Abstract
Background
CETP inhibitors block the transfer of cholesteryl ester from HDL-C to VLDL-C and LDL-C, thereby raising HDL-C and lowering LDL-C. In this study, we explored the effect of CETP inhibitors on hepatic LDL receptor (LDLR) and PCSK9 expression and further elucidated the underlying regulatory mechanism.
Results
We first examined the effect of anacetrapib (ANA) and dalcetrapib (DAL) on LDLR and PCSK9 expression in hepatic cells in vitro. ANA exhibited a dose-dependent inhibition on both LDLR and PCSK9 expression in CETP-positive HepG2 cells and human primary hepatocytes as well as CETP-negative mouse primary hepatocytes (MPH). Moreover, the induction of LDLR protein expression by rosuvastatin in MPH was blunted by cotreatment with ANA. In both HepG2 and MPH ANA treatment reduced the amount of mature form of SREBP2 (SREBP2-M). In vivo, oral administration of ANA to dyslipidemic C57BL/6J mice at a daily dose of 50 mg/kg for 1 week elevated serum total cholesterol by approximately 24.5% (p<0.05%) and VLDL-C by 70% (p<0.05%) with concomitant reductions of serum PCSK9 and liver LDLR/SREBP2-M protein. Finally, we examined the in vitro effect of two other strong CETP inhibitors evacetrapib and torcetrapib on LDLR/PCSK9 expression and observed a similar inhibitory effect as ANA in a concentration range of 1–10 µM.
Conclusion
Our study revealed an unexpected off-target effect of CETP inhibitors that reduce the mature form of SREBP2, leading to attenuated transcription of hepatic LDLR and PCSK9. This negative regulation of SREBP pathway by ANA manifested in mice where CETP activity was absent and affected serum cholesterol metabolism.
Keywords: CETP inhibitors, LDL receptor, PCSK9, SREBP2, Hyperlipidemia
1. Background
Epidemiological studies have strongly suggested that elevated plasma concentrations of low-density lipoprotein-cholesterol (LDL-C) and reduced concentrations of high-density lipoprotein-cholesterol (HDL-C) are independent risk factors for developing cardiovascular disease (CVD) [1]. The plasma LDL-C concentrations are primarily controlled by expression levels of hepatic LDL receptor (LDLR) [2–4]. Hepatic LDLR mediates the uptake of LDL particles from the circulation and delivers the receptor-bound LDL to the endosomal system for degradation while the LDLR returns to the cell surface. Statins are competitive inhibitors of HMG-CoA reductase (HMGCR), the rate-limiting enzyme in the cellular cholesterol biosynthetic pathway. The inhibition of cholesterol de novo synthesis leads to increased numbers of cell surface LDLR by activation of LDLR gene transcription. Thus, statins are the most widely prescribed drugs to treat hypercholesterolemia and combined hyperlipidemia [5].
The plasma concentrations of HDL-C are modulated by several proteins including plasma protein cholesteryl ester transfer protein (CETP), which is a hydrophobic glycoprotein secreted from liver. CETP mediates the equal molar transfer of CE from HDL to apoB containing lipoproteins VLDL and LDL and the equimolar transfer of triglycerides (TGs) from VLDL and LDL to HDL. Thus, inhibition of CETP activity raises plasma HDL-C and lowers LDL-C, which favorably reduces both CVD risk factors simultaneously. Over the last decade, a great deal of efforts has been put into the development of CETP inhibitors as new therapy to raise HDL-C [6–9]. Thus far, four CETP inhibitors have been tested in human clinical studies including torcetrapib (TOR) [7] dalcetrapib (DAL) [10–13], anacetrapib (ANA) [14–17] and evacetrapib (EVA) [18]. The TOR program was terminated early due to its off-target effects on inducing aldosterone and cortisol production that were the underline causes for excess CVD endpoints and mortality in the TOR group versus placebo [19]. The DAL program was discontinued in 2012 due to the lack of its efficacy in reducing the risk of recurrent cardiovascular events despite the elevation of plasma HDL-C levels [20,21].
ANA is a potent CETP inhibitor that is currently undergoing Phase III clinical trials. In a clinical study of 589 hyperlipidemic patients, ANA monotherapy increased HDL-C up to 139% and reduced LDL-C up to 40%. When added to atorvastatin, ANA 150 mg daily produced a statistically significant 20% reduction in Friedewald equation-calculated LDL-C [22]. With regard to its effect on LDL-C reduction, a recent new study to compare different methods to determine LDL-C levels in placebo and ANA treated patients suggested that the true LDL-C reductions with this CETP inhibitor may have been less than reported, while its inductions on HDL-C were unaffected by different measurements [23].
The primary functionality of HDL-C rising by CETP inhibitors is the enhanced reverse-cholesterol transport (RCT) from extra periphery tissues to the liver. Due to the lack of CETP activity in mice and rats, hamsters [24–27] and CETP-transgenic mice [28] have been used as animal models for evaluations of effects of CETP inhibitors on RCT.
Besides CETP, PCSK9 is another promising therapeutic target [29,30]. Plasma PCSK9 binds to hepatic LDLR, promoting its degradation, and consequently raising plasma LDL-C. Owing to the critical function of PCSK9 in the control of protein levels of LDLR, currently many approaches have been taken to either block its interaction with LDLR by anti-PCSK9 antibodies [31] or to reduce PCSK9 expression by antisense oligonucleotides [32] or small interference RNAs (siRNAs) [33]. Interestingly, it was recently reported that a new CETP inhibitor (K-312) exhibited negative effects on PCSK9 expression in HepG2 cells at the level of gene transcription [34,35].
