Abstract
Loss of protein homeostasis (proteostasis) is a common feature of aging and disease that is characterized by the appearance of nonnative protein aggregates in various tissues. Protein aggregation is routinely suppressed by the proteostasis network (PN), a collection of macromolecular machines that operate in diverse ways to maintain proteome integrity across subcellular compartments and between tissues to ensure a healthy life span. Here, we review the composition, function, and organizational properties of the PN in the context of individual cells and entire organisms and discuss the mechanisms by which disruption of the PN, and related stress response pathways, contributes to the initiation and progression of disease. We explore emerging evidence that disease susceptibility arises from early changes in the composition and activity of the PN and propose that a more complete understanding of the temporal and spatial properties of the PN will enhance our ability to develop effective treatments for protein conformational diseases.
Keywords: chaperones, protein folding, stress response, neurodegenerative disease, protein misfolding, aggregation
Introduction
Proteome fidelity is maintained by the protein homeostasis (proteostasis) network (PN), a multi-compartmental system that coordinatesprotein synthesis, folding, disaggregation, and degradation (1). Despite the common factors required for protein synthesis and maintenance, the expression of many PN components is tailored to the specific proteomic demands of different cells and tissues (1). Furthermore, the activity of the PN can be altered permanently or transiently by development and aging, alterations in physiology, or exposure to environmental stress (1). As PN activity changes, so too does the capacity of cells to buffer against the accumulation of misfolded and damaged proteins. Therefore, temporal and spatial fluctuations in the PN could have profound consequences for disease presentation and progression.
Whereas the classical view of the PN in relation to aging and disease has been described as “young versus old” or “before and after,” there has been considerably less attention devoted to the malleability and dynamism of PN properties and composition with respect to different tissue types and life stages; however, it is likely that the relationship between the PN and disease encompasses these complexities. Here, we review the intra- and intercellular organization of the PN and discuss how temporal and spatial changes in PN composition and activity can influence proteostasis, aging, and disease. We focus on the relationship between the PN and neurodegenerative disorders such as Huntington's disease (HD), Alzheimer's disease (AD), Parkinson's disease (PD), and amyotrophic lateral sclerosis (ALS) (for reviews of these diseases, see References 2–5). Note, however, that the importance of proteostasis extends to many diseases (Table 1) (1, 6).
Table 1. Disease-associated mutations of proteostasis network components.
| Gene | Protein | Function | Disease | Reference |
|---|---|---|---|---|
| SOD1 | SOD1 | Superoxide dismutase | ALS | 155 |
| UBQLN2 | Ubiquilin-2 | Ubiquitin-like protein | ALS | 156 |
| CRYAA | α-Crystallin | Small HSP | Early-onset cataracts | 157 |
| HSPD1 | HSP60 | Mitochondrial Chaperone | Hereditary spastic paraplegia | 158 |
| ATXN3 | Ataxin-3 | Deubiquitylase | SCA3/MJD | 159 |
| PARK1 | Parkin | E3 ubiquitin ligase | PD | 160 |
| CRYAB | α-Crystallin | Small HSP | Desmin-related myopathy, cardiomyopathy | 161 |
| SACS | Sacsin | DNAJ chaperone | Spastic ataxia | 162 |
| HSPB1 | HSP27 | Small HSP | Charcot–Marie–Tooth disease | 163 |
| VCP | VCP | AAA ATPase involved in ERAD | Paget disease and FTD | 164 |
| SIL1 | SIL1 | ER-associated nucleotide exchange factor | Marinesco–Sjogren syndrome | 165, 166 |
| HSPB8 | HSP22 | Small HSP | Charcot–Marie–Tooth disease type 2L | 167 |
| DNAJC19 | TIMM14 | Mitochondrial import | DCMA syndrome | 168 |
| HSPA9 | Mortalin | Mitochondrial HSP70 | PD | 169 |
| HSPA8 | HSC70 | Protein folding | Cardiovascular disease | 170 |
| PSMB8 | PSMB8 | Proteasomal subunit | Nakajo–Nishimura syndrome | 171 |
| DNAJB2 | HSJ1 | Neuronal DNAJ chaperone | Distal hereditary motor neuropathy | 172 |
| DNAJC6 | Auxilin | Clathrin uncoating | Juvenile parkinsonism | 173 |
| DNAJB6 | DNAJB6 | Protein folding | Dominantly inherited myopathy | 174 |
| SQSTM1 | p62 | Autophagy | Paget's disease, ALS | 175 |
| DNAJC13 | RME8 | Receptor-mediated endocytosis | PD | 176 |
| P4HB | PDI | Protein disulfide isomerase | ALS | 177 |
| UBE3A | UBE3A | E3 ubiquitin ligase | Angelman syndrome | 178 |
Abbreviations: ALS, amyotrophic lateral sclerosis; ERAD, endoplasmic reticulum (ER)-associated degradation; FTD, frontotemporal dementia; HSP, heat shock protein; MJD, Machado–Joseph disease; PD, Parkinson's disease; SCA, spinocerebellar ataxia; VCP, valosine-containing protein.
Defining the Proteostasis Network
Proteostasis is achieved by the coordinated action of many proteins known collectively as the PN (1). Here, we define the PN as a protein network with an immediate role in protein synthesis, folding, disaggregation, or degradation. This definition encompasses the translational machinery, molecular chaperones and cochaperones, the ubiquitin–proteasome system (UPS), and the autophagy machinery (Figure 1). Although essential for PN function and proteostasis, transcription factors (TFs), chromatin remodelers, structural components, signaling pathways, metabolic factors, general import/export machinery, and regulators of posttranslational modifications (PTMs) are considered auxiliary to the PN. Likewise, we consider stress response pathways such as the heat shock response (HSR) (7) and unfolded protein response (UPR) (8) to be critical and essential modifiers of PN composition rather than direct components of the PN.
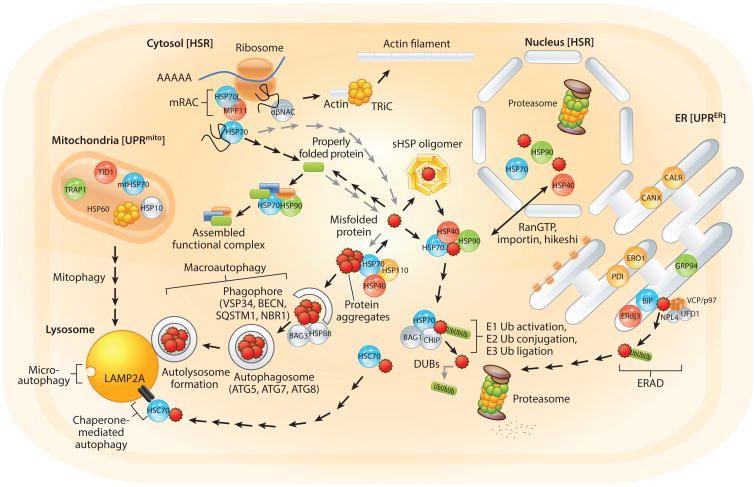
Figure 1.
Overview of the proteostasis network (PN). The human PN must ensure proteome stability across cellular compartments (not drawn to scale). Molecular chaperones of the HSP70 (blue spheres), HSP40/DNAJ (red spheres), and HSP90 (green spheres) families are found in all major cellular compartments and cooperate with cochaperones (gray spheres) to promote folding of nascent chains, assembly of protein complexes, refolding of misfolded clients (serrated red spheres), or degradation of terminally misfolded substrates by the proteasome. Small heat shock protein (sHSP) homo-oligomers bind misfolded proteins and maintain them in a folding-competent state for the HSP70 machinery. If refolding is unsuccessful, cooperation with the nucleotide exchange factor Bcl2-associated athanogene 1 (BAG1) and the E3 ubiquitin ligase C terminus of HSC70-interacting protein (CHIP) can direct substrates to the proteasome. The HSP60 family of chaperones is crucial for mitochondrial proteostasis (in conjunction with the “lid” cochaperone HSP10) and for folding of cytoskeletal components via the TCP-1 ring complex (TRiC). Certain chaperones and cochaperones perform specialized or compartment-specific functions (orange spheres). For example, in the endoplasmic reticulum (ER), protein disulfide isomerase (PDI) and ER oxidoreductin 1 (ERO1) cooperate to promote disulfide bond formation, while calnexin (CANX) and calreticulin (CALR) perform calcium-dependent folding of substrates. Upon protein misfolding, specific stress response pathways such as the heat shock response (HSR) and unfolded protein responses of the ER (UPRER) and mitochondria (UPRmito) can boost chaperone levels. Occasionally, misfolded proteins can form aggregates that can be deleterious to cells. HSP110 cooperates with HSP70/HSP40 to act as a disaggregase. Alternatively, large aggregates can be degraded by the lysosome through autophagy. A modification of this pathway (mitophagy) can also be used to remove old or damaged mitochondria. Gray arrows indicate pathways that should occur only at low levels in healthy cells. Abbreviations: DUB, deubiquitinating enzyme; ERAD, ER-associated degradation; mRAC, mammalian ribosome-associated complex; NAC, nascent polypeptide chain–associated complex; Ub, ubiquitin.
The PN spans all subcellular compartments and is integral for cell viability and organismal health (1). Within the cell, compartmental subnetworks of the PN exist that are uniquely tailored both to the specific biochemical and functional properties of the proteome that they encounter and to the cellular environments they must inhabit (9). Importantly, these subnetworks cooperate substantially to monitor and maintain proteostasis across the cell (9).
Molecular Chaperones
Molecular chaperones are central to the function of the PN and can be broadly grouped into the HSP70, HSP90, DNAJ/HSP40, chaperonin/HSP60, and small HSP (sHSP) families (10, 11). Chaperones can act alone or in various combinations with different cochaperones to regulate client–substrate interactions, folding, disaggregation, degradation, and trafficking within the cell (11). Many molecular chaperones are highly conserved and pivotal, but the best studied are members of the HSP70 and HSP90 families. Although more than 15 mammalian HSP70 homologs and 4 HSP90 homologs exist, most of our understanding of HSP70 and HSP90 function comes from studies of the cytosolic/nuclear HSC70, HSP90, and inducible HSP70 (HSPA1A/B). However, homologs of HSP70 and HSP90 are also integral to endoplasmic reticulum (ER) (BiP and GRP94) and mitochondrial (Mortalin and TRAP1) function (12).
