked on 1.4% agarose gels and numbers and sizes of the amplicons were determined. Finally, ten loci producing single fragments were chosen.
Including six already published loci, a total of two chloroplast and 14 nuclear loci were amplified by PCR (Supplementary Table S2). PCR amplifications for all loci were performed in 30 reaction volume containing approximately 10–50 ng of genomic DNA, 1x Phusion HF Buffer, 0.2 mM each dNTP, 0.5 M of each primer, and 1 U Phusion Hot Start DNA polymerase, a proofreading enzyme (Finnzymes OY). The amplification program consisted of initial denaturation for 3 min at 98 °C, followed by 35 cycles of 30 sec at 98 °C, 1 min at 59 °C, 1 min at 72 °C, and a final extension of 10 min at 70 °C. Variations of this protocol with regards to the primer annealing temperatures for each locus are summarized in Supplementary Table S2. PCR amplification of one chloroplast region, ndhF, failed in more than a third of the individuals and thus ndhF was not included in the following steps.
Two sequencing approaches were used: (i) The amplicons of 96 individuals (all ploidy levels) were 454 sequenced. (ii) Amplicons of eight diploid individuals and one autotetraploid (H. bulbosum F2142) were cloned and eight clones per individual were sequenced following Brassac et al. (2012). The cloned amplicons were used as a control for the NGS approach and to provide reference sequences for assembling of 454 reads. All amplicons were purified using NucleoFast 96 Spin Plates (Macherey-Nagel) according to the protocol of the manufacturer and eluted in 20 of TE buffer.
For the NGS set, the concentrations of each amplicon were quantified using a Quant-iT Picogreen dsDNA (Molecular Probes) assay with a standard curve ranging from 1.25 pg/L to 40 pg/L. Fluorescence at 520 nm was measured on a Tecan Infinite M200 plate reader. The quantification of each amplicon was replicated three times and the average value was used to calculate the concentration of each sample.
Library preparation was performed as described in Meyer et al. (2008b) with the modifications defined below. For each individual, all amplicons were pooled in roughly equimolar ratio with a total amount of 1.5 g of DNA in a reaction volume of 130 . Each pool was sheared from 700–2500 bp to a targeted size of 500–1000 bp via sonication in a microTUBE using a Covaris S220 (Covaris).
For barcoding, we used 7-nucleotide barcodes differentiated from each other by at least two substitutions on a maximum of 400 ng of DNA per individual to limit chimera formation. Tagging efficiency was verified by comparing the lengths of an untagged and a tagged fragment of a known size (200 bp) on a 2% agarose gel.
The tagged samples were quantified using a Quant-iT Picogreen dsDNA (Molecular Probes) assay as described earlier. After a sequencing trial of a diploid (Hordeum bogdanii BCC2070) and a hexaploid individual (H. arizonicum BCC2054), we decided to standardize the DNA input according to the ploidy of the individuals in order to achieve sufficient coverage for the different alleles of the polyploids. Hence, twice the DNA amount of a diploid was used for a tetraploid and three times for a hexaploid individual.
All individually tagged sheared fragments were pooled, concentrated using MinElute PCR purification kit (Qiagen) and loaded on a SYBR Safe (Invitrogen) stained 1.5% agarose gel for size selection. The part of the gel lane comprising the fragments between 500 and 1000 bp was cut and DNA was extracted and purified using NucleoSpin Gel Clean-up (Macherey-Nagel) according to the manufacturer's protocol. DNA concentration of the pool was determined using a Qubit High Sensitivity (Invitrogen) assay. The quality of the pool was also checked using an Agilent High Sensitivity Chip assay. The DNA pool was dephosphorylated using the FastAP Thermosensitive Alkaline Phosphatase (Fermentas) and cut using the SrfI restriction enzyme, leaving the 5′ phosphates free for the ligation of universal adapters for sequencing. To assess the efficiency of adapter ligation and quantify DNA concentration a quantitative PCR was performed as described in Meyer et al. (2008a) using the emPCR primers.
The pool of DNA fragments from 96 individuals was sequenced in a single run on Roche's 454 sequencing platform using a PicoTiterPlate and the GS FLX Titanium XL+ chemistry to obtain long reads (up to 1000 bp) facilitating the identification of alleles and homoeologues. With an estimated length of 16 kbp amplicon lengths per haploid genome and 2- and 3-fold this amount for the polyploids and considering a typical output of 700 Mbp for a 454 run, we calculated that on average 270-fold coverage should be reached.
Quality Control and Haplotype Phasing
Barcode deconvolution, that is, sorting fragments according to the single individuals, was performed with a custom script in Perl, and the online tool TagCleaner (Schmieder et al. 2010) was used to detect and trim adapters and barcodes. Geneious r6.1 (Biomatters Ltd.) was used for quality control and downstream analyses. All alignments were performed in Geneious using Mafft 6.814b (Katoh et al. 2002; Katoh and Toh 2008) with the E-INS-i algorithm (with default settings) and manually checked. First, the sequences obtained from the diploid individuals via cloning were used to map the reads obtained from NGS with the High Sensitivity parameter (maximum mismatches at 40%) available in Geneious. For loci with large insertions/deletions (indels) relative to the reference sequences, an iterative procedure was used to assemble the reads to one of the primer sequence until the second primer-binding site was retrieved. To check for potential extra copies all the reads were remapped to the loci obtained via this initial method with High Sensitivity. Each assembly was then carefully inspected, especially regarding the coverage profile and the presence of single nucleotide polymorphisms (SNPs), trimmed and a consensus sequence was created.
Assemblies for the polyploids contained a mixture of reads belonging to the different homoeologues. To disentangle these copies we used an ad hoc method consisting of de novo assembling of the reads mapped to a particular locus with Low Sensitivity (<10% mismatches allowed). This created a set of smaller assemblies with a more homogenous read content. The assemblies with the most reads (more than 20) were carefully checked for coverage and SNP presence, and their consensuses aligned together to identify the different copies. Homoeologous copies were identified as unique combinations of SNPs. Two copies were expected for the tetraploids and three for the hexaploids. Finally, all the reads were mapped simultaneously, with Medium Sensitivity (20% mismatches), to confirm the different copies. To verify this method we compared the outcome with results obtained using NextAllele, a script for haplotype phasing for NGS data described in O'Neill et al. (2013), on a tetraploid individual (H. depressum BCC2047). Although the method was not designed for polyploids, the results were completely concordant due to the heterozygous-like behavior of the tetraploids. It appeared, however, that NextAllele was sensitive to the ratio between the reads corresponding to the two genomes resulting in ambiguities at the SNPs when this ratio departed from 50%, and, as expected, it could not deal with hexaploid individuals because of its limitation to two alleles. The single-copy status and the location of all nuclear loci were checked by blasting the sequences obtained for H. vulgare against its genome database (http://webblast.ipk-gatersleben.de/barley/viroblast.php, last accessed 19/06/2015; The International Barley Genome Sequencing Consortium 2012).
Phylogenetic Analyses
To infer the phylogeny of Hordeum we adopted the following analysis approach consisting of nine steps. After aligning the sequences from all loci, (i) models of sequence evolution were determined for each locus. Gene trees were calculated for each locus with (ii) the sequences derived from the diploid taxa by Bayesian phylogenetic inference (BI), and (iii) sequences from all diploid plus, consecutively, single polyploid individuals were clustered by neighbor-joining analysis to determine phylogenetic affiliation (phasing) of the homoeologous gene copies found in polyploid taxa. Concatenated sequences from all loci (supermatrices) were used for BI of (iv) diploid and (v) diploid plus phased homoeologs of polyploid taxa. (vi) A MSC-based analysis was conducted to infer species trees from gene trees for the diploid individuals. (vii) To date nodes within the Hordeum phylogeny a molecular clock approach was conducted together with the MSC. (viii) A BCA was conducted on the diploid taxa to estimate gene tree incongruences. Finally, (ix) chloroplast matK sequences were analyzed by BI to detect the maternal lineages in allopolyploids. These analysis steps are detailed below.
Model of sequence evolution
The best-fitting model of sequence evolution for each locus was identified with jModelTest 2.1.4 (Guindon and Gascuel 2003; Darriba et al. 2012) using default parameters. The best partitioning scheme for the concatenated nuclear loci was identified with PartitionFinder 1.1.1 (Lanfear et al. 2012) using the greedy algorithm. In both cases the Bayesian information criterion (BIC, Schwarz 1978) was used for model choice because of its high accuracy (Darriba et al. 2012) and its tendency to favor simpler models than the Akaike information criterion (Posada and Crandall 2001). The preferred partitioning scheme and models of evolution are summarized in Table 3.
Table 3.