It is well established that transcription of PCSK9 and LDLR genes shares one common regulatory mechanism mediated by sterol-regulatory element binding proteins (SREBPs) [36–38]. SREBPs are members of the basic helix-loop-helix leucine zipper family of transcription factors. SREBPs contain 2 transmembrane domains and are located to the endoplasmic reticulum (ER) after synthesis. In the inactive state within ER, the C-terminal domains of the SREBPs interact with another membrane protein SREBP-cleavage-activating protein (SCAP), which functions as a sterol sensor. In sterol-depleted cells, SCAP escorts the SREBPs from the ER to the Golgi, where they are processed by two membrane-associated proteases, the site 1 (S1P) and site 2 (S2P) proteases, which release the NH2-terminl transcription-activation domain of the SREBPs (mature forms of SREBPs) from the precursor proteins. The active forms of the SREBPs translocate to the nucleus, where they bind to the promoters of SREBP target genes, including genes involved in the synthesis and metabolism of cholesterol [39]. In addition, transcription of the genes encoding SREBP 2 and SREBP1c is enhanced by SREBPs by a feedforward mechanism through SREBP binding sites in the promoters of these genes.
PCSK9 and LDLR both contain an SRE-1 motif in their proximal promoters and thus are coordinately upregulated by statins through activation of SREBP [37,40]. In addition to SREBP2, our laboratory has identified HNF1α as a critical transcription factor for PCSK9 through its binding to HNF1 motif located 28 bp upstream to SRE-1 site of the PCSK9 gene promoter [41]. Because HNF1 site is not present in the LDLR promoter, modulations of PCSK9 transcriptions through HNF1 sequence will not affect LDLR gene expression. Indeed, we have shown that the natural cholesterol lowering compound berberine suppressed PCSK9 gene expression without an effect on LDLR promoter activity. We further demonstrated that berberine inhibits PCSK9 transcription by reducing cellular HNF1α protein levels in liver cells [41].
In light of the new reports of PCSK9 regulation by a CETP inhibitor and the critical role of PCSK9 in LDL-C metabolism via LDLR, we decided to investigate whether other potent CETP inhibitors possess similar functions and what are the underlying regulatory mechanisms. Initially, we utilized both CETP-positive human liver cells and CETP-negative mouse primary hepatocyes (MPH) to examine effects of ANA and DAL on PCSK9 and LDLR gene expression. We did not detect effects of DAL up to the tolerable concentration of 10 µM. In contrast, ANA showed significant effects within the concentration range of 1–10 µM in a dose dependent manner. Thus, we carried out further investigations on ANA to examine its in vivo effect on liver tissue of mice fed a high cholesterol diet or a normal chow diet. Our studies obtained compelling results that ANA negatively regulates both Ldlr and Pcsk9 gene expression via the common regulatory SREBP pathway by a mechanism that inhibits the processing of SREBP2, independent of its inhibitory activity to CETP. Finally, we extended this study to EVA and TOR to determine their effects on SREBP2-mediated transcription of LDLR and PCSK9 in HepG2 cells and demonstrated a similar inhibitory effect as ANA.
2. Materials and methods
2.1. Animals diet and drug treatment
All animal experiments were performed according to procedures approved by the VA Palo Alto Health Care System Institutional Animal Care and Use Committee (IACUC). Eight-week old male C57BL/6J mice were purchased from Jackson Labs (Bar Harbor, Maine). Mice were housed (4 animals/cage) under controlled temperature (72°F) and lighting (12 h light/dark cycle). Animals had free access to autoclaved water and food.
In the first in vivo study, after an acclimatization period of 7 days, mice were fed a high-fat high-cholesterol diet (HFHC) containing 35% calories from fat and 1.25% cholesterol (#D12336, Research Diets, Inc., New Brunswick, NJ) for two weeks. Mice were then divided into two groups (n = 8 per group) and were given a daily dose of ANA at 50 mg/kg by oral gavage. The control group received vehicle (0.5% methyl cellulose). The drug treatment lasted 7 days.
In the second in vivo study, mice fed a normal chow diet were treated with ANA (50 mg/kg, n=8) or vehicle (n=8) for 10 days.
Serum samples were collected after a 4 h fasting before and after the drug treatment. After the last dosing, all animals were sacrificed for collection of serum and liver tissues. Livers were immediately removed, cut into small pieces, and stored at −80°C for RNA and protein isolations and cholesterol measurement.
2.2 Cells and Reagents
The human hepatoma cell line HepG2 was obtained from ATCC. HepG2-B11 is a HepG2 derived cell line expressing human LDLR promoter construct pLDLR-234 [42]. HepG2-CL26 cells express human PCSK9 promoter construct pGL3-PCSK9-D1[41]. Human primary hepatocytes were obtained from Invitrogen. MPH were isolated from male C57BL/6J mouse at San Francisco General Hospital Liver Center. A rabbit anti-LDLR antibody was obtained from Biovision (Mountain View, CA). Rabbit anti-SREBP2 and anti-PCSK9 antibodies were generously provided by Dr. Sahng Wook Park (Yonsei University College of Medicine, Seoul, Korea) and were used as previously described [41]. Monoclonal anti-β-actin antibody was obtained from Sigma-Aldrich. ANA and DAL were purchased from Selleckchem (Houston, TX) with purities over 99%. EVA was purchased from Medchem Express (Princeton, NJ) with purity of 99.33%. TOR was purchased from Sigma-Aldrich with 98% purity. Rosuvastatin (RSV) calcium was purchased from AK Scientific (Union City, CA). Mouse and human PCSK9 Quantikine ELISA Kits were purchased from R&D system (Minneapolis, MN). Methyl Cellulose was purchased from Sigma-Aldrich (Cat. No. M0512).
2.3 Culture of primary hepatocytes
Primary human and mouse hepatocytes were seeded on collagen coated plates at a density of 0.5–1×106 cells/well in 12-well plate in Williams E Medium supplemented with a Cell Maintenance Cocktail (Cell Maintenance Supplement Pack, Invitrogen). After overnight seeding, cells were treated with CETP inhibitors for 24 h in Williams E Medium or DMEM supplemented with 10% FBS.