HSP70 and HSP90 are highly abundant proteins with levels estimated to make up 1–2% of the total protein in some cells (12). ATP hydrolysisis essential for the chaperone activity of HSP70 and HSP90, causing conformational changes that result in substrate binding (11). Nucleotide exchange releases bound substrates; several rounds of binding and release by HSP70 are sometimes required for complete client refolding (11, 13). When protein refolding is inefficient, the cochaperones C terminus of HSC70-interacting protein (CHIP) and Bcl2-associated athanogene 1 (BAG1) can interact with HSP70 and HSP90 complexes to promote substrate ubiquitylation, thereby redirecting HSP70 and HSP90 clients to the proteasome for degradation (14, 15). As such, HSP70 and HSP90 are central to the process of triaging proteins for refolding or elimination. The client specificity and functional properties of HSP70 and HSP90 are strongly influenced by interactions with a range of cochaperones (16). HSP90 associates with more than 20 cochaperones, the combinations of which dictate HSP90 function, whereas HSP70 is regulated principally by HSP40/DNAJ chaperones (12, 13). Although distinct, the HSP90 and HSP70 machines also cooperate, most notably in steroid hormone and signal transduction kinase maturation through the cochaperone HSC70/HSP90 organizing protein (HOP/STI1). HOP/STI1 simultaneously binds HSC70 and HSP90 through multiple tetratricopeptide repeat (TPR) domains, thereby facilitating maturation of clients through the sequential action of both HSP70 and HSP90 (12). HSP70 is also recruited to newly synthesized polypeptides by the ribosome-associated complex (RAC), which in mammals is formed from the HSP40/DNAJ protein MPP11 and HSP70L1 (17). RAC binds near the ribosome exit tunnel and stimulates HSP70 binding to nascent polypeptides, thus complementing the role of the nascent polypeptide chain–associated complex (NAC), the initial factor that binds and protects newly synthesized proteins, maintaining them in a folding-competent state for HSP70 (17).
HSP70 and HSP90 homologs are exceeded by the number of HSP40/DNAJ chaperones. HSP40/DNAJ chaperones contain a conserved J domain that mediates interactions with HSP70 but otherwise exhibit heterogeneity in structure, localization, and function. Due to this diversity, HSP40/DNAJ chaperones can be thought of as adaptors that provide versatility to HSP70 function (13). HSP40/DNAJ chaperones assist protein refolding by presenting misfolded proteins to HSP70 and by stimulating HSP70 ATPase activity (13). Efficient protein folding by HSP70 is also influenced by nucleotide exchange factors (NEFs), such as BAG1, that promote ADP/ATP exchange at the N terminus of HSP70 and form distinct functional complexes with the HSP70/HSP40 machinery (13). Perhaps the most significant example of this process is that the HSP110 molecular chaperone, long considered simply a NEF, can catalyze protein disaggregation as part of a complex containing HSP70 and DNAJ/HSP40 (18). This capability provides an extra mechanism by which mammalian cells can eliminate protein aggregates and suppress proteotoxicity, a function carried out by the dedicated chaperones HSP104 and ClpB in yeast and bacteria, respectively (19).
In addition to the HSP40/DNAJ family and cochaperones, the sHSP family facilitates client refolding by HSP70. There are 10 mammalian sHSPs, all of which are cytosolic and most of which contain an α-crystalline domain (10). sHSPs influence protein folding by forming large homo-oligomeric cages that trap misfolded clients and prevent them from forming undesirable intra-or intermolecular interactions in the cytosol. This process is ATP independent and presumed to create a reservoir of refolding competent clients for the HSP70 machinery. Although the full repertoire of sHSP clients is still unclear, it is assumed that sHSPs act broadly, although some degree of selectivity is expected between family members (10).
In addition to the more general chaperone functions carried out by the HSP70/HSP40 and HSP90 machines, some proteins require the specialized functions afforded by the HSP60/chaperonin family. Chaperonins consist of two large hexameric ring complexes that form a cylindrical structure whose core provides a protected environment for protein refolding (11). Two classes of chaperonins exist, the class I mitochondrial HSP60, which is similar to bacterial GroEL, and the class II TCP-1 ring complex (TRiC), which is found in the eukaryotic cytosol (11). HSP60 is essential for maturation and maintenance of the mitochondrial proteome and is therefore intimately linked to energy production. TRiC is essential for proper posttranslational folding of the cytoskeletal components actin and tubulin and is therefore essential for cell structure, division, and cargo delivery (11).
Protein Degradation Pathways
When protein functionality cannot be restored from misfolded and aggregated states, chaperones help redirect nonnative clients toward degradation pathways. Proteins can be degraded either individually or en masse by proteasomes (20) or lysosomes (21), respectively. Proteasomes are large multisubunit complexes that consist of a 19S regulatory cap and a 20S proteolytic core (22). The 19S regulatory particle recognizes ubiquitylated substrates, removes ubiquitin chains, and unfolds the client to allow entry into the 20S core, where it is rapidly degraded into peptides (20, 23). This process is initiated by the addition of polyubiquitin chains through the stepwise activity of E1 ubiquitin–activating enzymes, E2 ubiquitin–conjugating enzymes, and E3 ubiquitin ligases (20).
Ubiquitin chains are formed through conjugation of ubiquitin monomers to clients via distinct lysine residues. This process can take the form of a variety of linkages; however, K48-linked ubiquitin chains are directed primarily to the proteasome (24). Once ubiquitylated, chaperone– cochaperone complexes direct clients to the proteasome, where deubiquitylating enzymes (DUBs) remove ubiquitin chains to allow substrate entry into the 20S core (20). DUBs can be associated with a commitment to degradation and bulk removal of ubiquitin, as is the case for PSMD14, or they can operate independently of client degradation through sequential “trimming” of ubiquitin chains, as is observed with UCHL37 and USP14 (25–28). These opposing activities are proposed to allow “tuning” of proteasomal degradation to be either general or selective as required (20). Despite their cytosolic and nuclear localization, proteasomes are an important site of ER protein quality control through the ER-associated degradation (ERAD) pathway (29). Misfolded proteins in the ER lumen are recognized by the ER membrane–associated HRD1–SEL1–HERP complex and retrotranslocated to the cytoplasm by the AAA ATPase VCP/p97–NPL4–UFD-1 complex (several client-specific forms of this process requiring specialized adapters of the VCP/p97 and HRD1 complexes have been described). The retrotranslocated protein is then ubiquitylated and delivered to the proteasome for degradation, thereby eliminating terminally misfolded proteins from the ER. Distinct ERAD mechanisms are utilized depending on whether protein misfolding is exposed to the ER lumen (ERAD-L), exposed within the ER membrane (ERAD-M), or exposed to the cytosolic face of the ER membrane (ERAD-C), demonstrating the degree to which quality-control pathways can be tailored to substrate type and location (29). A recent study showed that some mutant forms of gonadotropin-releasing hormone receptor (GnRHR) are resistant to ERAD and are instead degraded by ER quality-control (ERQC) autophagy, a complementary pathway modulated by DNAJB12 when ERAD fails (30).
Although the proteasome is the primary source of protein degradation in the cell, biophysical limitations of the central pore of the 20S core do not permit the degradation of unfolded or large protein complexes (20). How, then, do cells remove large protein aggregates? As discussed above, one approach is to employ specialized molecular chaperone machines to release misfolded proteins from aggregates and direct them to the proteasome for degradation (19). Alternatively, bulkier substrates, such as large inclusions, can be directed to the lysosome, a membrane-bound organelle containing a host of nonspecific proteases that can degrade a wide range of substrates (21). Proteins and organelles are directed to lysosomes as the terminal step of autophagy. Autophagy complements the UPS in three mechanistically distinct forms: macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy (21). Macroautophagy is the best-studied form and entails the sequestration of organelles or regions of the cytosol into a double-membrane vesicle structure known as an autophagosome. The resulting autophagosome is then transported to, and fuses with, the lysosome, thereby delivering its cargo for degradation (21). In contrast, microautophagy occurs by direct engulfment of the cytosol at the lysosome membrane, and CMA occurs through HSC70-mediated delivery of proteins across the lysosomal membrane via the LAMP2A receptor (21, 31). Autophagy is regulated by the mammalian target of rapamycin complex 1 (mTORC1) and mTORC2, thereby integrating the PN with the nutritional status of the organism, the metabolic state of the cell, and rates of protein synthesis (21).
Stress Response Pathways Alter Proteostasis Network Composition
The composition of the PN is highly dynamic; the levels of molecular chaperones, cochaperones, and proteasomal subunits can be increased globally or in a compartment-specific manner to provide additional protection against acute and chronic protein misfolding in the cell (1). Malleability of the PN is provided by dedicated stress responsive TFs with distinct and complementary transcriptional targets. Although a number of stress response pathways can greatly influence proteostasis, aging, and disease (see the sidebar), here we focus our attention on the HSR, which is regulated by heat shock transcription factors (HSFs) and augments the cytosolic/nuclear arm of the PN (7) and the UPRs of the ER (UPRER) and mitochondria (UPRmito), which respond to protein misfolding through the TFs XBP1, ATF6, and ATF4 (8) and through ATFS1, respectively (32).
Stress response TFs are maintained in a repressed or inactive state that is distinct for each stress response pathway. In mammalian cells, despite the existence of multiple HSFs, HSF1 is considered the master regulator of the HSR (7). HSF1 is maintained as an inactive monomer in the cytosol through transient interactions with HSP70 and HSP90 (33, 34). Upon increased levels of nonnative proteins, HSF1 is released from its repressive complex, acquires DNA-binding activity through homotrimerization, and rapidly translocates to the nucleus to induce expression of genes encoding molecular chaperones (7, 35). Once stress has been relieved, HSF1 activity is repressed through acetylation and binding to molecular chaperones (34, 36, 37).
In contrast, the UPRER is a more elaborate process that involves three stress responsive arms. IRE1 is a transmembrane protein with kinase and endoribonuclease (RNase) activity that senses misfolding in the ER directly, leading to autophosphorylation, oligomerization, and acquisition of RNase activity (8). This process allows active IRE1 to cleave XBP1 messenger RNA (mRNA), thereby generating a spliced transcript (XBP1s) that encodes a stable form of XBP1 that binds DNA and induces transcription of UPR target genes (8). In parallel, ER stress promotes the relocation of ATF6 from the ER membrane to the Golgi apparatus, where it is cleaved by SP1 and SP2 proteases. The cytosolic N-terminal fragment of ATF6 that is generated translocates to the nucleus, binds DNA, and drives expression of a complementary set of UPR genes (8). Finally, a third ER transmembrane protein, PERK, promotes translation of the TF ATF4 by phosphorylating the translation initiation factor eIF2α in response to ER stress. Under these conditions, ATF4 mRNA is preferentially translated, leading to selective expression of the proapoptotic TF CHOP, which elicits apoptosis if ER stress is not resolved, presumably to ensure that irreversibly damaged cells are removed from the population.