Information on individual data sets and setting used in the analyses
| Locus | Alignment length | Parsimony-informative sites | Variable sites | Model of evolutiona | Clock for *BEAST |
|---|---|---|---|---|---|
| TNAC1035 | 752 (753) | 123 (184) | 147 (250) | HKY+G (1) | RLC |
| TNAC1142 | 4408 (4603) | 238 (387) | 310 (524) | HKY+G (2) | RLC |
| XYL | 915 (1121) | 138 (307) | 199 (448) | HKY+G (1) | RLC |
| TNAC1364 | 1631 (1631) | 260 (409) | 293 (584) | HKY+G (2) | RLC |
| NUC | 844 (889) | 99 (176) | 128 (268) | K80+G | b |
| TNAC1403 | 2094 (2707) | 268 (367) | 327 (562) | HKY+G (3) | RLC |
| TNAC1463 | 4913 (5163) | 519 (993) | 838 (1333) | HKY+G (4) | RLC |
| BLZ1 | 1490 (1495) | 119 (211) | 249 (412) | HKY+G (1) | RLC |
| TNAC1610 | 1647 (1736) | 104 (236) | 183 (379) | HKY+G (3) | RLC |
| TOPO6 | 1092 (1101) | 129 (211) | 164 (303) | HKY+G (1) | RLC |
| TNAC1497 | 1256 (2134) | 142 (225) | 165 (325) | HKY+G (1) | RLC |
| TNAC1740 | 1487 (1509) | 207 (309) | 254 (465) | HKY+G (1) | RLC |
| matK | 2606 (2628) | 95 (100) | 203 (210) | HKY+G | RLC |
| Supermatrix | 25135 (24996) | 2852 (3633) | 4915 (5469) | c | b |
Notes: Values in brackets correspond to alignments including polyploids; RLC, random local clock.
aModels used for single locus analyses, in brackets models linked in the *Beast analysis.
bData set not included in the *Beast analysis.
cData set consisting of all loci and divided in five partitions.
Homoeologue identification
Homoeologous copies in polyploid individuals were identified from clustering of the sequences derived from polyploids with those derived from diploid taxa. For each locus and each polyploid species separately the copies from the different individuals were aligned with the sequences obtained from the diploid taxa. The neighbor-joining (NJ) method (Saitou and Nei 1987) with HKY distances (Hasegawa et al. 1985) was used to build a tree with 100 bootstrap replicates (not shown) in Geneious using the Geneious Tree Builder option, as initial tests showed that NJ was able to safely discern and place copies and resulted in similar topologies as BI. The closest neighbor of the polyploids' sequences were identified and coded (A, B, or C) in the same way across the 12 different loci, and sequences were concatenated according to the designation as A, B, or C copies. When more copies than expected were retrieved, all sequences were included in the phylogenetic analysis per locus (diploids and polyploids) and a consensus sequence was created for the one clustering in the same clade, while the most distant copies (compared to the other individuals) were excluded from the supermatrices and the MSC-based analysis. From the prior knowledge acquired during the analysis of TOPO6 (Brassac et al. 2012), we included the sequences of tetraploid Hordeum jubatum, which is assumed to be one of the parents of the American hexaploids, when analyzing the sequences from hexaploid species.
Bayesian phylogenetic inference
All BI analyses were performed in MrBayes 3.2.2 (Ronquist et al. 2012) using the models and partitioning inferred by jModelTest and PartitionFinder. Each analysis consisted of two independent analyses each running four sequentially heated chains (temperature set at 0.05) for 10 million generations and sampling a tree every 1000 generations. Convergence of the runs was assessed in Tracer 1.5 (Rambaut and Drummond 2007) and the online application Awty (Nylander et al. 2008), which plots posterior probabilities of clade support values for both runs against each other. The first 25% of sampled trees were discarded as burn-in and a consensus tree was computed in MrBayes. For the supermatrix analyses with all loci concatenated, the data matrix was partitioned applying the respective model of sequence evolution for each locus/partition.
Sequences derived from all loci were first analyzed for just the diploid individuals using the following approaches: (i) all sequences belonging to single loci were analyzed separately in MrBayes to infer locus phylogenies (gene trees); (ii) the trees sampled from the MrBayes analyses were analyzed with BUCKy 1.4.2 to obtain concordance factors (CF) for all clades; (iii) the sequences from all loci were concatenated (supermatrix) and analyzed in MrBayes with partitions for all loci applying their respective models of sequence evolution. On a data set reduced to all Hordeum species plus Dasypyrum villosum and Brachypodium distachyon, (iv) the MSC was generated with *Beast (Heled and Drummond 2010) as part of the cross platform Beast 1.8.1 (Drummond and Rambaut 2007; Drummond et al. 2012).
Incongruences between gene trees
Concordance among gene trees was estimated with BUCKy 1.4.2 (Ané et al. 2007; Larget et al. 2010) on the diploids plus D. villosum. CF can be interpreted as the proportion of loci supporting a specific topology (Ané et al. 2007; Baum 2007). BUCKy computes the CF for all potential topologies supported by the loci and provides a primary concordance tree displaying relationships supported by the largest proportion of the loci (Larget et al. 2010). The trees sampled for each locus by the MrBayes analyses were summarized discarding 50% of each chain as burn-in. CF were estimated using the default a priori level of discordance () and 1000,000 generations. For the diploid taxa pairwise comparisons of single loci phylogenies as well as multilocus phylogenies were carried out using the Compare2Trees tool (Nye et al. 2006) and summarized in a heat map.
Dating of nodes and MSC
Species trees and clade dating were estimated in *Beast. The analysis was performed on a data set corresponding to all loci except NUC (high amount of missing data) consisting of the diploid Hordeum individuals plus D. villosum, a species nested in a clade with Triticum (Escobar et al. 2011), and B. distachyon as primary out-group. Sequences for B. distachyon were retrieved by performing a Blastn search (Altschul et al. 1990) of our loci on the B. distachyon genome database (http://www.brachypodium.org/, last accessed June 19, 2015). Priors on the root age (normal distribution; 44.4 Ma ± 3.53) and on the Triticeae crown age (normal distribution; 15.32 Ma ± 0.34) were set as inferred by Marcussen et al. (2014). Monophyly was constrained for the Hordeum clade and for the Triticeae clade. The analysis was run using the partitioning scheme and models of sequence evolution identified by PartitionFinder, the Yule species tree prior, as well as the piecewise linear and constant root population model. As rate constancy was systematically rejected for all loci based on the likelihood-ratio test, as proposed by Huelsenbeck and Rannala (1997), a random local clock model (uniform clock rate; min 0, max 1.0) was used. To speed up the analysis we used a starting tree obtained by NJ of the supermatrix. Three independent analyses were computed for 300 million generations each, sampling the states every 3000 generations. Effective sample size (ESS) and convergence of the analyses were assessed using Tracer. Appropriate burn-ins were estimated from each trace file, discarded and all analyses were combined with LogCombiner. A maximum clade credibility (MCC) tree was summarized with TreeAnnotator (part of the Beast package).
Maximum parsimony
To see if the phylogeny obtained by BI is robust regarding different analysis algorithms, a parsimony analysis (MP) of the supermatrix of diploid taxa was conducted in Paup* 4.0b10 (Swofford 2002) using a heuristic search with 200 random sequence additions and tree bisection and reconnection (TBR) branch swapping, saving all shortest trees. Node support was evaluated by 500 bootstrap resamples with the same settings but without random addition sequences.
Inference of parental progenitors of polyploids
To infer parental species of polyploids, the supermatrix of the concatenated sequences derived from each individual, including polyploids, was analyzed by BI as previously described. If homoeologues of polyploids fell within clades with diploid species these were interpreted as progenitor taxa. Clades containing polyploid-only sequences were interpreted as indication of extinct lineages (Blattner 2004; Brassac et al. 2012). Species relationships were summarized in a schematic tree consisting of a backbone as inferred from the MSC obtained from the diploids, and the polyploids were connected to their closest relative as revealed by the supermatrix analysis.
Inference of maternal progenitors
BI of chloroplast matK sequences was used to determine the direction of crosses resulting in allopolyploid taxa, that is, to infer the maternal progenitor, by comparing the position of chloroplast haplotypes derived from polyploids with the respective positions of parents in the phylogenetic tree derived from nuclear loci.
Results
Sequencing and Sequence Assembly
The 454 sequencing of the DNA library combining barcoded PCR amplicons from one chloroplast and 12 nuclear loci (Table 2) analyzed for 96 individuals resulted in 1,170,496 sequence reads, of which 999,492 (85%) were assembled to reference sequences, obtained from nine individuals by direct Sanger sequencing (matK) or cloning and Sanger sequencing of PCR amplicons (nuclear loci) of all analyzed loci. The average read length was 472.1 (± 23.8) bp. On average the number of sequence reads per individual (Supplementary Table S1) was high but uneven. As expected, hexaploid individuals generated more reads than tetraploids and diploids (Supplementary Fig. S1). The number of reads per locus and coverage was high for all loci but TOPO6 and NUC. For these two loci only 66% and 77% of the individuals respectively received sufficient sequence reads mapping to the reference. For TOPO6, sequences from our previous study (Brassac et al. 2012) were used to complete the data set. Two loci, namely TNAC1577 and TNAC1781, were sequenced but not further analyzed as they did not appear to be single copy in the diploid individuals and were difficult to assemble because of many homopolymer regions. Two Hordeum brevisubulatum individuals (PI229753 and GRA2230/97) were excluded from the analyses because of generally bad quality sequences.
Table 2.