2.4 Transient transfection and luciferase reporter assays
The plasmid pLDLR234-WT was constructed by subcloning a 177-bp fragment of the LDLR promoter obtained by HindIII digestion of pLDLR-CAT 234 into HindIII-digested pGL3-basic vector (Promega). The plasmid pLDLR234-SRE-mu was obtained by site-directed mutation [43]. The plasmid pGL3-PCSK9-D4 contains 5’ flanking region of the PCSK9 gene from −440 to −94, relative to the ATG start codon in front of the luciferase coding sequence [38]. The plasmid pGL3-PCSK9-D4-SRE-mu was obtained by site-directed mutation [38]. PCSK9 or LDLR promoter reporters of wild-type and SRE-1 mutation were cotransfected with pRL-SV40, a renilla luciferase vector, into HepG2 cells seeded in 96 well plates in MEM medium containing 10% FBS. One day post-transfection, cells were treated with indicated compounds for 24 h prior to cell lysis. Dual luciferase activities were measured and the firefly luciferase activity was normalized to the renilla luciferase activity. Triplicate wells were assayed for each transfection condition.
2.5 Cell viability assay
Cells were seeded in a 96 well plate overnight prior to the treatment by different concentrations of CETP inhibitors for 24 h. Cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay kit from Promega according to vendor’s instruction. Four wells were evaluated under each experimental condition.
2.6 RNA isolation and real time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells or liver tissue using the Quick RNA mini Prep kit (Zymo Research) and was reverse-transcribed into cDNA as described previously [44]. Real-time qRT-PCR was performed with 50 ng of cDNA template and specific primers using a SYBR Green PCR Kit (power SYBR® Green PCR Master Mix) and an ABI Prism 7700 system (Applied Biosystems® Life Technologies) according to the manufacturer’s protocols. qRT-PCR primers for each gene is listed in Table 1. Target mRNA expression in each sample was normalized to the housekeeping gene GAPDH. The 2”AACt method was used to calculate relative mRNA expression levels.
Table 1.
qRT-PCR primer sequences
| Forward | Reverse | |
|---|---|---|
| Human | ||
| GAPDH | ATGGGGAAGGTGAAGGTCG | GGGGTCATTGATGGCAACAATA |
| CETP | TGCCAAAACAAGGGAGTCGT | AATAGGAGGCCTGGACGGTA |
| LDLR | GACGTGGCGTGAACATCTG | CTGGCAGGCAATGCTTTGG |
| PCSK9 | AGGGGAGGACATCATTGGTG | CAGGTTGGGGGTCAGTACC |
| SREBP1 | CCTTGCATTTTCTGACACGCT | TCCCCATCCACGAAGAAACG |
| SREBP2 | GACGCCAAGATGCACAAGTC | ACCAGACTGCCTAGGTCGAT |
| ACLY | ATCGGTTCAAGTATGCTCGGG | GACCAAGTTTTCCACGACGTT |
| FASN | CAGGAGTTCTGGGACAACCTC | TTGGGGTGGACTCCGAAGA |
| HMGCR | TGATTGACCTTTCCAGAGCAAG | CTAAAATTGCCATTCCACGAGC |
| HMGCS1 | CATTAGACCGCTGCTATTCTGTC | TTCAGCAACATCCGAGCTAGA |
| HNF1A | TGGCGCAGCAGTTCACCCAT | TGAAACGGTTCCTCCGCCCC |
| INSIG2 | ACCCCTGCATTGACAGACAT | TCCACTTTAGCACTGGCATGA |
| SCAP | TATCTCGGGCCTTCTACAACC | GGGGCGAGTAATCCTTCACA |
| S1P | AAACACAAGGCAGTGGTGGA | CCTTCATACAGGCCATCGCT |
| Mouse | ||
| GAPDH | AACTTTGGCATTGTGGAAGG | GGATGCAGGGATGATGTTCT |
| LDLR | ACCTGCCGACCTGATGAATTC | GCAGTCATGTTCACGGTCACA |
| PCSK9 | TTGCAGCAGCTGGGAACTT | CCGACTGTGATGACCTCTGGA |
| SREBP2 | CCAAAGAAGGAGAGAGGCGG | CGCCAGACTTGTGCATCTTG |
| HMGCR | CTTTCAGAAACGAACTGTAGCTCAC | CTAGTGGAAGATGAATGGACATGAT |
| FASN | GGCATCATTGGGCACTCCTT | GCTGCAAGCACAGCCTCTCT |
| IDOL | AGGAGATCAACTCCACCTTCTG | ATCTGCAGACCGGACAGG |
| ABCA1 | AACAGTTTGTGGCCCTTTTG | AGTTCCAGGCTGGGGTACTT |
| ABCG1 | GAGGACCTTCCTCAGCATCA | AGGACCTTCTTGGCTTCGTT |
2.7 Small interference RNA (siRNA) transfection
A pool of four pre-designed siRNAs targeted to human CETP mRNA (Cat. No. L-009485-00) were obtained from Dharmacon (Lafayette, CO). The silencer negative control siRNA was obtained from Applied Biosystem. 4×10 cells were mixed with 50 nM siRNA using siPORT NeoFX siRNA transfection reagent (Ambion) and plated in 6-well plates. Next day, fresh medium was added to the transfected cells and then cells were treated with ANA for 24 h prior to isolation of total RNA.
2.8 Western blot analyses of LDLR, PCSK9 and SREBP2 in total lysates of hepatic cells and LDLR in mouse liver tissue
Approximately 50 mg of frozen liver tissue from individual hamster were homogenized in 0.3 ml RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, pH 7.4) containing 1 mM PMSF and protease inhibitor cocktail (Roche). Hepatic cells in culture dishes were washed by cold PBS and lysed by incubation in RIPA buffer for 5 min on ice, followed by a brief sonication using Bioruptor 300 (Diagenode Inc. Denville, NJ). After protein quantification using BCA™ protein assay reagent (PIERCE), 50 µg of homogenate proteins from individual liver samples or 30 µg protein of total cell lysates from HepG2 cells were separated on SDS-PAGE, transferred to nitrocellulose membranes, and blotted with specific antibodies. LDLR protein was detected with a rabbit anti-LDLR antibody (Biovision, Mountain View, CA). Intracellular PCSK9 in HepG2 cell lysates were detected by rabbit anti-human PCSK9 polyclonal antibody as previously described [41]. Membranes were reprobed with anti-β-actin antibody (Santa Cruz Biotechnology). Immunoreactive bands of predicted molecular mass were visualized using SuperSignal West Substrate (Thermo Scientific) and quantified with the Alpha View Software with normalization by signals of β-actin.