Although the UPRmito has been less extensively studied than the HSR and UPRER, a mechanistic basis for this process is emerging. In the absence of stress, the TF ATFS1 is transported into the mitochondria and degraded by LON protease (32). However, upon mitochondrial stress, import is impaired, allowing ATFS1 to accumulate in the cytosol and translocate to the nucleus, where it regulates transcription of genes encoding mitochondrial chaperones, mitochondrial import machinery, and glycolysis components (32).
As a complement to stress-inducible transcription, global reductions in RNA splicing and translation are also observed upon stress. These changes suppress the de novo synthesis of the majority of the proteome and prioritize the expression of chaperones and other stress response proteins until more favorable conditions are achieved (38, 39). In yeast, the expression of chaperones involved in nascent chain folding is reduced upon stress, consistent with a global repression of protein synthesis (40). Similarly, in mammalian cells, heat shock results in a global pausing of translation elongation due to reduced association of HSP70 with translating ribosomes (39). Interestingly, a recent screen for regulators of HSF1 activity in yeast suggests that stalled ribosomes can also signal back to the PN through the ribosome-associated quality-control complex (RQC) (41). Therefore, stress responses are tightly coupled to the translational state of the cell in order to coordinate changes in PN composition with the quality and quantity of protein biogenesis. Although the extent to which stress responses communicate with one another remains unknown, it is clear that they are integrated into a network that enshrouds the PN to influence proteostasis across subcellular compartments.
Organismal Connectivity of the Proteostasis Network and Stress Responses
With the evolution of multicellular organisms, regulation of the PN has acquired additional layers of complexity to ensure optimal proteostasis both within the cell (cell-autonomous control) and between cells and tissues (cell-nonautonomous control) (42). To this end, cells have evolved the ability to communicate local environmental and proteostasis states to distal cells and tissues. This ability has been demonstrated primarily in the nematode Caenorhabditis elegans, in which genetic disruption of AFD thermosensory neurons alters the organismal response to acute or chronic stress (43, 44). Subsequent complementary studies investigating the role of the UPRER and reduced mitochondrial function in C. elegans life-span extension revealed that XBP1 overexpression or disruption of mitochondrial homeostasis, specifically in neurons, enhances the PN throughout the organism through activation of the UPRER or UPRmito, respectively (45, 46).
Cell-nonautonomous control of the PN and stress responses does not occur solely through neuronal signaling. Altered expression of HSP90 in muscle cells, intestinal cells, or neurons influences the folding environment and stress responses in unperturbed tissues through the TF PHA-4/FOXA (47), whereas overexpression of dFOXO in the muscle tissue of flies can influence protein aggregation in neurons and the retina (48). Furthermore, removal of germ-line stem cells (GSCs) enhances the PN and organismal proteostasis, whereas damage of GSC DNA leads to enhanced somatic stress resistance in worms (49–52).
Together, these findings reveal that the PN and stress response pathways are organized to sense and respond to both intercellular and intracellular stress signals in metazoans. Although the extent to which these observations extend to vertebrate biology is unknown, it is tempting to speculate that in mammals, local signals from affected cells could act to galvanize entire tissues against imminent proteostasis threats such as infection or infarction, or that signals from one tissue could prime the PN of a distal tissue in preparation for periods of intense activity and chronic stress.
Differential Regulation of the Proteostasis Network Across Tissues and Cell Types
The preceding section describes the composition of the PN (and its subnetworks) in terms of the major individual components and their function, an approach that has been adopted by previous reviews (1, 9). However, the inconvenient truth is that the PN is unlikely to exist as a single identity across all cell and tissue types. Certain observations support the hypothesis that the PN and stress responses are not equivalent between cell types. For instance, differential requirements for PN components can be inferred from the disease pathology associated with mutations in genes encoding PN components (Table 1).
Microarray profiling across 80 human tissues revealed that the expression of PN genes is highly heterogeneous, even for central chaperones such as HSC70 and HSP90 (1). PN heterogeneity was not restricted to specific components or chaperone families; instead, all classes of chaperone, autophagy mediators, UPS components, and stress response regulators exhibited profoundly altered expression patterns between tissues. Significantly altered PN profiles were also observed with age and development, as exemplified by the finding that adult liver was more similar to almost every other tissue tested than to its fetal form (1). Differential PN requirements have also been linked to developmental potential through studies in human embryonic stem cells (hESCs), in which maintenance of pluripotency and efficient differentiation require high levels of proteasome activity through increased expression of the 19S subunit PSMD11 (53).
Such differences in the PN are further supported by the Allen Brain Atlas, a catalog of gene expression based on in situ hybridization studies in 17 mouse brain regions (54). HSP70, HSP90, and HSP110 family members were found to be expressed at high levels throughout the brain; however, the expression of DNAJ/HSP40 chaperones and cochaperones was highly heterogeneous, suggesting that differences in the levels of these proteins may tailor HSP70 and HSP90 function to accommodate region-specific clients (54). Although these findings are in keeping with expected differences in the composition of the transcriptome and proteome between tissues, the degree of heterogeneity in the PN remains surprising, given the common folding requirements of proteins (1).
A genetic investigation of HSR regulation in C. elegans also supports the hypothesis that different cell types have different PN requirements. A genome-wide RNA interference (RNAi) screen for genes that influence induction of a fluorescent reporter of the HSR [hsp-70 promoter driving green fluorescent protein (GFP) expression] identified 52 genes whose knockdown resulted in reporter activation, of which 39 genes encode PN components (55). Whereas knockdown of HSC70 or HSP90 resulted in reporter activation in all tissues, knockdown of other PN components resulted in tissue-specific patterns of reporter activation, despite the ubiquitous expression of these genes. For example, knockdown of TRiC subunits activated the HSR exclusively in muscle, whereas knockdown of a mitochondrial HSP70 homolog induced the HSR reporter only in the intestine. Conversely, depletion of proteasomal subunits or an ER HSP70 homolog induced the HSR in muscle and the spermatheca but not in the intestine. These findings indicate that different tissues likely have distinct PN composition and requirements (55).
Studies to elucidate the relationship between chaperone levels, regulation of stress responses, and proteotoxicity have hinted at how PN heterogeneity can influence disease presentation and progression. An investigation of the HSR in cultured spinal cord cells revealed that compared with glial cells, motor neurons do not robustly upregulate HSP70 in response to heat shock due to an inability to activate HSF1 (56). The basis for this is unknown; however, these findings suggest that an inability to activate the HSR in motor neurons may contribute to ALS.
A complementary investigation of the selective pathology associated with polyglutamine (polyQ) disease conducted microarray profiling of primary cortical, striatal, and cerebellar neurons expressing either mutant huntingtin (mHTT) or mutant ataxin-1 (mAtaxin-1). Cerebellar granular neurons were found to express high levels of HSP70 in response to mHTT expression but not mAtaxin-1. This effect was independent of HSF1 and did not occur in striatal and cortical neurons (57). Furthermore, increased levels of HSP70 were found in the cerebellar cortex of human HD brain samples but not in tissue from unaffected individuals or from brains of PD patients (57). A more recent study investigating mHTT turnover in different neuronal types found that the ability to efficiently degrade mHTT correlates strongly with neuronal health and can vary among individual neurons (58). Turnover of mHTT was faster in cerebellar neurons than in cortical or striatal cells, suggesting that the degradative capacity of the PN can also contribute to the differential toxicity of mHTT associated with neuronal subtypes (58). Although limited in scope, these studies act as a proof of principle that tissue- and cell-specific regulation of the PN is entwined with cellular function, disease presentation, and progression.
Proteostasis Network Disruption and Proteostasis Collapse in Neurodegenerative Diseases
Whether due to the chronic expression of mutant proteins with altered stability or mutations that arise in PN genes, a large number of human disorders can be categorized as PN disruptions. This observation highlights the general importance of the PN in human health (Table 1) (59, 60).
Protein aggregation is recognized as a hallmark of neurodegenerative disease by the consistent appearance of detergent-insoluble inclusions and aggregates in the nucleus and cytoplasm of neurons. These structures contain amyloid, large-ordered fibrils of cross-β-sheet-enriched proteins, and form in HD, PD, AD, and ALS despite the lack of sequence similarity between the respective disease-causing proteins. Similar aggregate structures have also been detected in type II diabetes and a range of amyloid disorders throughout the periphery; some 50 human diseases have been linked to amyloid formation (59). Note that protein aggregates can also take many amyloid-free forms, each of which may have a unique role in disease presentation and progression, with some aggregates proposed to represent active compartmentalization of misfolded species by the PN (59–61).
The commonality of these observations has led to fundamental questions of how and why proteostasis collapse occurs and how this affects disease onset and progression. Does chronic expression of misfolding-prone proteins overwhelm or progressively disrupt the PN, or do age-related changes in the PN compromise its robustness, leading to accelerated misfolding, aggregation, and progression of disease? Intriguingly, there is evidence to support both disease models.
The expression of polyQ fused to yellow fluorescent protein (polyQ∷YFP) in C. elegans muscle, intestine, or neurons leads to aggregate formation in a polyQ length- and age-dependent manner (62). In the presence of endogenous metastable proteins, polyQ disrupts the folding capacity of the PN, enhances the misfolding of metastable proteins, and causes cellular dysfunction and specific loss-of-function phenotypes (63). In return, aggregation and toxicity of polyQ∷YFP are further intensified by the presence of metastable proteins, providing support for a “feed-forward” amplification of protein misfolding toxicity (63). These findings suggest that the chronic expression of aggregation-prone proteins reduces the folding capacity of the PN, resulting in the misfolding of metastable proteins across the proteome. Although these observations support PN dysregulation as a pivotal feature of neurodegenerative disease, these studies did not address which components of the PN are compromised.
Dysregulation of Molecular Chaperones
Studies of chaperone levels in tissue culture and mouse models of polyQ disease showed that the levels of HSP70 (HSPA1A/B) and DNAJ/HSP40 (DNAJB1), as well as some cochaperones, decline with protein aggregation (64, 65). This reduction in chaperone levels can occur through a combination of transcriptional dysregulation and sequestration of chaperones by mHTT or polyQ aggregates (64–66). Sequestration of chaperones by insoluble aggregates also leads to reduced levels of soluble HSP70, HSP40, HSP90, and sHSPs in mouse and C. elegans models of AD, tissue culture and Drosophila models of PD (67–70), and tissue culture and mouse models of spinocerebellar ataxia 3 [SCA3; also known as Machado–Joseph disease (MJD)] (71). These observations suggest that perturbation of molecular chaperones may be central to global proteostasis collapse in disease. How, then, might dysregulation of molecular chaperones contribute to neuronal dysfunction?