Loci initially explored in the study and location on barley chromosomes
| Locus name | RAP2 description | Chromosomea | No. of reads | Coverage mean ± 1 SD |
|---|---|---|---|---|
| TNAC1035 | Kinesin, motor region domain containing protein | 1H | 62,836 | 518 ± 340.0 |
| TNAC1142 | Similar to COP9 signalosome complex subunit 5b | 2H (?) | 51,409 | 305 ± 214.4 |
| XYL | Xylose isomerase | 2HS | 87,926 | 547 ± 329.8 |
| TNAC1364 | Ubiquitin domain containing protein | 3HL | 53,286 | 334 ± 243.1 |
| NUC | Nucellin | 4HL | 31,916 | 260 ± 275.4 |
| TNAC1403 | Similar to SAC domain protein 1 (FIG4-like protein AtFIG4) | 4HL | 151,385 | 490 ± 334.9 |
| TNAC1463 | Proteasome subunit beta type 2 | 4HS | 97,610 | 380 ± 389.5 |
| BLZ1 | Barley leucin zipper | 5HL | 56,064 | 376 ± 236.8 |
| TNAC1610 | Peptidase S16, ATP-dependent protease La family protein | 5HL | 71,133 | 374 ± 342.0 |
| TOPO6 | Topoisomerase VI subunit B | 5HL | 27,416 | 226 ± 159.1 |
| TNAC1577b | Conserved hypothetical protein | 5HL (?) | Na | Na |
| TNAC1497 | Similar to Nucleoside diphosphate kinase II, chloroplast precursor | 5HS | 71,439 | 502 ± 327.2 |
| TNAC1740 | Heat shock protein Hsp70 family protein | 6HL | 49,843 | 297 ± 241.7 |
| TNAC1781b | Beta 5 subunit of 20S proteasome | 7HS (?) | Na | Na |
| matK | Maturase K | cp-LSC | 179,382 | 426 ± 284.2 |
| ndhFb | Subunit 6 of NADH-dehydrogenase | cp-SSC | Na | Na |
aChromosome locations were checked by blasting sequences obtained against the barley genome, no significant result was obtained from TNAC1142, locations for this locus and the other loci missing were inferred from synteny with rice and wheat. Questionmarks refer to not safely determined positions.
bndhF was excluded because of large amount of missing data and TNAC1577 and TNAC1781 were not single-copy loci and difficult to sequence (large homopolymer regions).
The alignment lengths for the 13 loci varied between 753 bp and 5189 bp with 210 to 1333 variable sites (average 466) and 100 to 993 parsimony-informative characters (average 316) per locus (Table 3). Some species showed >1000 bp insertions in TNAC1142 (in Bromus tectorum and Triticum monococcum) and TNAC1463 (in H. murinum and H. gussoneanum). A Blastn search of these insertions returned only a highly similar match for the H. murinum element identified as a partial non-long terminal repeat retrotransposon. Concatenation of the 12 nuclear loci and chloroplast matK in a supermatrix resulted in an alignment of 25,135 bp.
NGS sequences of the TOPO6 locus, which were included as a control to compare the 454 results with those obtained by traditional Sanger sequencing, were longer than those obtained previously (Jakob and Blattner 2010; Brassac et al. 2012). This was due to high confidence in the bases close to PCR primer binding sites provided by NGS compared to traditional Sanger sequencing, where bases adjacent to sequencing primers were often not reliably recovered. Overall, the similarity between sequences obtained by 454 and cloning plus Sanger sequencing was high (99.81% ± 0.67 pairwise identity).
For most of the nuclear loci, the expected two homoeologous copies for a tetraploid individual and three copies for a hexaploid individual were retrieved. Notable exceptions were for tetraploids H. capense and H. secalinum (only one of the two copies at NUC and TNAC1142) and for hexaploid H. arizonicum (only two of the three copies at BLZ1). All tetraploid and hexaploid H. murinum cytotypes possessed only one of the two XYL copies. The high sequence coverage obtained allowed us to recover very rare copies (e.g., for TNAC1610 in H. capense BCC2062) occurring with only about 3% of reads mapping to a locus. Two copies, separated by a couple of nucleotides per locus, were systematically observed for diploid H. pusillum CN27877 and were analyzed as such for each locus. However, these alleles could not be phased between loci and were then merged into consensuses for the final analysis. No allelic variation among homoeologous copies was recorded besides polyploids H. bulbosum and H. brevisubulatum for which multiple copies were retrieved and dealt with as described earlier (see Section “Homoeologue identification”). All sequences obtained in this study were submitted to the NCBI nucleotide database (accession numbers KM039139–KM040760).
Phylogenetic Relationships
To arrive at a species-level phylogeny of Hordeum we initially analyzed sequences from all loci separately for diploid taxa and then repeated the analysis including the sequences from the polyploid taxa. The corresponding single-locus trees for all individuals are available as Supplementary Materials online (Supplementary Figs. S2–S14). Sequence data for the diploid taxa were analyzed in four ways to overcome inconsistencies due to different evolutionary histories of the analyzed loci. (i) BCA allowed us to evaluate those inconsistencies and to estimate the proportion of loci supporting different topology hypotheses. (ii) All loci were concatenated to obtain a supermatrix that was subjected to BI and MP. Moreover, we (iii) checked for topological stability if loci number was further increased with a BI of a supermatrix of all nuclear loci of our study plus the loci published by Petersen et al. (2011) for a data set reduced to one individual per species. Finally (iv) the MSC of gene trees was inferred to arrive at a species tree based on the individual gene trees.
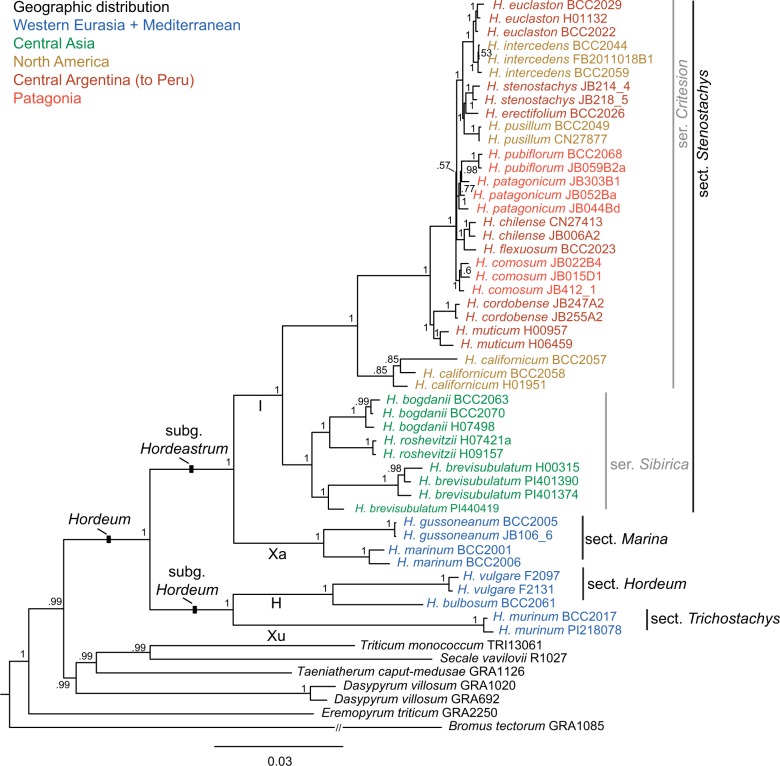
The BUCKy CF obtained from the BCA computed on the single locus trees sampled by BI are relatively low confirming a general discrepancy between loci. All species, with the exception of one, appeared monophyletic, usually with a higher CF than groups of species. Hordeum patagonicum, a diverse species, was paraphyletic, including its sister species H. pubiflorum (CF of 0.13) in the primary concordance tree (Supplementary Fig. S15), or monophyletic (CF of 0.04) in an alternative topology.
The BI of the supermatrix (Fig. 1) produced a completely congruent tree topology with the primary concordance tree (Supplementary Fig. S15). Within Hordeum all clades were highly supported except for H. californicum, which was only moderately supported (0.85 pp). As in the primary concordance tree, H. patagonicum was found paraphyletic with H. pubiflorum nested within the H. patagonicum grade. Within the South American clade, characterized by short internal branches, the nucleotide diversity was too low to resolve all sister relationships. The exclusion of NUC, the locus with the most missing data, from the supermatrix had no effect on the topology of the resulting tree. The consensus tree of 98 MP trees derived from the supermatrix (Supplementary Fig. S16) was very similar to the BI tree. Exceptions included the non-monophyly of H. comosum, placed on a polytomy within the series Critesion, and the paraphyly of H. brevisubulatum, with one individual (PI440419) clustering with low support as sister to the two other Asian species. The inclusion of seven additional nuclear loci and six chloroplast loci (Petersen et al. 2011), although for a data set with only one individual per diploid species, resulted in a highly supported topology (lowest pp 0.94) and confirmed generally the topology obtained with the supermatrix (Supplementary Fig. S17). Hordeum pusillum clustering as sister to the two sister species H. intercedens and H. euclaston, and H. chilense/H. flexuosum forming the sister clade to the one containing H. pusillum were the only species differentially affected in the 26-loci analysis.
Figure 1.
Phylogenetic tree derived from the concatenated supermatrix consisting of 13 loci of the diploid Hordeum taxa and six out-group species calculated with Bayesian inference. Posterior probability values of the clades are indicated close to the nodes. Infrageneric treatment of the genus and genome denominations (bold letters along branches) in Hordeum follow Blattner (2009).