2.9 Western blots of SREBP2 in cytoplasmic extracts and nuclear extracts
Cytoplasmic and nuclear protein extracts were isolated from 2×106 HepG2 cells or 20 mg liver tissue using NE-PER nuclear and cytoplasmic extraction reagents from Thermo Scientific according to the vendor’s protocol. Aliquots of each sample (30 µg protein) were separated by denaturing SDS-PAGE (10%), transferred onto a nitrocellulose membrane and blotted with a rabbit anti-SREBP2 antibody [41]. Immunoreactive bands of predicted molecular mass were visualized using SuperSignal West Substrate (Thermo Scientific) and quantified with the Alpha View Software with normalization by signals of HDAC1 for the mature form of SREBP2 (SREBP2-M) [41] or by signals of GAPDH for SREBP2 precursor form (SREBP2-P). Different exposure times were used to detect SREBP2-P (30 to 40 sec) and SREBP2-M (up to 10 min).
In addition to using the NE-PER nuclear/cytoplasmic extraction reagents, we used the dounce-homogenization method to obtain nuclear extracts from individual liver samples as previously described [45]. Briefly, ~ 100 mg frozen hamster liver tissue were dounce-homogenized 15 times in buffer A (10 mM KCl, 1.5 mM MgCl2, 10 mM Hepes, pH 7.9, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Sigma)). After centrifugation at 2,000g for 10 min at 4°C, the pellet was resuspended in the same buffer and incubated on ice for 10 min, followed by dounce-homogenization of 10 times and centrifugation at 2,000g for 10 min at 4°C. The nuclei-containing pellet was resuspended in buffer B (420 mM NaCl, 10 mM KCl, 20 mM Hepes, pH 7.9, 20% glycerol, 1 mM DTT, 1 mM PMSF, protease inhibitor cocktail and phosphatase inhibitor cocktail) and extracted for 30 min at 4°C on a shaking rotor. After centrifugation at 16,000g for 15 min at 4°C, the supernatant was collected and stored in −80°C. The protein concentration was determined using BCA protein assay kit (Thermo Scientific).
2.10 LDL uptake assay
HepG2 cells in 6-well culture plates were treated with ANA for 24 h. The fluorescent DiI-LDL (Biomedical Technologies, Stoughton, Massachusetts) at a concentration of 2 µg/ml was added to the cells at the end of treatment for 4 h and cells were trypsinized. The mean red fluorescence of 1×104 cells was measured using FACScan (filter 610/20 DF, BD LSRII, Becton Dickinson).
2.11 Serum isolation and cholesterol determination
Mice were fasted for 4 h before blood collection at the beginning and the end of drug treatment. Serum was isolated at room temperature and stored at −80°C. Standard enzymatic methods were used to determine TC, LDL-C and HDL-C with commercially available kits purchased from Stanbio Laboratory (Texas, USA). Each sample was assayed in duplicate.
2.12 HPLC analysis of lipoprotein-cholesterol profiles
Fifty µl of serum sample from two serum samples of the same treatment group were pooled together and a total of 4 pooled samples from vehicle group and 4 pooled samples from ANA-treated group were analyzed for cholesterol levels of each of the major lipoprotein classes including chylomicron (CM), VLDL, LDL, and HDL with a dual detection HPLC system consisting of two tandem connected TSKgel Lipopropak XL columns (300 × 7.8-mm; Tosoh, Japan) at Skylight Biotech, Inc. (Tokyo, Japan).
2.13 Assay of secreted PCSK9
Secreted PCSK9 in culture medium of HepG2, primary mouse hepatocytes and in serum of C57BL/6J mice were measured using human or mouse PCSK9 ELISA kit obtained from R&D System according to the instruction.
2.14 Measurement of hepatic cholesterol
Fifty mg of frozen mouse liver tissue was thawed and homogenized in 1 ml chloroform/methanol (2:1). After homogenization, lipids were further extracted by rocking samples overnight at room temperature, followed by centrifugation at 5000g for 10 min. One ml lipid extract was dried under nitrogen stream and redissolved in 0.25 ml isopropanol containing 10% triton X-100. Total cholesterol mass was measured using a kit from Stanbio Laboratory.
2.15 Statistical analysis
Values are presented as mean ± SEM. Significant differences between control and treatment groups were assessed by One-way ANOVA with proper posttest or Student two-tailed t-test. Statistical significance is displayed as p < 0.05 (one asterisk), p < 0.01 (two asterisks) or p < 0.001 (three asterisks).