Sequestration of HSC70 by intracellular aggregates interferes with clathrin-mediated endocytosis (CME) in mammalian cells, including neurons (72). These effects were observed in cells expressing aggregation-prone forms of mHTT, mAtaxin-1, and SOD1, suggesting that protein aggregation and chaperone sequestration may contribute to a range of neurodegenerative disease phenotypes (72). These observations haveled to a so-called chaperone competition model, in which the compulsive requirement of misfolded proteins for HSC70/HSP70, and likely other chaper-ones, reduces the chaperone pool available to maintain normal cellular function. This model could explain the pleiotropic effects observed in all neurodegenerative diseases; however, it is unknown whether chaperone sequestration causes a gradual or sudden impairment of cellular pathways, which chaperones are affected, or whether all chaperone-regulated processes are equally affected.
Disruption of Protein Degradation Pathways
One of the principal features of protein folding diseases is the ubiquitination of proteins within aggregates. The presence of ubiquitinated inclusions suggests that disease-related proteins marked for degradation may be inefficiently targeted to proteasomes or are resistant to degradation. Alternatively, investigators have reasoned that impairment of the UPS could underlie the accumulation of ubiquitylated proteins and therefore significantly contribute to multiple neurodegenerative diseases. Experiments examining the effects of Aβ on proteasomal activity in vitro revealed an inhibitory effect on the chymotrypsin-like properties of the 20S core (73), consistent with observations of impaired proteasome function in AD patient brains (74). Further evidence for UPS dysfunction in AD comes from studies showing that a mutant form of ubiquitin (UBB+1) accumulates in AD brain tissue and causes proteasome inhibition and neurotoxicity via the activity of the E2 ubiquitin–conjugating enzyme HIP-2 (75). The expression of mutant SOD1 also causes dys-regulation of the UPS in mammalian cells and in spinal cord motor neurons of mice (76, 77). This effect is proposed to occur through transcriptional dysregulation and sequestration of proteasomal subunits by mutant SOD1 aggregates (76, 77). Similarly, turnover of a fluorescent proteasome reporter (GFPU degron) is impaired in mammalian cells expressing mutant α-synuclein, and reduced proteasome activity has been reported in the substantia nigra of PD patients compared with that of unaffected individuals (78). This reduction in activity may be exacerbated in familial forms of PD caused by mutations in the E3 ubiquitin ligase parkin (79).
Although linked to AD, PD, and ALS, UPS dysfunction has been most intensely studied in the context of polyQ disease. Initial biochemical studies suggested that expansions of polyQ may directly inhibit or “choke” the proteasome (80), causing the accumulation of lysine 48– and lysine 63–linked ubiquitin chains in brain tissue from HD patients and mice (81). However, more recent data have shown that the inhibitory effect of mHTT on the proteasome is not associated with increased polyQ length or aggregation state, suggesting that ubiquitin chain accumulation in HD does not occur due to direct negative interactions between mHTT and the proteasome (82). Recent findings in yeast and HEK293 cells support this idea by demonstrating that sequestration of Sis1p (human DNAJB1) by mHTT or mAtaxin-3 aggregates decreases the trafficking of misfolded proteins to nuclear proteasomes for degradation, thus exacerbating proteostasis collapse (83). This finding suggests that UPS impairment arises from a progressive titration of chaperones away from clients, thereby interfering with the movement of ubiquitylated clients to the proteasome (82). Thus, it is the growing “queue” of proteasomal substrates that causes an accumulation of ubiquitin conjugates in HD (82), indicating that, like CME, dysregulation of the UPS is a symptom of chaperone competition in disease. Given that chaperone sequestration is also observed in AD, PD, and ALS models, this mechanism may contribute to proteostasis collapse in multiple neurodegenerative diseases.
In addition to their role in proteasomal impairment, alterations in autophagy have been linked to health and disease progression. Mice deficient for the autophagy-related genes Atg5 and Atg7 exhibit severe neurodegeneration (84, 85), and the expression of disease-associated proteins is reported to exert differential inhibitory effects on autophagic pathways. Consistent with a role of autophagy in disease, AD patient tissues exhibit impaired initiation of macroautophagy and an excess of autophagic vacuoles in dystrophic neurites, possibly due to impaired targeting of the vacuolar ATPase to the lysosome (86, 87). In contrast, α-synuclein overexpression impairs autophagy in mammalian cells and mice through reduced expression of RAB1A, thereby inhibiting autophagosome formation (88). In addition to macroautophagy, mutant α-synuclein may impair CMA, suggesting that defects in multiple autophagic arms could contribute to PD (89, 90). In HD, autophagosome formation and lysosomal fusion appear to be unaffected by the expression of mHTT; instead, it has been proposed that the loading of cytosolic cargo, particularly organelles, is inefficient, possibly due to aberrant interactions between p62, ubiquitin chains, and mHTT (91). In contrast to findings in HD, AD, and PD, a recent study has suggested that autophagy is enhanced in ALS mice. Reduced levels of lipofuscin, LC3, and p62 have been observed in motor neurons of SOD1G85R mice (92). Treatment with the autophagy inhibitor chloroquine restored lipofuscin, LC3, and p62 levels in motor neurons, suggesting that mutant SOD1 causes hyperactive autophagy in mice (92). Together, these findings support the idea that impairment of protein degradation pathways is likely fundamental to much of the cellular disruption observed in neurodegenerative diseases. However, as reported for molecular chaperones, it is clear that PN disruption can have disease-specific components.
Stress Response Pathway Impairment
Dysregulation of the HSR with disease progression is associated with toxicity in tissue culture, Drosophila melanogaster, and mouse models of HD, SCA-17, and SCA-3/MJD, as well as in mammalian cells expressing a synthetic amyloid-forming peptide (93–99). Impaired binding of HSF1 to DNA is observed genome wide in HD cells and is reported to affect the expression of many important nonchaperone genes (100). Although the mechanism of HSR impairment is unclear, it is possible that changes in HSF1 activation, reduced HSF1 levels, and altered chromatin architecture underlie transcriptional dysregulation observed at HSP genes (95, 97, 98). Therefore, disease-causing proteins robustly impair the PN by sequestering existing chaperones while simultaneously preventing their replacement through the activation of the HSR.
Surprisingly few studies have investigated the relationship between the UPR and neurodegenerative diseases, although several groups have reported that ER stress is an early feature of AD, PD, HD, ALS, and prion diseases (101). Disruption of the ATF6 arm of the UPRER is reported to occur in mouse models of HD and a VAPB cell model of ALS, suggesting that differential changes in UPR arms may be a feature of disease progression (102, 103). These observations point to the progressive loss of stress response pathways as a possibly crucial feature of multiple neurodegenerative diseases. The loss of the HSR may leave neurons increasingly vulnerable to transient environmental insults and to the chronic presence of aggregation-prone proteins, thereby exacerbating disease progression.
Spreading of Protein Aggregates
In addition to cell-autonomous disruptions of the PN, several observations indicate that α-synuclein, polyQ, mHTT, and mutant SOD1 aggregates exhibit spreading behavior in tissue culture cells and rodent and patient brains. Postmortem analyses of fetal mesencephalic dopaminergic neurons transplanted into PD patients, and neuronal transplants grafted into the striatum of HD patients, revealed disease-like degeneration of healthy tissues accompanied by Lewy body formation in PD patient grafts (104, 105). Transmission and internalization of α-synuclein and mHTT aggregates can be recapitulated in cell culture and rodent models of PD and HD, respectively, and are also observed in mammalian cells expressing mutant SOD1 and in C. elegans expressing a yeast prion domain protein (106–110). Although aggregate spreading appears to be a common feature of these diseases, the mechanism by which internalization and transmission occur may be disease specific and has been proposed to occur through endosomal pathways, secretory vesicles, and macropinocytosis in models of PD, HD, and ALS, respectively. Furthermore, the incorporation of aggregates from culture media or neighboring cells initiates nucleation and aggregation of otherwise-soluble proteins in the cytosol, raising the possibility that proteostasis collapse and PN dysregulation can be propagated between neurons. Collectively, these findings illustrate that the presence of aggregation-prone proteins can disrupt the PN and drive proteostasis collapse both within and between cells in, but not limited to, numerous neurodegenerative diseases (Figure 2).
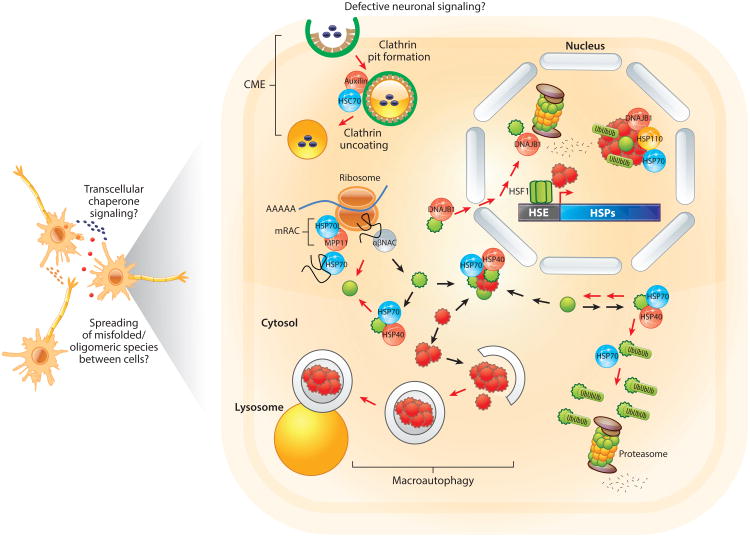
Figure 2.
Disruption of the proteostasis network (PN) in aging and disease. Numerous mechanisms contribute to proteostasis collapse in neurodegenerative disease and aging. Principal among these are reduced levels of soluble chaperones and disruption of protein degradation pathways. The action of HSP70 (blue spheres), HSP40/DNAJ (red spheres), and cochaperones (gray spheres) ensures that newly synthesized proteins fold properly (green spheres). Upon protein misfolding (serrated green spheres), molecular chaperones can either refold substrates to a functional conformation or direct substrates to the proteasome for degradation in the cytosol or nucleus. The presence of aggregation-prone proteins (serrated red spheres) sequesters chaperones and impairs the heat shock response (HSR). Reduced chaperone levels impair clathrin-mediated endocytosis (CME) (actually, HSC70 and auxilin reside within the clathrin cage) and inefficient degradation of ubiquitin (Ub)-conjugated substrates. As chaperone levels become compromised, the likelihood of protein misfolding and aggregation is exacerbated, driving neuronal dysfunction and disease. Protein aggregates can also spread between cells, thereby propagating proteostasis collapse. Red arrows denote pathways dysregulated in aging and/or neurodegenerative disease. Abbreviations: HSF1, heat shock factor 1; HSE, heat shock element; HSP, heat shock protein; mRAC, mammalian ribosome-associated complex; NAC, nascent polypeptide chain–associated complex.