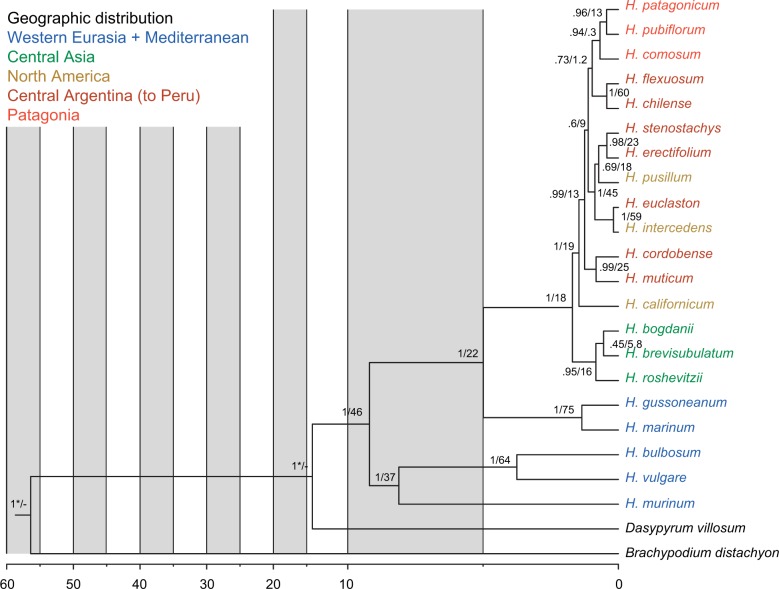
The MSC species tree (Fig. 2) resulted in a slightly different topology providing a better resolution for the closely related American species. The main disagreement concerned the three southern Patagonian species (H. comosum, H. patagonicum, and H. pubiflorum) found to be monophyletic and H. chilense/H. flexuosum as sister clade to the Patagonian clade. A second disagreement occurred with the Asian clade and the relationships within this clade where MSC provided high support (0.95 pp) for the monophyly of the clade but with H. roshevitzii outside although without any significant support within the clade (0.45 pp for H. brevisubulatum and H. roshevitzii). However, the differences are minor and do not influence the recognition of the major clades within Hordeum. In all analyses the recently proposed infrageneric taxonomic groups within Hordeum (Blattner 2009) were found to be monophyletic, for the first time with high support values (Fig. 1).
Figure 2.
Calibrated multispecies coalescent of the diploid Hordeum species as calculated with *Beast from all loci excluding NUC. Numbers along the branches are posterior probability values/concordance factors of the clades. Ages (in millions of years) were inferred from the crown clade age of B. distachyon + Triticeae at 44.4 Ma and from the divergence between Hordeum and Triticum lineages at 15.32 Ma (Marcussen et al. 2014); calibration points are depicted by asterisks, divergence dates are reported in Table 4.
Incongruence Among Loci and Between Methods
The gene trees obtained varied substantially among loci. For example, Hordeum appeared non-monophyletic in five cases (TNAC1142, TNAC1364, TNAC1403, TNAC1463, and TNAC1497), and the I-genome taxa (section Stenostachys) were split in six cases in two clades (TNAC1035, TNAC1142, TNAC1403, TNAC1463, TNAC1497, and XYL). However, the phylogenetic positions of closely related diploid species were mostly conserved among loci while intermediate and larger clades were often disrupted (Table 4). Notable exceptions concern the clade of H. cordobense/H. muticum, present in 25% of the gene trees, and the H. bogdanii/H. roshevitzii clade, occurring in only 13% of them. Single-locus phylogenies received, in pairwise comparisons, similarity scores ranging from 53.6% to 84.6% (Supplementary Fig. S18). The topology of TNAC1035 was the most similar (97.1%) to the one obtained from the concatenated data set. The clade of the Patagonian species H. comosum, H. patagonicum, and H. pubiflorum, as inferred by the MSC, was not recovered in any single-locus phylogeny.
Table 4.
Ages of the clades in the MSC analysis and CF
| Clade | (MSC/BI/MP) | Ages Myr (95% HPD) | Sample-wide CF (95% CI) |
|---|---|---|---|
| H. erectifolium/stenostachys (0.98/1/95) | 0.43 (0.17,0.68) | 0.230 (0.083,0.417) | |
| H. erectifolium/pusillum (0.69/1/93) | 0.73 (0.51,0.97) | 0.176 (0.000,0.333) | |
| H. euclaston/intercedens (1/1/100) | 0.19 (0.09,0. 30) | 0.586 (0.417,0.667) | |
| H. euclaston/pusilluma | 0.082 (0.000,0.250) | ||
| H. erectifolium/intercedens (1/1/100) | 0.87 (0.66,1.09) | 0.448 (0.333,0.583) | |
| H. pubiflorum/patagonicum (0.96/1/85) | 0.44 (0.23,0.65) | 0.127 (0.000,0.250) | |
| H. pubiflorum/comosum (0.94/-/-) | 0.69 (0.44,0.95) | 0.003 | |
| H. flexuosum/chilense (1/1/100) | 0.43 (0.17,0.68) | 0.596 (0.500,0.667) | |
| H. pubiflorum/chilense (0.82/-/-) | 0.97 (0.74,1.23) | 0.088 (0.000,0.167) | |
| H. muticum/cordobense (0.99/1/99) | 0.82 (0.51,1.14) | 0.255 (0.167,0.333) | |
| H. pubiflorum/erectifolium (0.6/1/100) | 1.12 (0.92,1.35) | 0.134 (0.083,0.167) | |
| New World I-clade taxa (0.98/1/94) | 1.46 (1.22,1.72) | 0.186 (0.083,0.250) | |
| H. bogdanii/brevisubulatumb(0.45/-/-) | 0.058 (0.000,0.083) | ||
| H. roshevitzii/PI440419c (-/-/61) | 0.072 (0.000,0.167) | ||
| H. bogdanii/roshevitziid(-/1/100) | 0.131 (0.000,0.250) | ||
| Old World I-clade taxa (0.66/1/94) | 0.89 (0.42,1.51) | 0.161 (0.083,0.250) | |
| I clade (1/1/100) | 1.71 (1.38,2.07) | 0.176 (0.083,0.333) | |
| Xa clade (1/1/100 | 1.37(0.71,2.06) | 0.746 (0.583,0.833) | |
| I plus Xa clade (1/1/100) | 5.01 (4.01,6.05) | 0.220 (0.083,0.333) | |
| H clade (1/1/100) | 3.74 (2.67,4.80) | 0.643 (0.500,0.833) | |
| H plus Xu clade (1/1/100) | 8.13 (6.96,9.40) | 0.373 (0.250,0.500) | |
| Hordeum (1/1/100) | 9.23 (8.07,10.45) | 0.919 (0.833,1.000) | |
| Triticeae | 14.79 (14.13,15.45) | e | |
| B. distachyon/Triticeae | 56.35 (51.08,61.49) | e | |
Notes: MSC, multispecies coalescence; BI, Bayesian inference; MP, maximum parsimony; Myr, millions of years; HPD, highest posterior density; CF, concordance factor; CI, credibility interval.
aNode present only in the 26 loci supermatrix (Supplementary Fig. S17).
bNode present only in the *Beast analysis (Fig. 2).
cNode present in the 13 loci supermatrix MP analysis (Supplementary Fig. S16).
dNode present in the 13 loci supermatrix analyses (Fig. 1, Supplementary Fig. S16).
eNo CF available.
Despite the general discrepancy between loci, the multilocus methods are congruent with the exception of the two clades harboring Asian and the South American species. In the case of the former, the four methods provided three different topologies (Table 4). BUCKy and the supermatrix (13% and 1 pp, respectively) favored H. brevisubulatum as sister to the two other species. The MSC topology is supported by only 5.8% of the loci. The MP topology with H. brevisubulatum paraphyletic is supported by 8.9% of the loci. For the South American species, and especially the Patagonian clade, only the MSC recovered its monophyly while BUCKy retrieved it only for 0.3% of the loci (Table 4).
Ages of Clades
Divergence times (Table 4) estimated with a random local clock model (Drummond and Suchard 2010) and the secondary calibration on the most recent common ancestor between B. distachyon and Triticeae and on the Triticeae crown clade in *Beast resulted in ages with relatively narrow 95% highest probability density (HPD) intervals. The posterior distribution for the age of the root was older than our prior (56.4 Ma, 95% HPD = 51–62), probably due to the fact that sequences from only one B. distachyon individual were used. The Triticeae crown clade age (14.8, 95% HPD = 14.1–15.5) fitted to our prior. The most recent common ancestor of Hordeum occurred 9.2 Ma (95% HPD = 8.1–10.5), while the split between the H and Xu-genome groups of subg. Hordeum happened 8.1 Ma (95% HPD = 7.0–9.4), and the divergence of Xa and I-genome lineages within subg. Hordeastrum started 5.0 Ma (95% HPD = 4.0–6.1). The colonization of the Americas occurred around 1.5 Ma (95% HPD = 1.2–1.7). The analysis revealed nearly contemporaneous speciation events for three pairs of species in southern South America: H. pubiflorum and H. patagonicum (0.44 Ma, 95% HPD = 0.23–0.65), H. erectifolium and H. stenostachys (0.43 Ma, 95% HPD = 0.17–0.68), and H. chilense and H. flexuosum (0.43 Ma, 95% HPD = 0.17–0.68) indicating a possible common climatic and/or geographic reason.