3. Results
3.1 SRE-1 dependent suppression of PCSK9 and LDLR promoter activities by ANA
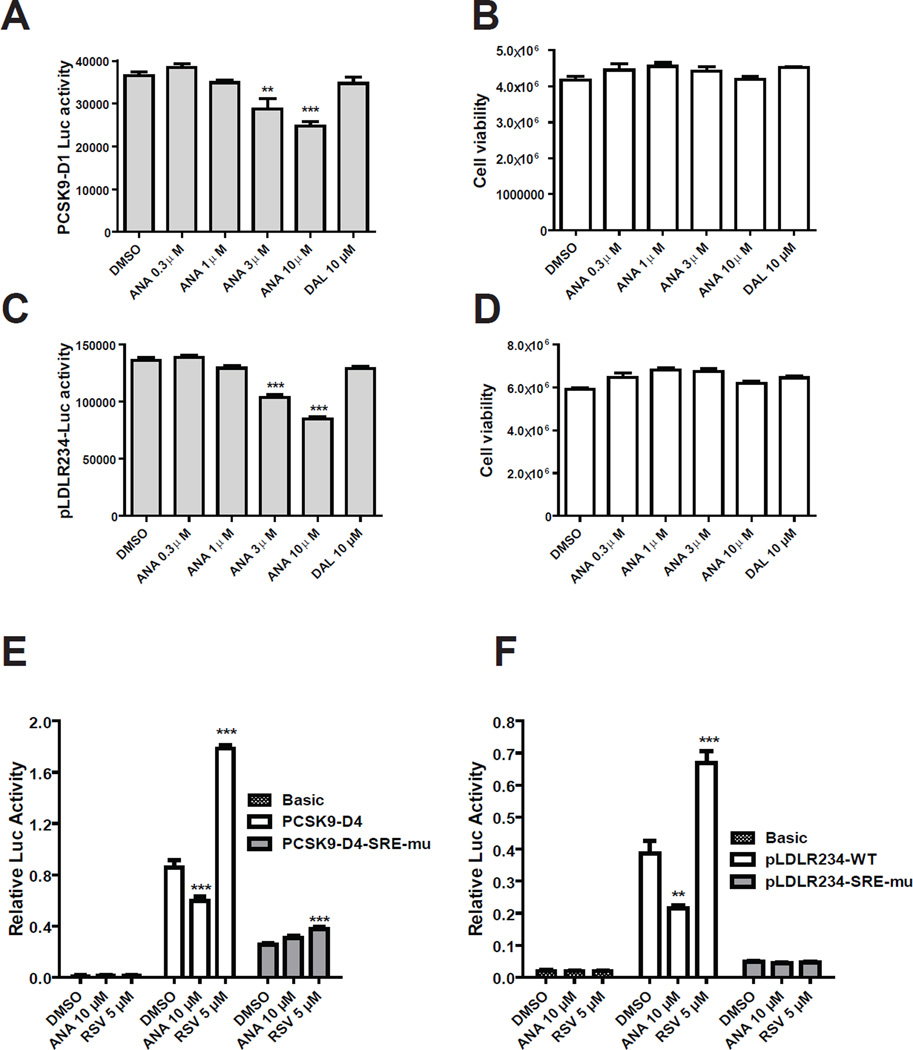
To determine whether CETP inhibitors affect PCSK9 and LDLR gene transcription, first we utilized two HepG2 derived cell lines CL26 and B11 that express a PCSK9 promoter luciferase reporter construct pGL3-PCSK9-D1 (CL26) [41] and LDLR promoter construct pGL3-LDLR234 (B11) [42]. These cells were treated with different doses of ANA or 10 µM of DAL for 24 h and luciferase activities were measured. A significant reduction of PCSK9 promoter activity by ANA was detected at 3 µM concentration (−22%, p<0.01) and further lowered to 68% of control at 10 µM (Fig. 1A). Likewise, luciferase activity of B11 cells were decreased by ANA at 3 µM concentration and reached to a maximal reduction of 38% of control at 10 µM (Fig. 1C). Up to a 10 µM concentration, DAL had no effect on LDLR or PCSK9 promoter activities. In parallel, we conducted cell viability assays and demonstrated that cell viability was not affected by ANA or DAL at these concentrations (Fig. 1B & D), indicating that decreased luciferase activities of CL26 and B11 upon ANA treatment were not caused by reduction of viable cell numbers.
Figure 1. Analysis of PCSK9 and LDLR promoter activities in stable and transiently transfected HepG2 cells without and with CETP inhibitors.
HepG2-CL26 cells (A, B) or HepG2-B11 (C, D) cells were incubated with ANA or DAL for 24 h. Luciferase activities were measured to determine PCSK9 promoter activity in CL26 cells and LDLR promoter activity in B11 cells. Cell viabilities in CL26 (B) and B11 cells (D) were determined as described in methods. HepG2 cells were transiently transfected with PCSK9 (E) or LDLR (F) wild-type and SRE-1 mutated reporters along with pRL-SV40 renilla control vector. One day post transfection, cells were treated for 24 h with 5 µM RSV or 10 µM ANA. Cells were harvested and dual luciferase activities were measured. Firefly luciferase activity was normalized with renilla luciferase activity. After normalization, the relative luciferase unit of each vector is expressed. Significant differences between control and treatments were assessed by One-way ANOVA with posttest of Dunnett’s Multiple Comparison Test. Data shown are representative of 2–4 separate experiments with similar results.
To determine whether ANA downregulates PCSK9 promoter and LDLR promoter via a common mechanism involving SREBP2, we transiently transfected HepG2 cells with the wild-type and SRE-1 mutated PCSK9 promoter constructs (Fig. 1E) and LDLR promoter constructs (Fig. 1F) along with the promoter-less vector pGL3-basic as a negative control. The plasmid pRL-SV40 was cotransfected to normalize variations in transfection efficiency. One day post transfection, cells were either treated with vehicle DMSO, ANA (10 µM), or HMGCR inhibitor RSV (5 µM). None of the drug treatments affected the activity of pGL3-basic. RSV and ANA produced opposite effects on LDLR and PCSK9 promoter activities. RSV increased the wild-type promoter activity of PCSK9 and LDLR whereas ANA reduced the promoter activities. Importantly, both RSV stimulation and ANA repression were not detected in SRE-1 mutated LDLR reporter. Likewise, ANA lost its effect on SRE-1 mutated PCSK9 promoter completely. Taken together, the results of promoter assays suggest that ANA inhibits SREBP-mediated transcription of LDLR and PCSK9 in HepG2 cells.