Could Programmed Changes in the Proteostasis Network Underlie Disease Onset and Progression?
Despite being expressed from birth, disease-associated proteins often trigger disease only later in life. This observation has led to the hypothesis that the pathways necessary to maintain proteostasis and suppress protein misfolding and aggregation are progressively compromised with age, eventually leading to disease onset. In support of this idea, reduced expression of molecular chaperones, altered proteasome activity, and disruption of stress responses have been observed in aged rodent tissues and senescent human cells (36, 111, 112). Additionally, an investigation of chaperone and cochaperone gene expression in young (36±4 years of age) and aged (73 ±4 years of age) human brain tissue revealed that of 332 genes examined, 101 are significantly repressed with age, including HSP70, HSP40, HSP90, and TRiC genes (113). Furthermore, 62 chaperone genes, including several small HSPs, were found to be significantly induced, likely as a result of the cellular response to accumulating protein damage with age (113). Concordant changes were observed in HD and AD brain tissues, and RNAi of genes dysregulated in human aging exacerbated polyglutamine aggregation and toxicity in C. elegans muscles and in HeLa cells (113). Together, these data suggest that the PN undergoes significant remodeling in human brain tissue during aging and that these changes have functional consequences for the onset and progression of neurodegenerative diseases. However, these findings did not address whether the age-associated compromise in PN function is late onset, gradual, or abrupt. It has been widely assumed that proteostasis decline with age is gradual; however, studies in invertebrates have begun to challenge this model and suggest that the initiating factors for proteostasis collapse and disease susceptibility may be part of programmed events that occur earlier in adulthood than anticipated (Figure 3).
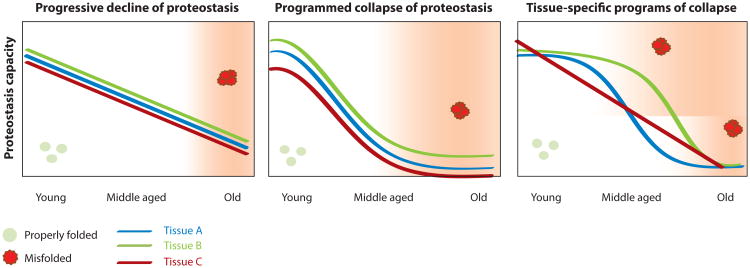
Figure 3.
Proposed models for the relationship between proteostasis collapse, aging, and disease. Proteostasis collapse is associated with aging and disease in multiple tissues. High proteostasis capacity early in life maintains proteome integrity (green spheres) and minimizes the risk of disease (unshaded areas). The conventional model of aging proposes that proteostasis capacity declines progressively with age, leading to increased incidence of protein misfolding (red spheres). Once proteostasis capacity falls below a certain threshold, proteome integrity is widely compromised and leads to disease (shaded areas). An alternate model emerging from studies in Caenorhabditis elegans is that proteostasis capacity collapses early in adulthood and may be part of a programmed event, thereby resulting in a longer window of disease susceptibility. It is possible that tissues (colored lines) exhibit the same or differential patterns of proteostasis collapse during life, which, coupled with specific mutations, result in a specific pattern of disease presentation and progression.
Changes in Proteostasis Capacity Occur in Early Adulthood
Protein folding capacity during aging was initially assessed using C. elegans expressing endogenous metastable temperature-sensitive proteins (myosin, perlecan, dynamin, ras, and acetylcholine receptor subunit) in various tissues (114). The metastability of temperature-sensitive protein folding sensors allows a prediction to be tested: If protein folding capacity declines during aging, then metastable proteins should misfold even in animals grown under permissive temperatures. Therefore, by monitoring the folding state of these proteins through life, one can pinpoint precisely when protein folding capacity declines.
Metastable proteins were found to mislocalize and aggregate between day 2 and day 5 of adulthood, indicating that, relative to the mean life span of C. elegans (∼21 days at 15°C), protein folding capacity declines in early adulthood. For example, the misfolding of temperature-sensitive paramyosin at day 4 of adulthood occurs long before the cytological decline of myofilament structure and the appearance of gross motility defects (114). The fact that these observations were made with multiple unrelated metastable proteins expressed in different tissues suggests that the early decline in proteostasis is not limited to one group of proteins or to one tissue or cell type, but rather reflects a general decline in the PN (114).
Complementary studies in C. elegans adopted an unbiased proteomics-based approach to characterize the composition of detergent-insoluble protein aggregates collected from worm lysates at different days of life (115, 116). A subset of the proteome was aggregated at day 3 of adulthood; however, little to no aggregation was reported at day 1 of adulthood, matching the observations described above using metastable proteins. Aggregation increased with age, occurred in both the soma and the germ line, and contained proteins with sequence and structural similarities (high β-sheet content), suggesting that protein aggregation in early adulthood is influenced by protein sequence (115). These findings suggest that protein aggregation is an early, nonrandom event and that early adulthood is the critical threshold for proteostasis collapse. However, a large fraction of the proteome is clearly protected against aggregation, and even among proteins that form aggregates the rate may be markedly different. Furthermore, even though all animals displayed aggregation in early adulthood, heterogeneity in the timing of aggregation was observed across the population (115).
Age-associated protein aggregates were consistently found to contain molecular chaperones, translation factors, and ribosomal subunits (115–117), perhaps explaining observations that translation, as measured by polysome profiling, also declines early in C. elegans adulthood (117). RNAi knockdown of proteins associated with aggregates was found to increase life span in ∼50% of cases, suggesting that these proteins contribute to normal aging, perhaps due to their propensity to form aggregates. Although informative, the approaches employed by this first wave of studies could not detect oligomers and small aggregates that remain detergent soluble. Therefore, protein misfolding may be even more widespread than suggested. Why, then, does early loss of proteostasis occur in C. elegans, and is this mechanism conserved?
Studies on normal aging in D. melanogaster indicate that protein aggregation with age is a conserved feature in both muscle and brain tissue (48). However, it is unclear how early in life it occurs. Ubiquitin-positive aggregates are observed in flight muscles, retina, brain, and adipose tissue of adult flies. These aggregates dramatically increase in size and shape between days 7 and 35 of adulthood; however, the precise timing, progression, and consequence of aggregation in flies have not been determined (48). Furthermore, the composition of aggregates in old Drosophila is currently unknown, and it will be interesting to determine whether they are also enriched for ribosomal subunits, translation factors, and other abundant proteins, as is observed in aged C. elegans (115–117). It will also be important to determine the extent to which protein aggregation contributes to normal aging in mammals.
Requirements of Stress Response Pathways Through Life
Stress response pathways are traditionally regarded as a safeguard against an increase in misfolded proteins due to translational errors, altered pH, inflammation, and fluctuating temperatures. However, stress response pathways should also be considered an important process by which cells can rapidly and transiently alter the composition of the PN and shift the proteostasis boundary to meet acute temporal demands (1). In this context, stress response arms of the PN are essential to all physiological cellular commitments, as demonstrated by the fact that HSF1 is essential for proper development and growth in yeast, C. elegans, D. melanogaster, and mice (118–121). However, inducing a fully fledged cell stress response represents a substantial cellular commitment. Constitutive activation of HSF1 is detrimental to cells and increased expression, and activity of HSF1 has been linked to multiple forms of cancer, highlighting the need for appropriate and balanced activation of stress response pathways as and when required throughout life (122).
How, then, are cell stress responses controlled to meet the changing demands of metazoans? Recent studies have shown that when challenged with a toxic insult such as heat shock or the ER stress inducer tunicamycin, the induction levels of HSR- and UPR-regulated genes is reduced throughout the soma by ∼80% by day 2 or 3 of adulthood in C. elegans (46, 114). This results in markedly reduced stress resistance and suggests that collapse of the HSR and UPR precedes proteostasis collapse. We do not know how these observations translate to a complex mammalian system with distinct sexes and different parent/progeny requirements following birth; however, a similarly timed collapse of restraint stress–induced Hsp70 induction occurs in the aging rat adrenal cortex, and both male and female Drosophila subjected to hyperthermia exhibit reduced tolerance and Hsp70 induction early in adulthood (123). These findings support the idea that early transcriptional dysregulation of stress responses may be a conserved event in metazoans, at least in some tissues.
The ability of HSF1 to modify aging and disease also depends on the life stage of the organism. RNAi of the phosphatidylinositol 3-kinase age-1 or the sole C. elegans insulin/insulin-like signaling (IIS) receptor daf-2 impairs IIS, extends life span, and suppresses the age-dependent aggregation and toxicity of polyQ and Aβ expressed in body wall muscle cells (62, 124). These effects are strictly dependent on HSF1 and DAF-16/FOXO3A, which minimize proteotoxicity by partitioning misfolded proteins into large nontoxic aggregates (DAF-16-regulated pathway) or by suppressing aggregation entirely (HSF1-controlled pathway) (124).
Experiments exploring the temporal requirements of HSF1 and DAF-16/FOXO3A for protection against protein misfolding and toxicity demonstrated that HSF1 and DAF-16/FOXO3A are strongly required early in life, suggesting that molecular events in early adulthood may be critical to disease suppression in later life (125, 126). Furthermore, RNAi of daf-2 extends life span most effectively when initiated before day 2 of adulthood, with the degree of life span extension progressively decreasing to day 8 of adulthood (127). Collectively, these findings suggest that in C. elegans, the PN is in its most robust state early in life and confers protection across the proteome. However, as animals commence reproduction, the proteostasis boundary shifts and protein aggregation accelerates.
Why, then, do stress responses decline in early adulthood, and does this decline represent a random, passive, or programmed event? If stress response collapse is programmed, what are the organismal benefits of such an event? Future experiments to determine the molecular basis of these events, as well as which other stress response pathways also decline early in life, will greatly enhance our understanding of aging and disease onset.
Temporal Requirements of Protein Degradation Pathways
Cells can sculpt proteome composition and functionality through protein degradation pathways; however, as described for neurodegenerative diseases, disruption of these pathways also leads to cellular dysfunction. As such, it is feasible that any reduction in the protein degradation capacity of a cell could contribute to proteostasis collapse and promote aging. Studies in C. elegans expressing a photoconvertible proteasomal substrate showed that by day 5 of adulthood, reporter turnover occurred at a much slower rate in dorsorectal neurons than was observed at day 2 of adulthood, suggesting a significant reduction in proteasome activity in these cells. In contrast, no such decline was observed in body wall muscle cells, suggesting that age-related changes in the UPS may be tissue specific (128).