Inference of Parental Progenitors of Polyploids
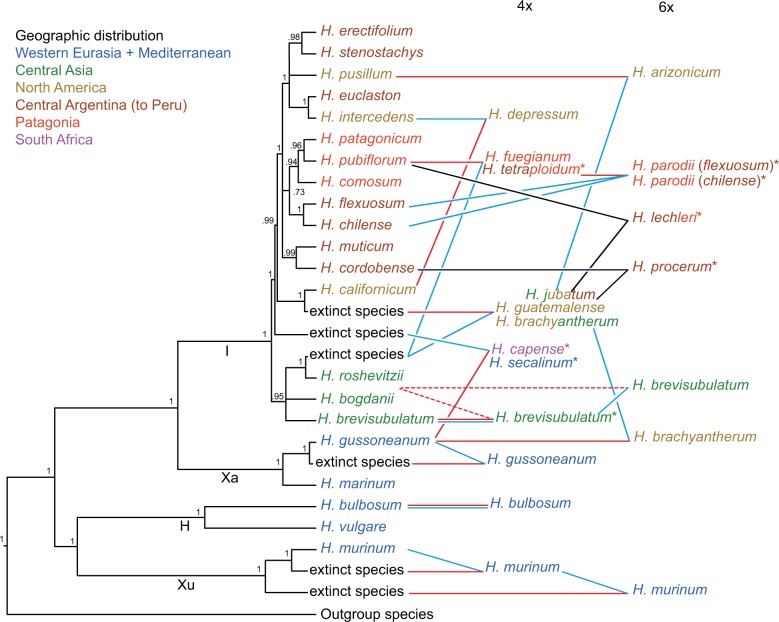
To identify the progenitors of the polyploid species, a nuclear loci supermatrix including the sequences derived from diploid and polyploid taxa was phylogenetically analyzed with BI (Supplementary Fig. S19). The positions of the different homoeologues of polyploids in relation to their closest relatives derived from diploids were used to infer the lineages contributing to polyploids. If sequences from a polyploid lineage grouped within different diploids, this was interpreted as an indication for allopolyploidy, while autopolyploidy was inferred if all sequences of a polyploid were in a clade with a single diploid. Clades consisting solely of polyploid-derived sequences were interpreted as indication of extinct progenitor lineages (Blattner 2004; Jakob and Blattner 2010; Brassac et al. 2012). The results of this analysis were summarized into a scheme where polyploids were integrated in the modified diploid species tree (Fig. 3). The MSC topology was modified to take into account the incongruences between the different methods and to integrate the inferred extinct lineages. The polyploid relationships could mostly be identified with confidence. The wide genetic variety found in some species probably indicates multiple origins of such polyploids. Hordeum parodii, a hexaploid species, as well as H. tetraploidum, together with H. fuegianum one of its potential tetraploid progenitors , appeared to be polyphyletic involving the two closely related diploid species H. chilense and H. flexuosum. The partially autopolyploid taxon H. brevisubulatum, containing both auto- and allopolyploid individuals (Brassac et al. 2012), was treated differently. The high diversity of the copies recovered for the different individuals and the difficulty to assign the parental species/individuals across loci made it difficult to create phased haplotypes. Only one tetraploid (PI401387) appeared autopolyploid with gene copies clustering essentially with the species' diploid cytotypes while sequences of other individuals clustered with species within the Asian Hordeum clade (H. roshevitzii and H. bogdanii). The position of many sequences (for example Bre_PI401376_C at BLZ1, Supplementary Fig. S2) retrieved for the polyploid taxa indicates ongoing intergenomic recombination as already suggested by the high proportion of chimerical sequences in cloned TOPO6 sequences (Brassac et al. 2012). Sequencing and analysis of the individual H00312 revealed it to be probably mislabeled and it was then excluded from our conclusions (see Section “Discussion”).
Figure 3.
Scheme summarizing phylogenetic relationships of species and cytotypes in the genus Hordeum based on 12 loci and one chloroplast region. Diploid taxa were drawn directly at the tree obtained from MSC analysis (Fig. 2) modified to reflect differences with BI analysis based on 13 (Fig. 1) and 26 loci (Supplementary Fig. S17) and position of extinct taxa inferred from the BI tree including diploid and polyploids (Supplementary Fig. S19). Tetra- and hexaploids were connected to their inferred progenitors. Double lines indicate autopolyploid origin, dashed lines mark uncertainties and colors refer to the direction of crosses resulting in allopolyploid taxa (blue for paternal, red for maternal parent, black for both observed) based on the chloroplast data (Supplementary Fig. S14). Asterisks indicate incongruent relationships of polyploids recovered by chloroplast sequences (Jakob and Blattner 2006) compared to nuclear sequences that probably indicate polyphyletic origins.
BI of the chloroplast matK sequences of all individuals (Supplementary Fig. S14) resulted in a poorly resolved phylogeny due to the small number of informative characters, and in an incongruent topology compared to the nuclear loci (see Jakob and Blattner 2006 for a larger sampling of chloroplast haplotypes). Despite the numerous individuals grouping in the basal polytomy and in single-species clades, chloroplast data identify H. roshevitzii as paternal progenitor of all the polyploids it is related to. The polyploid cytotypes of H. murinum carry the chloroplast type of their inferred extinct progenitors. The two sister species H. secalinum and H. capense possess a Eurasian chloroplast. The first one is closer to H. brevisubulatum and the latter to H. marinum, while their paternal progenitor H. gussoneanum possesses a very different type (Petersen and Seberg 2003; Jakob and Blattner 2006). The American hexaploid H. arizonicum falls in a clade with H. pusillum, its diploid progenitor. At least three polyploid species (H. lechleri, H. procerum, and H. tetraploidum) were found polyphyletic regarding their chloroplast types, with haplotypes occurring in different clades.
Discussion
Combining PCR Amplification with Second-Generation Sequencing
In this study, we took advantage of the long reads of 454 sequencing together with parallel barcoding to sequence one chloroplast and 12 nuclear single-copy loci, distributed among nearly all barley chromosomes, in 96 individuals representing all species and cytotypes of the genus Hordeum. In addition we cloned and sequenced these loci in eight diploids and one autotetraploid to serve as references for mapping the reads of the 454-sequenced individuals. To evaluate the capacity of our assembling strategy to recover all true haplotypes, a de novo-assembly approach was applied to some individuals. Although successful, this method can be very time consuming and tedious due to the high number of contigs produced and thus was not further used.
The cloned sequences together with previously published sequences (Petersen et al. 2011; Brassac et al. 2012) for some of the loci were used as a control to compare the results of traditional Sanger sequencing and NGS. Apart from the lengths of the obtained sequences, due to the high-quality base calls close to the PCR priming sites with NGS, hardly any differences occurred between both sequencing approaches. High-coverage NGS allowed us to recover rare copies that would have otherwise required sequencing of at least an order of magnitude more clones than traditionally used in phylogenetic studies. The amount of chimerical sequences was lower compared to our previous analysis (Brassac et al. 2012) due to the exclusive use of a proofreading DNA polymerase and could only be seen in the polyploids H. bulbosum and H. brevisubulatum, for which the sequences were either excluded or merged through consensus formation. The obtained unambiguous sequences allowed us to safely exclude one accession (H00312) because of its peculiar placement. First, described as an Iranian H. bogdanii for which the Giemsa-C banding pattern was analyzed (Linde-Laursen et al. 1980), it was then assumed to be a tetraploid H. brevisubulatum because of its genome size (own unpublished result). This analysis revealed its close relationship with sequence copies of hexaploid H. brachyantherum, a cytotype only known from a very small population in California (Komatsuda et al. 2009). Based on our results we now believe that this individual does not represent a true Hordeum taxon but was instead either mislabeled or hybridized during ex situ propagation (Jakob et al. 2014), and is therefore removed from our conclusions. Uncertainty regarding the correct resolution of mononucleotide repeats, an error specific for pyrosequencers like the 454-sequencing platform, was minor. This was likely due to the very high sequence coverage (270-fold on average) that we aimed for.
Due to the lack of dedicated bioinformatic tools that separate homoeologues (see O'Neill et al. 2013; Ranwez et al. 2013), we used a method based on a combination of reference-based mapping and de novo assembly to disentangle the sequence reads from polyploid individuals. We were able to successfully reconstruct phased haplotypes for all the individuals analyzed by NGS, resulting in the most comprehensive phylogenetic analysis of the genus Hordeum to date. For this approach, the long reads of the 454 platform in combination with a relatively high coverage were favorable. The recent progress in NGS, with increasing read lengths and paired-end sequencing of libraries, means that sequencing of long PCR amplicons and reconstructing phased haplotypes is no longer restricted to the long contiguous reads obtained by 454 sequencing, and therefore will become much cheaper.