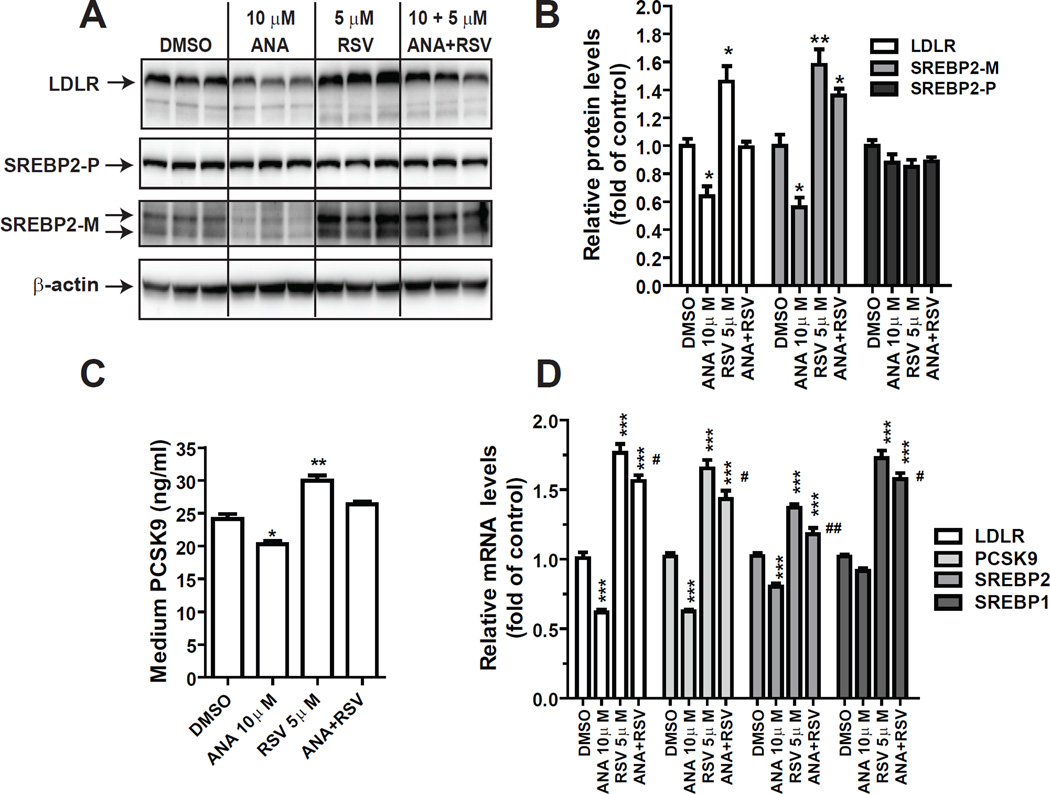
3.2 Reduction of mature SREBP2 cellular levels by ANA treatment
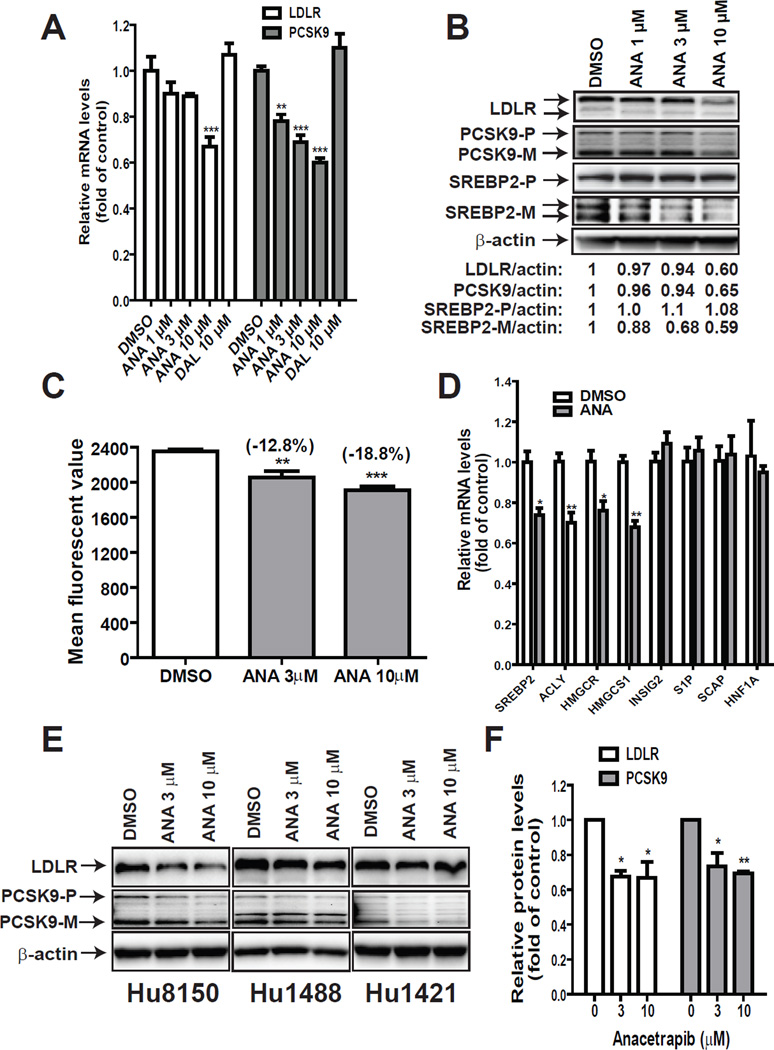
Next, the effect of ANA and DAL on PCSK9 and LDLR mRNA expression were examined in HepG2 cells by qRT-PCR (Fig. 2A). Consistent with the promoter analysis, ANA but not DAL exhibited a dose-dependent suppression on mRNA levels of PCSK9 with a 22% reduction by 1 µM (p<0.01). At 10 µM concentration, ANA lowered PCSK9 mRNA level to 60% of control and LDLR mRNA level to 67% of control. Western blot detections of LDLR and PCSK9 proteins in total cell lysates showed that in spite of the reduction of PCSK9, LDLR protein levels were still lowered by ANA treatment, particularly at 10 µM concentration (Fig. 2B).
Figure 2. Downregulation of the mature form of SREBP2 by ANA attenuated LDLR and PCSK9 expression in HepG2 and primary human hepatocytes.
(A, B) HepG2 cells were treated with vehicle DMSO, ANA or DAL for 24 h. Total RNA was isolated and mRNA levels of LDLR and PCSK9 were quantified by qPCR in A. In B, total cell lysates were isolated and protein levels of LDLR, PCSK9 and SREBP2 were examined by Western blotting. PCSK9-P refers to the PCSK9 proprotein and PCSK9-M refers to the processed mature protein. SREBP2-P refers to the precursor form and SREBP-M refers to the processed and active mature form of SREBP2.