Another study that monitored proteasome activity between day 1 and day 2 of adulthood found a dramatic increase in ubiquitin G76V∷GFP turnover in all somatic tissues as animals began reproducing (129). This phenomenon correlated with a global increase in levels of K48-linked polyubiquitylated proteins, indicating that increased activity of the UPS as a whole occurs in early adulthood, as opposed to a specific increase in the proteolytic properties of the proteasome (129). Together, these findings suggest that the overall activity of the UPS increases substantially in the soma as animals reach reproductive maturity and declines thereafter in a tissue specific manner. Reduced proteasome activity has also been reported in early adulthood in D. melanogaster heads (between day 1 and day 5 of adulthood) and in rat spinal cord (between 3 and 12 months of age), indicating that early change in UPS activity may be a conserved feature of aging in some tissues (130, 131).
Pharmacological Enhancement of the Proteostasis Network as a Modifier of Aging and Disease
If the programmed collapse of proteostasis has a role in disease presentation, is it possible to intervene? If so, what are the consequences? Genetically enhancing the expression or activity of individual PN components suppresses disease onset and progression in a multitude of cell and animal models. Overexpression of many PN components can suppress protein folding toxicity, an observation consistent with the importance of all PN nodes for proteostasis. Although genetic studies have been invaluable in developing the paradigm that enhancing the PN is a viable approach to treating neurodegenerative disease, future therapies will require the identification and development of small-molecule enhancers of PN function. In this section, we focus on efforts to develop small molecules that can enhance various aspects of the PN, as well as their effects on disease (Table 2).
Table 2. Effects of pharmacological proteostasis network (PN) modifiers on animal models of neurodegenerative disease.
| Compound | Effect on PN | Disease model | Effect | Reference |
|---|---|---|---|---|
| Radicicol | Activates HSF1 | HD mouse brain slices | Delays aggregation | 64 |
| Geldanamycin | Activates HSF1 | PD flies | Suppresses loss of dopaminergic neurons | 135 |
| 17-AAG | Activates HSF1 | SCA3/MJD flies | Suppresses compound eye degeneration and SCA3 aggregation, improves survival | 134 |
| 17-AAG | Activates HSF1 | HD flies | Suppresses degeneration of photoreceptor neurons | 134 |
| HSP990 | Activates HSF1 | HD mice | Transiently suppresses aggregation and motor decline | 95 |
| Arimoclomol | Coactivates HSF1 | ALS mice (G93A) | Suppresses motor neuron death, rescues hind-limb function, and increases life span | 179 |
| NG-094 | Coactivates HSF1 | Worms expressing polyQ in body wall | Reduces aggregation and suppresses motility defects | 180 |
| Dexamethasone | Increases HSF1 levels | HD mice | Suppresses aggregation and improves motor performance | 97 |
| Dexamethasone | Increases HSF1 levels | HD flies | Suppresses aggregation and improves behavioral phenotypes | 97 |
| Geranylgeranyl-acetone | Increases HSP70 levels | SBMA mice | Suppresses aggregation and muscle wasting and improves motor function | 181 |
| HSF1A | Activates HSF1 | SCA3/MJD flies | Restores eye morphology, size, and pigment color | 137 |
| F1 | Activates multiple stress responses | Worms expressing polyQ in body wall muscle | Suppresses aggregation and movement defects | 136 |
| Rapamycin | Increases autophagy | HD flies | Suppresses photoreceptor neuron death | 141 |
| CCI-779 (rapamycin ester) | Increases autophagy | HD mice | Reduces striatal aggregation and improves motor function | 141 |
| SMER10 | Increases autophagy | HD flies | Suppresses photoreceptor neuron degeneration | 146 |
| SMER18 | Increases autophagy | HD flies | Suppresses photoreceptor neuron degeneration | 146 |
| SMER28 | Increases autophagy | HD flies | Suppresses photoreceptor neuron degeneration | 146 |
| Verapamil | Increases autophagy | HD zebrafish | Reduces aggregation and restores rhodopsin expression | 182 |
| Clonidine | Increases autophagy | HD zebrafish | Reduces aggregation and restores rhodopsin expression | 182 |
Abbreviations: ALS, amyotrophic lateral sclerosis; HD, Huntington's disease; HSF1, heat shock factor 1; PD, Parkinsons disease; SBMA, spinobulbar muscular atrophy; polyQ, polyglutamine; SMER, small-molecule enhancer of rapamycin.
One strategy is to directly activate HSF1, thereby increasing the expression of multiple molecular chaperones simultaneously. This approach has been traditionally achieved by inhibition of HSP90 with compounds that bind the N-terminal ATP-binding pocket, such as radicicol, geldanamycin, or 17-AAG (64, 132–134). Treatment with HSP90 inhibitors activates HSF1 and increases chaperone levels, suppressing polyQ aggregation and toxicity in cell, Drosophila, C. elegans, and mouse models of disease (95, 132, 134–136). Although these studies are an important proof of principle, inhibition of HSP90 is not a viable long-term option given the central role of HSP90 in numerous essential cellular processes (12). Therefore, we must look beyond these studies to new classes of HSP90-independent HSF1 activators for future therapeutics.
High-throughput screens in yeast and HeLa cells identified HSF1A and F1, respectively, as two small molecules that activate HSF1 independently of HSP90 inhibition (136, 137). HSF1A suppresses toxicity in cell and tissue culture models of HD and SCA-3/MJD and appears to activate HSF1 by impairing the activity of TRiC, a recently discovered negative regulator of HSF1. HSF1A treatment blocks the direct interaction between TRiC and HSF1, thereby eliciting an HSR (137, 138). Although the precise mechanism by which F1 activates HSF1 is unknown, F1 treatment increases the expression of chaperones and antioxidant enzymes and markedly suppresses polyQ toxicity in C. elegans and tissue culture models of HD. Despite only modest induction of stress response genes, F1 is able to correct protein misfolding and toxicity in diverse models of protein conformational disease, suggesting that subtle, holistic changes to the PN could be a powerful way to manage protein folding disorders (136). However, sustained activation of the HSR can be detrimental in the context of some protein misfolding diseases. For example, a recent study of the HSR in the context of cell models of α1-antitrypsin deficiency, Niemann–Pick type C1 disease, and cystic fibrosis revealed that the HSR is hyperactive in these diseases and that reduced HSF1 activity suppresses toxicity (139).
An alternative and complementary approach to the treatment of neurodegenerative disease could be to enhance degradation pathways and thus the turnover of misfolded proteins. Treatment of cell, Drosophila, and mouse models of HD, SCA3/MJD, AD, PD, and ALS with the mTOR inhibitor rapamycin (or a derivative) reduces aggregation and suppresses disease (140– 143). However, similar to HSP90 inhibitors, the role of rapamycin as an immunosuppressant means that lifelong treatment of patients may not be practical. As such, small-molecule screens have attempted to identify new classes of autophagy inducers.
An in silico screen based on the structure of 10-NCP, an Akt inhibitor that potently induces autophagy (144), identified the molecules FPZ and MTM as potent activators of autophagic flux and clearance of TDP-43 in mammalian cells (145). Treatment of induced pluripotent stem cell–derived motor neurons and astrocytes with these molecules suppressed mutant TDP-43-dependent cell death (145). A screen for autophagy inducers in yeast identified the molecules SMER-10, -18, and -28 as TOR-independent activators of autophagy (146). Treatment of PC12 cells stably expressing mutantα-synuclein (A53T) with SMER-10, -18, or-28 significantly reduced levels of mutant α-synuclein, an effect that was enhanced by cotreatment with rapamycin (146). In addition, SMER-10, -18, and -28 were effective at suppressing mHTT aggregation and toxicity in COS-7 cells and flies (146). Together, these results suggest that novel small-molecule modifiers of autophagy have promise as future disease therapeutics.
In addition to autophagy activators, small molecules that affect the activity of the proteasome represent an effective means to clear misfolded proteins from cells. The DUB USP14 suppresses turnover of Tau and TDP-43 in mouse embryonic fibroblasts (MEFs) by impairing the protea-some; therefore, small-molecule inhibitors of USP14 could help clear these toxic proteins from cells (147). To identify small-molecule inhibitors of USP14, investigators set up an in vitro system containing proteasomes, reconstituted USP14, and a 7-amido-4-methylcoumarin (Ub-AMC) fluorogenic degradation substrate. By screening ∼63,000 molecules for their ability to restore proteasomal substrate degradation, these authors identified the potent and selective USP14 inhibitor IU1. Treatment with IU1 reduced the levels of Tau, TDP-43, and ataxin-3 in MEFs in a USP14-dependent manner and independently of changes in proteasome levels or composition (147). These findings suggest that small molecules that target the PN have promise in the fight against age-related disease. Future studies to identify further small-molecule regulators of the PN and to elucidate to what extent existing PN modifiers can act synergistically to suppress proteotoxicity will pave the way for exciting new therapeutic options for largely untreatable diseases.
The Future of the Proteostasis Network in Health and Human Disease
Maintaining proteostasis is a demanding job that must be accomplished efficiently by every cell to avoid dysfunction. Efforts to understand the fundamental components involved in protein synthesis, folding, and degradation, and the extent to which they are connected, have resulted in the concept of the PN and recognition of the high degree of cooperation required to ensure proteome stability (1, 9). Unraveling the complete repertoire of PN function and connectivity will be a demanding endeavor; however, such studies will elucidate the extent to which the PN and auxiliary factors can regulate aging and age-related disease. A more challenging aspect of PN biology is to integrate these findings with an understanding of the extent to which the PN is tailored to individual cells, tissues, and life stages and the potential implications of this understanding to aging and disease. Given the complexity of this scenario, the temporal and spatial particulars of the PN have been only tentatively addressed to date. Nevertheless, we are discovering that many long-held assumptions regarding proteostasis control and decline are incorrect, and that wide-scale changes in the composition and function of the PN are precise early events, likely coordinated with reproduction. Although we do not know how these observations will extend to complex mammalian systems, a complete understanding of the signals and mechanisms responsible for PN reprogramming will undoubtedly provide important new insights into the means and necessity for organismal control of the PN. As we continue to unravel the foundations of proteostasis biology, we predict that future discoveries encompassing spatial and temporal control of the PN will allow us to develop new combinations of small molecules with the potential to provide long-term effective treatment for a range of human diseases.
The Oxidative Stress Response in Aging and Disease.