Allopolyploid species possess merged genomes and thus require special care when sequencing nuclear loci. This is traditionally done by cloning of PCR amplicons and sequencing of a certain number of clones. Moreover, obtaining sufficient resolution in recently diverged species requires many characters, and longer loci might be favorable for reconstructing species trees from gene trees. The required locus length, together with sequencing of many clones per locus, makes molecular phylogenies of even a medium-sized genus with many polyploid species time consuming and expensive. Here we combined traditional methods, that is, amplifying long loci not necessarily designed to fit the NGS platform's size optimum, with the capacity of these new sequencing techniques to handle a mixture of sheared and barcoded PCR amplicons, extending the method proposed by Griffin et al. (2011). Moreover, in certain cases the high throughput of NGS allowed us to overcome what might be the result of PCR drift (Wagner et al. 1994), where an allelic variant is randomly favored during the PCR, and thus to explore a potentially more complete set of allelic diversity in comparison to a cloning approach.
A major benefit of phylogenetic studies in grasses is the availability of genomic information for Brachypodium, rice, sorghum, barley, and many other species. This makes design of PCR primers for a set of nuclear single-copy loci relatively easy. We took advantage of the rice PLUG system (Ishikawa et al. 2007) that lists a high number of potentially single-copy loci and their chromosomal position in the rice genome. With rapidly increasing genomic information for many plant taxa (Van Bel et al. 2012) it is expected that similar analyses are already or will soon be possible in taxonomic groups throughout the angiosperms.
Phylogeny of Hordeum
The phylogenetic relationships obtained from the set of 12 nuclear single-copy loci and one chloroplast locus (Fig. 1) is in accord with the recently proposed new infrageneric treatment of Hordeum (Blattner 2009). Compared to previous studies of the genus a better resolution was obtained particularly in the MSC analysis (Fig. 2). The main differences appear in the grouping of the recently diverged American species. Thus, we were for the first time able to show the monophyly of the Patagonian diploid species H. comosum, H. patagonicum, and H. pubiflorum using nuclear sequence data, although this relationship was already deduced from the distribution pattern of shared ancient chloroplast haplotypes (Jakob et al. 2009) and an AFLP-based phylogeny (Pleines and Blattner 2008). However, long-term large population sizes and young species ages in this clade resulted in far reaching ILS that impedes phylogenetic analysis (Jakob and Blattner 2006) particularly when nuclear loci are analyzed (Petersen and Seberg 2003, Petersen et al. 2011, Brassac et al. 2012). Taking into account population genetic processes, as done by MSC, results in more plausible phylogenetic relationships in comparison to single gene and supermatrix analyses. Also the three Central Asian diploids could be confirmed to be monophyletic, with high support values in the supermatrix analyses and in MSC.
The increased resolution resulting from this study allowed us to both confirm and more precisely define the progenitor-derivative relationships of polyploids previously identified (Blattner 2004; Brassac et al. 2012). The importance of the Asian diploid H. roshevitzii, or a genotype very similar to that of the extant species, in the evolution of the American polyploids is emphasized here, and the matK analysis showed that this species was never a maternal progenitor. Jakob and Blattner (2006) did not discuss this particular result when analyzing chloroplast relationships in Hordeum, but the two studies are completely concordant. An extinct close relative of H. californicum was confirmed as a second key lineage (Brassac et al. 2012) in the evolution of most American polyploids, and chloroplast sequences indicated that this species functioned as the maternal progenitor of polyploids. Additionally, the extinct lineages contributing to polyploids within the H. murinum taxon complex (Jakob and Blattner 2010; Tanno et al. 2010; Ourari et al. 2011) and tetraploid H. gussoneanum (Brassac et al. 2012; Carmona et al. 2013) could be safely confirmed, as well as the probably extinct Central Asian paternal parent of tetraploid H. capense and H. secalinum (Brassac et al. 2012). We assume that at least six now-extinct lineages contributed to the formation of the polyploid taxa in Hordeum (Fig. 3). Although it may be impossible to prove that something does not exist (Popper 1935), extinct progenitors of allopolyploids have been repeatedly inferred in a wide range of plant genera (Roelofs et al. 1997; Hoot et al. 2004; Lihová et al. 2006). Prerequisite for this deduction is the representative sampling of genetic diversity of all possible progenitor taxa. In the case of Hordeum, we base our assumption on sequence data of more than 2000 individuals (partly unpublished), which up to now provided no indication for the presence of the deduced progenitor alleles in a population of a diploid Hordeum species. In all cases where we inferred extinct lineages, one homoeologous copy was (nearly) identical with an allele of a diploid while the second (extinct) was grouping in a clade different from such diploid-derived copies. Thus, an alternative explanation for differences between one of the homoeologs found in allopolyploids and the respective alleles of extant diploid taxa could be elevated evolutionary rates for a copy that is released from selection or gains new function due to the buffering presence of the homoeologs in the polyploid. However, the probability of observing such a pattern consistently across 12 randomly selected loci we deem rather low. Thus, until we find polyploid homoeologs grouping with diploid-derived alleles for at least some of the loci analyzed, we regard such progenitors as probably extinct.
For the two tetraploid sister species H. fuegianum and H. tetraploidum, we could identify H. pubiflorum as the second progenitor in addition to the H. roshevitzii-like taxon. However, the chloroplast sequences of H. fuegianum clustered with those of H. pubiflorum, while the sequences derived from H. tetraploidum were placed in the basal polytomy. This suggests that H. tetraploidum either evolved independently or that gene flow and chloroplast capture might still be ongoing for these species (Jakob and Blattner 2006). The recurrent formation of polyploids has been extensively demonstrated (Soltis et al. 1993; Soltis and Soltis 1999) partially explaining the high diversity and complexity of polyploid genomes. Based on our analysis, the hexaploid species H. parodii seems to be a good example of recurrent formation involving two diploid species, H. chilense and H. flexuosum, and the tetraploids H. fuegianum or H. tetraploidum. The H. brevisubulatum species complex (di-, tetra-, and hexaploid cytotypes), an obligate outcrossing taxon distributed between Iran and northeastern Siberia, shows signs of recurrent auto- and allopolyploidization involving the entire Asian genepool (including H. roshevitzii and H. bogdanii) as well as H. californicum or its Asian progenitor. To understand the evolution of this very diverse lineage extensive sampling and extended population-based studies would be necessary. It could also be interesting to test the strategy suggested by Marcussen et al. (2015) to evaluate potential network topologies for such a particularly complex polyploid taxon.
Our dated phylogeny provided younger and more accurate age estimations within Hordeum than previously inferred (Blattner 2006). However, within the American species group, such ages are surprisingly low taking into account their clear ecological diversification (Jakob et al. 2010) and that the respective polyploids could only have evolved after the origin of their diploid progenitors. The entire American clade seems to be only about 1.5 Myr old. This rapidly speciating clade (Jakob and Blattner 2006) seems to have been shaped at least in part by the repeated glaciations of the Pleistocene (2.6–0.01 Ma). We obtained no fewer than three nearly contemporaneous speciation events (H. pubiflorum and H. patagonicum, H. erectifolium and H. stenostachys, and H. chilense and H. flexuosum) in South America coinciding with the glacial period 0.40–0.50 Ma which seems to have left no major geological evidence in Patagonia (Rabassa et al. 2011). The effects of ice ages on speciation and divergence are complex processes (Hewitt 1996) but have been shown to be of major importance in shaping current biodiversity in general (Comes and Kadereit 1998) and specifically in Hordeum (Jakob et al. 2007, 2009, 2010).
Incongruence Among Loci and Between Methods
Our analysis showed that even at the scale of a single genus, single nuclear loci exhibit quite different histories. Sister species relationships are generally conserved between loci except when ILS and/or hybridization disrupt the signal. Interestingly, the locus (TNAC1035) resulting in a gene tree topology closest to the one from the supermatrix is not the locus with the highest number of parsimony informative sites. The use of different methods of multilocus phylogenetic inference allowed us to test the performance of those methods in the presence of ILS and potential hybridization.
The concatenation approach is known to potentially lead to high support for incorrect species trees compared to coalescent-based approaches (Kubatko and Degnan 2007; Xi et al. 2014). However, it can be used as a null hypothesis for comparison to methods explicitly modeling the biological processes that are resulting in gene tree incongruences. Despite the general discrepancy between loci, relationships within only 2 three-species clades appeared incongruent between methods: the Asian clade and the Patagonian clade (Figs. 1 and 2). In the former, the pattern observed is most probably due to ILS and hybridization, especially involving the H. brevisubulatum species complex. The coalescent model implemented in *Beast does not take into account gene flow between species (Heled and Drummond 2010), which in turn can potentially disrupt the signal. BUCKy's performance decreases with increasing number of taxa and/or when loci have few informative sites (Chung and Ané 2011). Thus, we interpret the relatively low CF for the Asian clade and its support in MSC (pp 0.95) as an indication of past hybridization. The Patagonian clade, characterized by far reaching ILS within young species (originating 0.23–0.95 Ma) with long-term large population sizes, seems to be better dealt with by the coalescent approach. Interestingly, increasing the number of loci in the supermatrix (Supplementary Fig. S17) did not recover the monophyly of the Patagonian clade, confirming the superiority of the coalescent approach to summarize conflicting phylogenies in the presence of ILS. More studies analyzing the discrepancy between multilocus phylogenetic inference methods such as Zwickl et al. (2014) are necessary to better understand their relative performance in the presence of natural processes leading to incongruence among loci.