(C) HepG2 cells cultured in 10% FBS medium on six-well culture plates were treated for 24 h with 3 mM or 10µM ANA. After treatment, fluorescent DiI-LDL at a concentration of 2 µg/ml was added to the medium and cells were trypsinized 4 h later. The uptake of DiI-LDL was measured by FACScan with 1×104 cells per sample. The data shown are mean ± SEM of four wells per treatment condition. Data shown are representative of 2 separate experiments with similar results.
(D) HepG2 cells were treated with 10 µM ANA for 24 h and total RNA was harvested for gene expression analysis. Significant differences between control and ANA treatment were assessed by two-tailed Student’s t-test.
(E, F) Primary human hepatocytes of three different donors (Hu8105, Hu1488 and Hu1421) were seeded in 12-well plates at a density of 1×106 cells/well in William E Medium supplemented with 10% FBS. After overnight seeding, cells were treated with 3 or 10 µM ANA for 24 h before cell lysis for Western blotting with anti-LDLR, anti-PCSK9, or anti-β-actin antibodies. The LDLR and PCSK9 bands were quantified with the Alpha View Software with normalization by signals of β-actin and were graphed relative to vehicle DMSO-treated cells.
In a pilot experiment to detect SREBP2 in total lysates using anti-SREBP2 antibody, we observed that the inactive precursor form of SREBP2 (SREBP2-P) was highly abundant in HepG2 cells and the amount of SREBP2-P did not change after ANA treatment. In contrast, the amount of active mature form of SREBP2 (SREBP2-M) was scarce and it was reduced in ANA-treated cells (Supplementary Figure 1). Therefore, using different exposure times, we separated detected SREBP2-M/P forms in HepG2 cells treated with different doses of ANA. We show that levels of SREBP2-P were comparable in different samples whereas the amount of SREBP2-M was decreased by ANA treatment dose-dependently (Fig. 2B). In these assays, DAL showed no effects at concentrations up to 10 µM (data not shown). To further determine whether the function of LDLR was affected by ANA treatment, LDL uptake assays with fluorescent DiI-LDL were conducted in HepG2 cells treated with ANA at 3 or 10 µM concentrations (Fig. 2C). The amount of fluorescent dye that accumulated inside the cells was reduced by approximately 12.8% (p<0.01) and 18.8% (p<0.01) when cells were treated with ANA at 3 and 10 µM doses. Altogether, these results confirm the negative effect of ANA on LDLR expression.
Since SREBP2-M is the active form for SREBP2 target genes, the reduction of SREBP2-M accounted for the coordinated repression of LDLR and PCSK9 gene expression. To confirm this, we analyzed a set of genes that are known targets of SREBPs or genes encoding for upstream regulators of SREBP processing. Fig. 2D shows that mRNA levels of SCAP, INSIG2 and S1P were unchanged by the drug treatment whereas mRNA levels of all four genes involved in cholesterol and fatty acids metabolisms including SREBP2 were downregulated in HepG2 cells treated with ANA. We did not observe changes in mRNA levels of HNF1α, the key transactivator for PCSK9.
Because HepG2 cells are derived from a human hepatoma, we wanted to know whether ANA treatment affects PCSK9/LDLR pathway in normal human liver cells. Thus, we treated different primary human hepatocytes derived from three individual donors with ANA at 3 and 10 µM concentrations. Total cell lysates were analyzed for LDLR and PCSK9 protein levels by Western blotting. Fig. 2E shows imaging results of individual blots and quantitative results after normalization with β-actin are presented in Fig. 2F. ANA at 3 µM lowered LDLR protein by 33% (p<0.05) and PCSK9 by 27% (p<0.05), respectively. The 10 µM dose produced similar effects as 3 µM, suggesting that ANA at 3 µM concentration reached a plateau in these primary liver cells. Importantly, the results of primary human hepatocytes were consistent with the observations made in HepG2 cells.
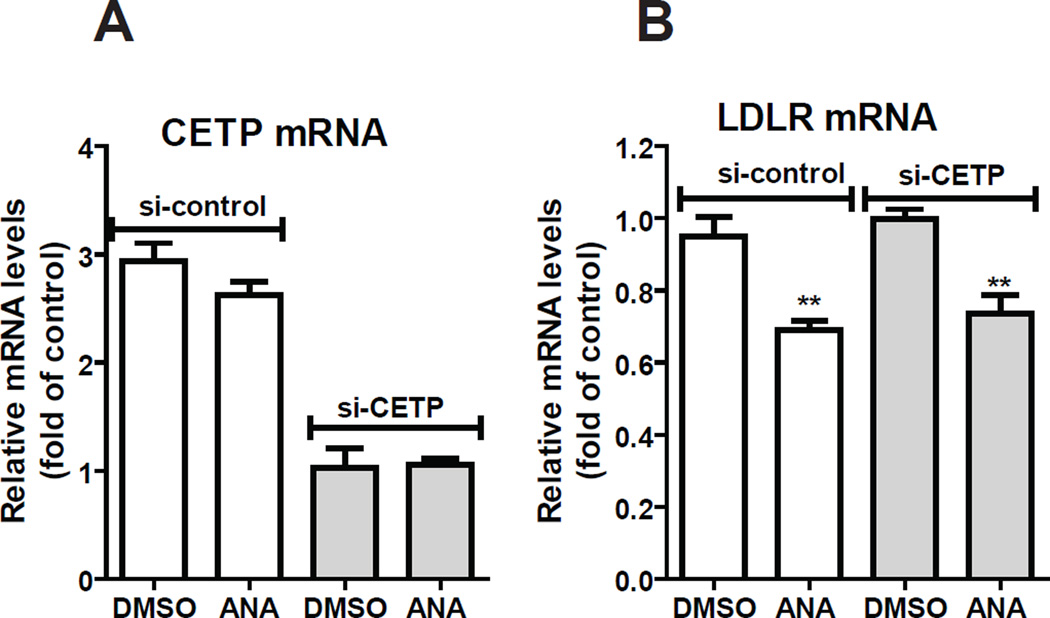
Next, we applied siRNA approach to address the question of whether the downregulation of LDLR/PCSK9 expression is related to the primary action of inhibition of CETP activity by this potent CETP inhibitor. HepG2 cells were transfected with 50 nM si-CETP or a control siRNA for two days before ANA treatment. Gene expression analysis by qRT-PCR showed that CETP mRNA levels were reduced by around 60–70% in cells transfected with si-CETP as compared to the control siRNA, and ANA treatment (10 µM) had no effect on CETP mRNA levels (Fig. 3A). In contrast, LDLR mRNA levels were reduced to similar degrees by ANA in si-CETP and sicontrol transfected cells (Fig. 3B), suggesting that the ANA-mediated suppression of SREBP activation is unrelated to its inhibitory activity to CETP.