Increased protein oxidation is strongly linked to aging and disease (148). Reactive oxygen species (ROS) are prevented from causing oxidative protein damage by antioxidant enzymes such as superoxide dismutases (SODs), glutamate cysteine ligases (GCLs), and glutathione S-transferases (GSTs). In response to ROS accumulation in cells, the TF SKN-1/NRF2 is activated and drives the oxidative stress response (OxSR) (149). Mouse striatal cells expressing mutant huntingtin (mHTT) exhibit a reduced OxSR when challenged with tert-butylhydroxyquinone (tBHQ), independent of changes in the level of Nrf2 (150). In contrast, mutant SOD1 impairs the OxSR in ALS mice through reduced expression of Nrf2 messenger RNA (mRNA) in motor neurons (151). Similarly, transcriptional dysregulation of Nrf2 and sequestration of Nrf2 protein into Aβ1–42 aggregates underlie dysregulation of the OxSR in the hippocampus of AD mice (152). During normal aging in flies, the level of carbonylated proteins is increased twofold by middle age (153). These effects were attributed to reduced expression of proteasomal subunits and impaired turnover of damaged proteins; however, it is likely that dysregulation of the OxSR also contributes to increased protein oxidation with age (153, 154). Although not a direct PN component or modifier, protein oxidation and its control are intimately linked to aging and age-related disease.
Summary Points.
The PN spans multiple cellular compartments and is coordinated to maintain organismal proteostasis.
The PN exhibits substantial heterogeneity in composition and capacity between cells and tissues.
Loss of proteostasis and dysregulation of the PN are common features of neurodegenerative disease and normal aging.
Changes in proteostasis occur earlier in life than anticipated.
Small-molecule enhancers of molecular chaperone activity or protein degradation pathways are effective in suppressing protein aggregation and related toxicity in cell and animal disease models.
Future Issues.
How do cells transmit proteostasis states to one another?
What is the mechanism behind the transmission of aggregates between cells?
What is the relationship between the PN and the proteome in different tissues?
What is the mechanistic basis for proteostasis collapse, and what are the consequences on organismal health if it is prevented?
Do all tissues exhibit identical patterns of proteostasis collapse during aging and disease? If not, which tissues are spared and why?
Acknowledgments
We apologize profusely to any authors whose important contributions were not cited in this review due to space limitations. Our studies were supported by a postdoctoral fellowship to J.L. from the ALS Association and grants from the National Institutes of Health (the National Institute of General Medical Sciences, the National Institute on Aging, and the National Institute of Mental Health), the Ellison Medical Foundation, and the Daniel F. and Ada L. Rice Foundation to R.I.M. We thank the members of the Morimoto laboratory for their support and critical reading of the manuscript.
Footnotes
Disclosure Statement: R.M. is a founder and member of the scientific advisory board of Proteostasis Therapeutics, Inc. J.L. is not aware of any affiliations, memberships, funding, or financial holdings that might affect the objectivity of this review.
Literature Cited
- 1.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 2.Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol. 2013;9:25–34. doi: 10.1038/nrneurol.2012.236. [DOI] [PubMed] [Google Scholar]
- 3.Labbadia J, Morimoto RI. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38:378–85. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–64. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 5.Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013;9:445–54. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 7.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–86. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 9.Powers ET, Balch WE. Diversity in the origins of proteostasis networks—a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–48. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–46. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 11.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 12.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–28. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 13.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 15.Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–77. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 16.Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–48. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci. 2012;37:274–83. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–35. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14:617–29. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 20.Finley D. Recognition and processing of ubiquitin–protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–47. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 23.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274:3363–71. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 24.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 25.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 26.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–15. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 27.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsässer S, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 28.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–40. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 29.Nakatsukasa K, Kamura T, Brodsky JL. Recent technical developments in the study of ER-associated degradation. Curr Opin Cell Biol. 2014;29C:82–91. doi: 10.1016/j.ceb.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, et al. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell. 2014;54:166–79. doi: 10.1016/j.molcel.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–17. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23:311–18. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–80. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–66. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993;13:2486–96. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–66. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raychaudhuri S, Loew C, Körner R, Pinkert S, Theis M, et al. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 2014;156:975–85. doi: 10.1016/j.cell.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 38.Shalgi R, Hurt JA, Lindquist S, Burge CB. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–70. doi: 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 39.Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. Widespread regulation of translation by elongation pausing in heat shock. Mol Cell. 2013;49:439–52. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 41.Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Oosten–Hawle P, Morimoto RI. Organismal proteostasis: role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 2014;28:1533–43. doi: 10.1101/gad.241125.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–14. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. PNAS. 2011;108:14204–9. doi: 10.1073/pnas.1106557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain– mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–47. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Oosten–Hawle P, Porter RS, Morimoto RI. Regulation of organismal proteostasis by trans-cellular chaperone signaling. Cell. 2013;153:1366–78. doi: 10.1016/j.cell.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–25. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shemesh N, Shai N, Ben-Zvi A. Germline stem cell arrest inhibits the collapse of somatic pro-teostasis early in Caenorhabditis elegans adulthood. Aging Cell. 2013;12:814–22. doi: 10.1111/acel.12110. [DOI] [PubMed] [Google Scholar]
- 50.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–68. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 51.Ermolaeva MA, Segref A, Dakhovnik A, Ou HL, Schneider JI, et al. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–20. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–14. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–8. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tebbenkamp AT, Borchelt DR. Analysis of chaperone mRNA expression in the adult mouse brain by meta analysis of the Allen Brain Atlas. PLOS ONE. 2010;5:e13675. doi: 10.1371/journal.pone.0013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLOS Genet. 2013;9:e1003466. doi: 10.1371/journal.pgen.1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batulan Z, Shinder GA, Minotti S, He BP, Doroudchi MM, et al. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J Neurosci. 2003;23:5789–98. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tagawa K, Marubuchi S, Qi ML, Enokido Y, Tamura T, et al. The induction levels of heat shock protein 70 differentiate the vulnerabilities to mutant huntingtin among neuronal subtypes. J Neurosci. 2007;27:868–80. doi: 10.1523/JNEUROSCI.4522-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–92. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–96. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 60.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;9:506–14. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Sontag EM, Vonk WI, Frydman J. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol. 2014;26:139–46. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. PNAS. 2002;99:10417–22. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–74. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 64.Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, et al. Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet. 2004;13:1389–405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- 65.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–31. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 66.Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, et al. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–39. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu G, Stevens SM, Jr, Moore BD, McClung S, Borchelt DR. Cytosolic proteins lose solubility as amyloid deposits in a transgenic mouse model of Alzheimer-type amyloidosis. Hum Mol Genet. 2013;22:2765–74. doi: 10.1093/hmg/ddt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–68. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 69.Fonte V, Kapulkin WJ, Taft A, Fluet A, Friedman D, Link CD. Interaction of intracellular β amyloid peptide with chaperone proteins. PNAS. 2002;99:9439–44. doi: 10.1073/pnas.152313999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLean PJ, Kawamata H, Shariff S, Hewett J, Sharma N, et al. TorsinA and heat shock proteins act as molecular chaperones: suppression of α-synuclein aggregation. J Neurochem. 2002;83:846–54. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 71.Chai Y, Koppenhafer SL, Bonini NM, Paulson HL. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J Neurosci. 1999;19:10338–47. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu A, Shibata Y, Shah B, Calamini B, Lo DC, Morimoto RI. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. PNAS. 2014;111:e1481–90. doi: 10.1073/pnas.1321811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gregori L, Fuchs C, Figueiredo-Pereira ME, Van Nostrand WE, Goldgaber D. Amyloid β-protein inhibits ubiquitin-dependent protein degradation in vitro. J Biol Chem. 1995;270:19702–8. doi: 10.1074/jbc.270.34.19702. [DOI] [PubMed] [Google Scholar]
- 74.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000;75:436–39. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 75.Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, et al. Inhibition of the ubiquitin– proteasome system in Alzheimer's disease. PNAS. 2000;97:9902–6. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheroni C, Marino M, Tortarolo M, Veglianese P, De Biasi S, et al. Functional alterations of the ubiquitin–proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:82–96. doi: 10.1093/hmg/ddn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis–associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McNaught KS, Jenner P. Proteasomal function is impaired in substantia nigra in Parkinson's disease. Neurosci Lett. 2001;297:191–94. doi: 10.1016/s0304-3940(00)01701-8. [DOI] [PubMed] [Google Scholar]
- 79.Um JW, Im E, Lee HJ, Min B, Yoo L, et al. Parkin directly modulates 26S proteasome activity. J Neurosci. 2010;30:11805–14. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–18. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 82.Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–87. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–45. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 84.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–84. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 85.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–89. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 86.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, et al. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023–37. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–95. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 90.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, et al. Dopamine-modified α-synuclein blocks chaperone-mediated autophagy. J Clin Investig. 2008;118:777–88. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bandyopadhyay U, Nagy M, Fenton WA, Horwich AL. Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. PNAS. 2014;111:11055–60. doi: 10.1073/pnas.1409314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan WM, Tsoi H, Wu CC, Wong CH, Cheng TC, et al. Expanded polyglutamine domain possesses nuclear export activity which modulates subcellular localization and toxicity of polyQ disease protein via exportin-1. Hum Mol Genet. 2011;20:1738–50. doi: 10.1093/hmg/ddr049. [DOI] [PubMed] [Google Scholar]