Conclusions
Our study of the phylogeny of Hordeum using second-generation sequencing of PCR amplicons obtained, for the first time, the species phylogeny and progenitor-derivative relationships of all di- and polyploid Hordeum taxa within a single analysis. We were also able to provide a time frame for the evolution of the genus. The general shift of paradigm toward multilocus analyses in phylogeny is still limited by the initial selection of enough single-copy loci. The resources available for grasses and especially Triticeae made it possible to use a PCR-based method, which considerably reduced the data complexity. A sequence-capture approach (Lemmon et al. 2012; Mascher et al. 2013) might result in much more demanding data, as a substantial amount of sequences may be off-target and therefore harder to handle (own unpublished data). We chose the 454 platform for its long reads, but with generally increasing read lengths in NGS, the possibility to sequence the ends of fragments up to .700 bp length in paired-end mode on an Illumina HiSeq (even larger on a MiSeq) might define the maximum distance of two polymorphic sites to be used to correctly phase alleles or homoeologues. Thus, long conserved stretches of DNA, separating more variable parts within a sequenced locus, can prevent phasing until the very long reads of single-molecule sequencing becomes widely available. Finally, it appears necessary to analyze multilocus data using different methods to disentangle biological and methodological biases.
Most of the open questions regarding relationships among di- and polyploid taxa of the genus were solved in this phylogenetic analysis. However, the H. brevisubulatum polyploid complex is still puzzling, and its evolution might only be apprehended by extensive population studies throughout the distribution area of the taxon. Also for the hexaploid H. parodii and the two closely related tetraploid species pairs H. fuegianum/H. tetraploidum and H. capense/H. secalinum some additional attention might be needed regarding their mode of evolution. This must, however, include many more individuals/populations in comparison to this study, whose aim was the overall phylogeny within Hordeum.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.fn2nt.
Acknowledgments
We like to thank A. Himmelbach, U. Beier, and A. Kusserow for support during 454-library construction, P. Oswald and B. Kraenzlin for technical help in the lab, and F.E. Anderson, N. Bernhardt, K. Herrmann, E.A. Kellogg, R.J. Mason-Gamer, and an anonymous reviewer for valuable remarks on the manuscript.
Funding
This work was supported by the German Research Foundation (DFG) through grant BL462/9 to FRB.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Álvarez I., Wendel J.F. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29:417–434. [DOI] [PubMed] [Google Scholar]
- Ané C., Larget B., Baum D.A., Smith S.D., Rokas A. 2007. Bayesian estimation of concordance among gene trees. Mol. Biol. Evol. 24:412–426. [DOI] [PubMed] [Google Scholar]
- Baum D.A. 2007. Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon 56:417–426. [Google Scholar]
- Blattner F.R. 2004. Phylogenetic analysis of Hordeum (Poaceae) as inferred by nuclear rDNA ITS sequences. Mol. Phylogenet. Evol. 33:289–299. [DOI] [PubMed] [Google Scholar]
- Blattner F.R. 2006. Multiple intercontinental dispersals shaped the distribution area of Hordeum (Poaceae). New Phytol. 169:603–614. [DOI] [PubMed] [Google Scholar]
- Blattner F.R. 2009. Progress in phylogenetic analysis and a new infrageneric classification of the barley genus Hordeum (Poaceae: Triticeae). Breed. Sci. 59:471–480. [Google Scholar]
- Blattner F.R., Pleines T., Jakob S.S.Glaubrecht M. 2010. Rapid radiation in the barley genus Hordeum (Poaceae) during the Pleistocene in the Americas. Evolution in action. Berlin, Heidelberg: Springer; p. 17–33. [Google Scholar]
- Bothmer R. von, Jacobsen N., Baden C., Jørgensen R., Linde-Laursen I. 1995. An ecogeographical study of the genus Hordeum. Rome, Italy: IPGRI. [Google Scholar]
- Brassac J., Jakob S.S., Blattner F.R. 2012. Progenitor-derivative relationships of Hordeum polyploids (Poaceae, Triticeae) inferred from sequences of TOPO6, a nuclear low-copy gene region. PLoS ONE 7:e33808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochmann C., Soltis P.S., Soltis D.E. 1992. Recurrent formation and polyphyly of Nordic polyploids in Draba (Brassicaceae). Am. J. Bot. 79:673–688. [Google Scholar]
- Buggs R.J.A., Renny-Byfield S., Chester M., Jordon-Thaden I.E., Viccini L.F., Chamala S., Leitch A.R., Schnable P.S., Barbazuk W.B., Soltis P.S., Soltis D.E. 2012. Next-generation sequencing and genome evolution in allopolyploids. Am. J. Bot. 99:372–382. [DOI] [PubMed] [Google Scholar]
- Carmona A., Friero E., Bustos A. de, Jouve N., Cuadrado A. 2013. The evolutionary history of sea barley (Hordeum marinum) revealed by comparative physical mapping of repetitive DNA. Ann. Bot. 112:1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Ané C. 2011. Comparing two Bayesian methods for gene tree/species tree reconstruction: simulations with incomplete lineage sorting and horizontal gene transfer. Syst. Biol. 60:261–275. [DOI] [PubMed] [Google Scholar]
- Comes H.P., Kadereit J.W. 1998. The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 3:432–438. [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan J.H., Rosenberg N.A. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24:332–340. [DOI] [PubMed] [Google Scholar]
- Doebley J., Bothmer R. von, Larson S. 1992. Chloroplast DNA variation and the phylogeny of Hordeum (Poaceae). Am. J. Bot. 79:576–584. [Google Scholar]
- Doyle J.J., Flagel L.E., Paterson A.H., Rapp R.A., Soltis D.E., Soltis P.S., Wendel J.F. 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42:443–461. [DOI] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A. 2010. Bayesian random local clocks, or one rate to rule them all. BMC Biol. 8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.V. 2009. Is a new and general theory of molecular systematics emerging? Evolution 63:1–19. [DOI] [PubMed] [Google Scholar]
- Escobar J., Scornavacca C., Cenci A., Guilhaumon C., Santoni S., Douzery E., Ranwez V., Glémin S., David J. 2011. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol. Biol. 11:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin P., Robin C., Hoffmann A. 2011. A next-generation sequencing method for overcoming the multiple gene copy problem in polyploid phylogenetics, applied to Poa grasses. BMC Biol. 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174. [DOI] [PubMed] [Google Scholar]
- Heled J., Drummond A.J. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G.M. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58:247–276. [Google Scholar]
- Hoot S.B., Napier N.S., Taylor W.C. 2004. Revealing unknown or extinct lineages within Isoëtes (Isoëtaceae) using DNA sequences from hybrids. Am. J. Bot. 91:899–904. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Rannala B. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science 276:227–232. [DOI] [PubMed] [Google Scholar]
- Ishikawa G., Nakamura T., Ashida T., Saito M., Nasuda S., Endo T.R., Wu J., Matsumoto T. 2009. Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 118:499–514. [DOI] [PubMed] [Google Scholar]
- Ishikawa G., Yonemaru J., Saito M., Nakamura T. 2007. PCR-based landmark unique gene (PLUG) markers effectively assign homoeologous wheat genes to A, B and D genomes. BMC Genomics 8:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob S.S., Blattner F.R. 2006. A chloroplast genealogy of Hordeum (Poaceae): long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for phylogenetic inference. Mol. Biol. Evol. 23:1602–1612. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Blattner F.R. 2010. Two extinct diploid progenitors were involved in allopolyploid formation in the Hordeum murinum (Poaceae: Triticeae) taxon complex. Mol. Phylogenet. Evol. 55:650–659. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Heibl C., Rödder D., Blattner F.R. 2010. Population demography influences climatic niche evolution: evidence from diploid American Hordeum species (Poaceae). Mol. Ecol. 19:1423–1438. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Ihlow A., Blattner F.R. 2007. Combined ecological niche modelling and molecular phylogeography revealed the evolutionary history of Hordeum marinum (Poaceae) — niche differentiation, loss of genetic diversity, and speciation in Mediterranean Quaternary refugia. Mol. Ecol. 16:1713–1727. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Martinez-Meyer E., Blattner F.R. 2009. Phylogeographic analyses and paleodistribution modeling indicate Pleistocene in situ survival of Hordeum species (Poaceae) in Southern Patagonia without genetic or spatial restriction. Mol. Biol. Evol. 26:907–923. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Meister A., Blattner F.R. 2004. The considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Mol. Biol. Evol. 21:860–869. [DOI] [PubMed] [Google Scholar]
- Jakob S.S., Rödder D., Engler J.O., Shaaf S., Özkan H., Blattner F.R., Kilian B. 2014. Evolutionary history of wild barley (Hordeum vulgare subsp. spontaneum) analyzed using multilocus sequence data and paleodistribution modeling. Genome Biol. Evol. 6:685–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286–298. [DOI] [PubMed] [Google Scholar]