- 94.Huang S, Ling JJ, Yang S, Li XJ, Li S. Neuronal expression of TATA box–binding protein containing expanded polyglutamine in knock-in mice reduces chaperone protein response by impairing the function of nuclear factor Y transcription factor. Brain: J Neurol. 2011;134:1943–58. doi: 10.1093/brain/awr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Investig. 2011;121:3306–19. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Silva-Fernandes A, Duarte-Silva S, Neves-Carvalho A, Amorim M, Soares-Cunha C, et al. Chronic treatment with 17-DMAG improves balance and coordination in a new mouse model of Machado–Joseph disease. J Am Soc Exp Neurother. 2014;11:433–49. doi: 10.1007/s13311-013-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maheshwari M, Bhutani S, Das A, Mukherjee R, Sharma A, et al. Dexamethasone induces heat shock response and slows down disease progression in mouse and fly models of Huntington's disease. Hum Mol Genet. 2014;23:2737–51. doi: 10.1093/hmg/ddt667. [DOI] [PubMed] [Google Scholar]
- 98.Chafekar SM, Duennwald ML. Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin. PLOS ONE. 2012;7:e37929. doi: 10.1371/journal.pone.0037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 100.Riva L, Koeva M, Yildirim F, Pirhaji L, Dinesh D, et al. Poly-glutamine expanded huntingtin dramatically alters the genome wide binding of HSF1. J Huntington's Dis. 2012;1:33–45. [PMC free article] [PubMed] [Google Scholar]
- 101.Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15:233–49. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 102.Gkogkas C, Middleton S, Kremer AM, Wardrope C, Hannah M, et al. VAPB interacts with and modulates the activity of ATF6. Hum Mol Genet. 2008;17:1517–26. doi: 10.1093/hmg/ddn040. [DOI] [PubMed] [Google Scholar]
- 103.Fernandez-Fernandez MR, Ferrer I, Lucas JJ. Impaired ATF6α processing, decreased Rheb and neuronal cell cycle re-entry in Huntington's disease. Neurobiol Dis. 2011;41:23–32. doi: 10.1016/j.nbd.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 104.Cicchetti F, Saporta S, Hauser RA, Parent M, Saint-Pierre M, et al. Neural transplants in patients with Huntington's disease undergo disease-like neuronal degeneration. PNAS. 2009;106:12483–88. doi: 10.1073/pnas.0904239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li JY, Englund E, Holton JL, Soulet D, Hagell P, et al. Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–3. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- 106.Hansen C, Angot E, Bergström AL, Steiner JA, Pieri L, et al. α-Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. J Clin Investig. 2011;121:715–25. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ren PH, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–25. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pecho-Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci. 2014;17:1064–72. doi: 10.1038/nn.3761. [DOI] [PubMed] [Google Scholar]
- 109.Nussbaum-Krammer CI, Park KW, Li L, Melki R, Morimoto RI. Spreading of a prion domain from cell-to-cell by vesicular transport in Caenorhabditis elegans. PLOS Genet. 2013;9:e1003351. doi: 10.1371/journal.pgen.1003351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Munch C, O'Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase 1 misfolding in neuronal cells. PNAS. 2011;108:3548–53. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carnemolla A, Labbadia JP, Lazell H, Neueder A, Moussaoui S, Bates GP. Contesting the dogma of an age-related heat shock response impairment: implications for cardiac-specific age-related disorders. Hum Mol Genet. 2014;23:3641–56. doi: 10.1093/hmg/ddu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blake MJ, Udelsman R, Feulner GJ, Norton DD, Holbrook NJ. Stress-induced heat shock protein 70 expression in adrenal cortex: an adrenocorticotropic hormone-sensitive, age-dependent response. PNAS. 1991;88:9873–77. doi: 10.1073/pnas.88.21.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014;3:1135–50. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. PNAS. 2009;106:14914–19. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLOS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11:120–27. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kirstein-Miles J, Morimoto RI. Ribosome-associated chaperones act as proteostasis sentinels. Cell Cycle. 2013;12:2335–36. doi: 10.4161/cc.25703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xiao X, ZuoX, Davis AA, McMillan DR, Curry BB, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–52. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–62. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–64. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 121.Walker GA, Thompson FJ, Brawley A, Scanlon T, Devaney E. Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J. 2003;17:1960–62. doi: 10.1096/fj.03-0164fje. [DOI] [PubMed] [Google Scholar]
- 122.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pappas C, Hyde D, Bowler K, Loeschcke V, Sørensen JG. Post-eclosion decline in ‘knock-down’ thermal resistance and reduced effect of heat hardening in Drosophila melanogaster. Comp Biochem Physiol A. 2007;146:355–59. doi: 10.1016/j.cbpa.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 124.Coh en E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 125.Cohen E, Du D, Joyce D, Kapernick EA, Volovik Y, et al. Temporal requirements of insulin/IGF-1 signaling for proteotoxicity protection. Aging Cell. 2010;9:126–34. doi: 10.1111/j.1474-9726.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Volovik Y, Maman M, Dubnikov T, Bejerano-Sagie M, Joyce D, et al. Temporal requirements of heat shock factor 1 for longevity assurance. Aging Cell. 2012;11:491–99. doi: 10.1111/j.1474-9726.2012.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–34. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 128.Hamer G, Matilainen O, Holmberg CI. A photoconvertible reporter of the ubiquitin–proteasome system in vivo. Nat Methods. 2010;7:473–78. doi: 10.1038/nmeth.1460. [DOI] [PubMed] [Google Scholar]
- 129.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin–proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Keller JN, Huang FF, Markesbery WR. Decreased levels of proteasome activity and proteasome expression in aging spinal cord. Neuroscience. 2000;98:149–56. doi: 10.1016/s0306-4522(00)00067-1. [DOI] [PubMed] [Google Scholar]
- 131.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, et al. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum Mol Genet. 2001;10:1307–15. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 133.Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–95. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 134.Fujikake N, Nagai Y, Popiel HA, Okamoto Y, Yamaguchi M, Toda T. Heat shock transcription factor 1–activating compounds suppress polyglutamine-induced neurodegeneration through induction of multiple molecular chaperones. J Biol Chem. 2008;283:26188–97. doi: 10.1074/jbc.M710521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Auluck PK, Bonini NM. Pharmacological prevention of Parkinson disease in Drosophila. Nat Med. 2002;8:1185–86. doi: 10.1038/nm1102-1185. [DOI] [PubMed] [Google Scholar]
- 136.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–96. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLOS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele D. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 2014;3:955–66. doi: 10.1016/j.celrep.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roth DM, Hutt DM, Tong J, Bouchecareilh M, Wang N, et al. Modulation of the maladaptive stress response to manage diseases of protein folding. PLOS Biol. 2014;11:e1001998. doi: 10.1371/journal.pbio.1001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 141.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 142.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 143.Menzies FM, Huebener J, Renna M, Bonin M, Riess O, Rubinsztein DC. Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3. Brain: J Neurol. 2010;133:93–104. doi: 10.1093/brain/awp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicityin aHuntington disease model. PNAS. 2010;107:16982–87. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–38. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee BH, Lee MJ, Park S, Oh DC, Elsässer S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, et al. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. PNAS. 1991;88:10540–43. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jin YN, Yu YV, Gundemir S, Jo C, Cui M, et al. Impaired mitochondrial dynamics and Nrf2 signaling contribute to compromised responses to oxidative stress in striatal cells expressing full-length mutant huntingtin. PLOS ONE. 2013;8:e57932. doi: 10.1371/journal.pone.0057932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kirby J, Halligan E, Baptista MJ, Allen S, Heath PR, et al. Mutant SOD1 alters the motor neuronal transcriptome: implications for familial ALS. Brain: J Neurol. 2005;128:1686–706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- 152.Kanninen K, Malm TM, Jyrkkänen HK, Goldsteins G, Keksa-Goldsteine V, et al. Nuclear factor erythroid 2–related factor 2 protects against βamyloid. Mol Cell Neurosci. 2008;39:302–13. doi: 10.1016/j.mcn.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 153.Tsakiri EN, Sykiotis GP, Papassideri IS, Gorgoulis VG, Bohmann D, Trougakos IP. Differential regulation of proteasome functionality in reproductive versus somatic tissues of Drosophila during aging or oxidative stress. FASEB J. 2013;27:2407–20. doi: 10.1096/fj.12-221408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rahman MM, Sykiotis GP, Nishimura M, Bodmer R, Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell. 2013;12:554–62. doi: 10.1111/acel.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 156.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–15. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human α crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–74. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 158.Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–32. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Takiyama Y, Nishizawa M, Tanaka H, Kawashima S, Sakamoto H, et al. The gene for Machado– Joseph disease maps to human chromosome 14q. Nat Genet. 1993;4:300–4. doi: 10.1038/ng0793-300. [DOI] [PubMed] [Google Scholar]
- 160.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 161.Rajasekaran NS, ConnellP, Christians ES, Yan LJ, Taylor RP, et al. Human αB-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–39. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Engert JC, Bérubé P, Mercier J, Doré C, Lepage P, et al. ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat Genet. 2000;24:120–25. doi: 10.1038/72769. [DOI] [PubMed] [Google Scholar]
- 163.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, et al. Mutant small heat-shock protein 27 causes axonal Charcot–Marie–Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–6. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 164.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–81. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 165.Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, et al. Mutations in SIL1 cause Marinesco–Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 2005;37:1312–14. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 166.Anttonen AK, Mahjneh I, Hämäläinen RH, Lagier-Tourenne C, Kopra O, et al. The gene disrupted in Marinesco–Sjägren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet. 2005;37:1309–11. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- 167.Tang BS, Zhao GH, Luo W, Xia K, Cai F, et al. Small heat-shock protein 22 mutated in autosomal dominant Charcot–Marie–Tooth disease type 2L. Hum Genet. 2005;116:222–24. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- 168.Davey KM, Parboosingh JS, McLeod DR, Chan A, Casey R, et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome–like condition. J Med Genet. 2006;43:385–93. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Burbulla LF, Schelling C, Kato H, Rapaport D, Woitalla D, et al. Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum Mol Genet. 2010;19:4437–52. doi: 10.1093/hmg/ddq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.He M, Guo H, Yang X, Zhou L, Zhang X, et al. Genetic variations in HSPA8 gene associated with coronary heart disease risk in a Chinese population. PLOS ONE. 2010;5:e9684. doi: 10.1371/journal.pone.0009684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, et al. Proteasome assembly defect due to a proteasome subunit βtype 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo– Nishimura syndrome. PNAS. 2011;108:14914–19. doi: 10.1073/pnas.1106015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Blumen SC, Astord S, Robin V, Vignaud L, Toumi N, et al. A rare recessive distal hereditary motor neuropathy with HSJ1 chaperone mutation. Ann Neurol. 2012;71:509–19. doi: 10.1002/ana.22684. [DOI] [PubMed] [Google Scholar]
- 173.Edvardson S, Cinnamon Y, Ta-Shma A, Shaag A, Yim YI, et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLOS ONE. 2012;7:e36458. doi: 10.1371/journal.pone.0036458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harms MB, Sommerville RB, Allred P, Bell S, Ma D, et al. Exome sequencing reveals DNAJB6 mutations in dominantly inherited myopathy. Ann Neurol. 2012;71:407–16. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Teyssou E, Takeda T, Lebon V, Boillée S, Doukouré B, et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125:511–22. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- 176.Vilarino-Guell C, Rajput A, Milnerwood AJ, Shah B, Szu-Tu C, et al. DNAJC13 mutations in Parkinson disease. Hum Mol Genet. 2014;23:1794–801. doi: 10.1093/hmg/ddt570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kwok CT, Morris AG, Frampton J, Smith B, Shaw CE, de Belleroche J. Association studies indicate that protein disulfide isomerase is a risk factor in amyotrophic lateral sclerosis. Free Radic Biol Med. 2013;58:81–86. doi: 10.1016/j.freeradbiomed.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 178.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 179.Kieran D, Kalmar B, Dick JR, Riddoch-Contreras J, Burnstock G, Greensmith L. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med. 2004;10:402–5. doi: 10.1038/nm1021. [DOI] [PubMed] [Google Scholar]
- 180.Haldimann P, Muriset M, Vigh L, Goloubinoff P. The novel hydroxylamine derivative NG-094 suppresses polyglutamine protein toxicity in Caenorhabditis elegans. J Biol Chem. 2011;286:18784–94. doi: 10.1074/jbc.M111.234773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Katsuno M, Sang C, Adachi H, Minamiyama M, Waza M, et al. Pharmacological induction of heat-shock proteins alleviates polyglutamine-mediated motor neuron disease. PNAS. 2005;102:16801–6. doi: 10.1073/pnas.0506249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]