- Komatsuda T., Salomon B., Bothmer R. von. 2009. Evolutionary process of Hordeum brachyantherum 6x and related tetraploid species revealed by nuclear DNA sequences. Breed. Sci. 59:611–616. [Google Scholar]
- Kotseruba V., Pistrick K., Blattner F.R., Kumke K., Weiss O., Rutten T., Fuchs J., Endo T., Nasuda S., Ghukasyan A., Houben A. 2010. The evolution of the hexaploid grass Zingeria kochii (Mez) Tzvel. (2n = 12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Mol. Phylogenet. Evol. 56:146–155. [DOI] [PubMed] [Google Scholar]
- Kubatko L.S., Degnan J.H. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56:17–24. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y.W., Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Larget B.R., Kotha S.K., Dewey C.N., Ané C. 2010. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics 26:2910–2911. [DOI] [PubMed] [Google Scholar]
- Lemmon A.R., Emme S.A., Lemmon E.M. 2012. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol. 61:727–744. [DOI] [PubMed] [Google Scholar]
- Lihová J., Shimizu K.K., Marhold K. 2006. Allopolyploid origin of Cardamine asarifolia (Brassicaceae): incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Mol. Phylogenet. Evol. 39:759–786. [DOI] [PubMed] [Google Scholar]
- Linde-Laursen I., Bothmer R. von, Jacobsen N. 1980. Giemsa C-banding in Asiatic taxa of Hordeum section Stenostachys with notes on chromosome morphology. Hereditas 93:235–254. [Google Scholar]
- Maddison W.P. 1997. Gene trees in species trees. Syst. Biol. 46:523–536. [Google Scholar]
- Marcussen T., Heier L., Brysting A.K., Oxelman B., Jakobsen K.S. 2015. From gene trees to a dated allopolyploid network: insights from the angiosperm genus Viola (Violaceae). Syst. Biol. 64:84–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Jakobsen K.S., Danihelka J., Ballard H.E., Blaxland K., Brysting A.K., Oxelman B. 2012. Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae). Syst. Biol. 61:107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Sandve S.R., Heier L., Spannagl M., Jakobsen K.S., Pfeifer M., The International Wheat Genome Sequencing Consortium. Wulff B.B.H., Steuernagel B., Mayer K.F.X., Olsen O.-A. 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092–1–1250092–4. [DOI] [PubMed] [Google Scholar]
- Mascher M., Richmond T.A., Gerhardt D.J., Himmelbach A., Clissold L., Sampath D., Ayling S., Steuernagel B., Pfeifer M., D'Ascenzo M., Akhunov E.D., Hedley P.E., Gonzales A.M., Morrell P.L., Kilian B., Blattner F.R., Scholz U., Mayer K.F.X., Flavell A.J., Muehlbauer G.J., Waugh R., Jeddeloh J.A., Stein N. 2013. Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 76:494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Briggs A.W., Maricic T., Höber B., Höffner B., Krause J., Weihmann A., Pääbo S., Hofreiter M. 2008a. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 36:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Stenzel U., Hofreiter M. 2008b. Parallel tagged sequencing on the 454 platform. Nat. Protoc. 3:267–278. [DOI] [PubMed] [Google Scholar]
- Nishikawa T., Salomon B., Komatsuda T., Bothmer R. von, Kadowaki K. 2002. Molecular phylogeny of the genus Hordeum using three chloroplast DNA sequences. Genome 45:1157–1166. [DOI] [PubMed] [Google Scholar]
- Nye T.M.W., Liò P., Gilks W.R. 2006. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics 22:117–119. [DOI] [PubMed] [Google Scholar]
- Nylander J.A.A., Wilgenbusch J.C., Warren D.L., Swofford D.L. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24:581–583. [DOI] [PubMed] [Google Scholar]
- O'Neill E.M., Schwartz R., Bullock C.T., Williams J.S., Shaffer H.B., Aguilar-Miguel X., Parra-Olea G., Weisrock D.W. 2013. Parallel tagged amplicon sequencing reveals major lineages and phylogenetic structure in the North American tiger salamander (Ambystoma tigrinum) species complex. Mol. Ecol. 22:111–129. [DOI] [PubMed] [Google Scholar]
- Ourari M., Ainouche A., Coriton O., Huteau V., Brown S., Misset M.-T., Ainouche M., Amirouche R. 2011. Diversity and evolution of the Hordeum murinum polyploid complex in Algeria. Genome 54:639–654. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Nei M. 1988. Relationships between gene trees and species trees. Mol. Biol. Evol. 5:568–583. [DOI] [PubMed] [Google Scholar]
- Petersen G., Aagesen L., Seberg O., Larsen I.H. 2011. When is enough, enough in phylogenetics? A case in point from Hordeum (Poaceae). Cladistics 27:428–446. [DOI] [PubMed] [Google Scholar]
- Petersen G., Seberg O. 2003. Phylogenetic analyses of the diploid species of Hordeum (Poaceae) and a revised classification of the genus. Syst. Bot. 28:293–306. [Google Scholar]
- Petersen G., Seberg O. 2004. On the origin of the tetraploid species Hordeum capense and H. secalinum (Poaceae). Syst. Bot. 29:862–873. [Google Scholar]
- Pleines T., Blattner F.R. 2008. Phylogeographic implications of an AFLP phylogeny of the American diploid Hordeum species (Poaceae: Triticeae). Taxon 57:875–881. [Google Scholar]
- Popper K. 1935. Die Logik der Forschung. Vienna: Julius Springer. [Google Scholar]
- Posada D., Crandall K.A. 2001. Selecting the best-fit model of nucleotide substitution. Syst. Biol. 50:580–601. [PubMed] [Google Scholar]
- Rabassa J., Coronato A., Martínez O. 2011. Late Cenozoic glaciations in Patagonia and Tierra del Fuego: an updated review. Biol. J. Linn. Soc. 103:316–335. [Google Scholar]
- Rambaut A., Drummond A. 2007. Tracer v1.5. Available from: http://tree.bio.ed.ac.uk/software/tracer/, last accessed June 19, 2015.
- Ranwez V., Holtz Y., Sarah G., Ardisson M., Santoni S., Glémin S., Tavaud-Pirra M., David J. 2013. Disentangling homeologous contigs in allo-tetraploid assembly: application to durum wheat. BMC Bioinformatics 14:S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud X., Zeyl C. 1994. Organelle inheritance in plants. Heredity 72:132–140. [Google Scholar]
- Roelofs D., van Velzen J., Kuperus P., Bachmann K. 1997. Molecular evidence for an extinct parent of the tetraploid species Microseris acuminata and M. campestris (Asteraceae, Lactuceae). Mol. Ecol. 6:641–649. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Mark P. van der, Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Salomon B., Bothmer R. von. 1998. The ancestry of Hordeum depressum (Poaceae, Triticeae). Nord. J. Bot. 18:257–265. [Google Scholar]
- Sang T. 2002. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit. Rev. Biochem. Mol. Biol. 37:121–147. [DOI] [PubMed] [Google Scholar]
- Sang T., Pan J., Zhang D., Ferguson D., Wang C., Pan K.-Y., Hong D.-Y. 2004. Origins of polyploids: an example from peonies (Paeonia) and a model for angiosperms. Biol. J. Linn. Soc. 82:561–571. [Google Scholar]
- Schmieder R., Lim Y.W., Rohwer F., Edwards R. 2010. TagCleaner: identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics 11:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann. Stat. 6:461–464. [Google Scholar]
- Small R.L., Cronn R.C., Wendel J.F. 2004. Use of nuclear genes for phylogeny reconstruction in plants. Aust. Syst. Bot. 17:145–170. [Google Scholar]
- Soltis D.E., Soltis P.S. 1999. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14:348–352. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Rieseberg L.H. 1993. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci. 12:243–273. [Google Scholar]
- Swofford D. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods). Version 4.0b10. Sunderland, MA: Sinauer Associates.
- Taketa S., Ando H., Takeda K., Ichii M., Bothmer R. von. 2005. Ancestry of American polyploid Hordeum species with the I genome inferred from 5S and 18S-25S rDNA. Ann. Bot. 96:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketa S., Nakauchi Y., Bothmer R. von. 2009. Phylogeny of two tetraploid Hordeum species, H. secalinum and H. capense inferred from physical mapping of 5S and 18S-25S rDNA. Breed. Sci. 59:589–594. [Google Scholar]
- Tanno K., Bothmer R. von, Yamane K., Takeda K., Komatsuda T. 2010. Analysis of DNA sequence polymorphism at the cMWG699 locus reveals phylogenetic relationships and allopolyploidy within Hordeum murinum subspecies. Hereditas 147:34–42. [DOI] [PubMed] [Google Scholar]
- The International Barley Genome Sequencing Consortium. 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716. [DOI] [PubMed] [Google Scholar]
- Triplett J.K., Wang Y., Zhong J., Kellogg E.A. 2012. Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS ONE 7:e38702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. 2012. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A., Blackstone N., Cartwright P., Dick M., Misof B., Snow P., Wagner G.P., Bartels J., Murtha M., Pendleton J. 1994. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43:250–261. [Google Scholar]
- Weiss-Schneeweiss H., Emadzade K., Jang T.-S., Schneeweiss G.M. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet. Genome Res. 140:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Liu L., Rest J.S., Davis C.C. 2014. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst. Biol. 63:919–932. [DOI] [PubMed] [Google Scholar]
- Zwickl D.J., Stein J.C., Wing R.A., Ware D., Sanderson M.J. 2014. Disentangling methodological and biological sources of gene tree discordance on Oryza (Poaceae) chromosome 3. Syst. Biol. 63:645–659. [DOI] [PubMed] [Google Scholar]