Figure 3. Examination of the involvement of CETP in ANA-mediated suppression of LDLR mRNA expression.
HepG2 cells were transfected with si-CETP or a control siRNA for 48 h, followed by ANA treatment of 24 h. qPCR was conducted to determine CETP mRNA levels (A) or LDLR mRNA levels (B). The figures shown are representatives of 2 separate transfection experiments with similar results.
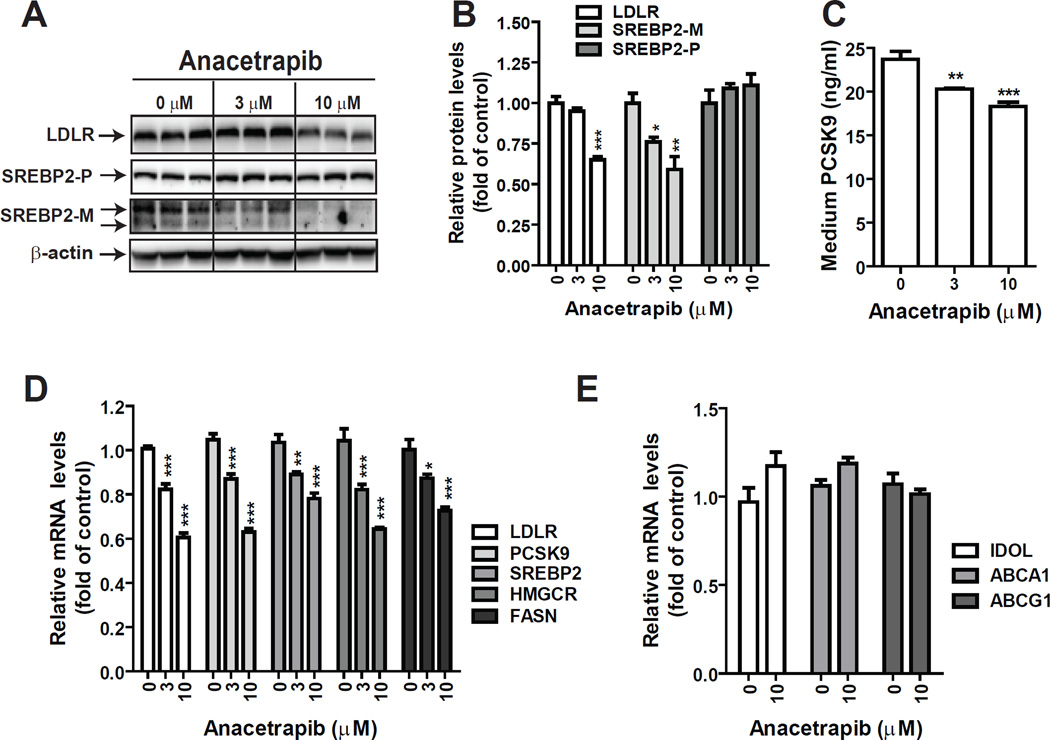
3.3 Reduction of SREBP2-M and LDLR protein levels in mouse primary hepatocytes by ANA
In order to absolutely rule out the involvement of CETP in ANA-mediated suppression of LDLR and PCSK9, we utilized primary hepatocytes isolated from a C57BL/6 mouse; a species naturally does not have CETP activity. MPH in triplicate wells were treated with different doses of ANA for 24 h before isolations of total RNA or cell lysates. The results shown in Fig. 4A are Western blots of individual lysate samples. Quantitative analyses of the results are presented in Fig. 4B. Western blotting with anti-SREBP2 showed that at 3 µM, ANA lowered the level of SREBP2-M by ~24% (p<0.05) and further reduced SREBP2-M by 41% at 10 µM as compared to control. LDLR protein levels were reduced approximately by 35% (p<0.001) at ANA 10 µM dose. We utilized a mouse PCSK9 ELISA kit to measure medium PCSK9 levels. As expected, the amount of PCSK9 secreted to cell culture medium was reduced in ANA-treated hepatocytes (Fig. 4C).
Figure 4. CETP-independent suppression of SREBP pathway by ANA in MPH.
MPH in triplicate wells were treated with ANA for 24 h. In A, total cell lysates were isolated and the protein abundance of LDLR or SREBP2 was examined by Western blotting and specific signals were quantified and presented in B. In C, concentrations of secreted PCSK9 in medium of different treatments were measured by a mouse PCSK9 ELISA kit. In D & E, total RNA was isolated and mRNA levels of SREBP-target genes (D) and LXR target genes (E) were measured and presented relative to control without ANA treatment. Data are mean ± SEM of triplicate RNA samples with duplicate measurements of each cDNA sample.
Gene expression analysis of qRT-PCR showed that starting at 3 µM ANA dose-dependently reduced LDLR, PCSK9, SREBP2, HMGCR and FASN (Fig. 4D), which was in line with its effects on lowering SREBP2-M protein levels. In contrast to SREBP pathway, ANA treatment did not affect mRNA levels of IDOL, ABCA1 and ABCG1 (Fig. 4E), whose transcriptions are commonly regulated by liver X receptor (LXR), thus indicating that the reduction of SREBP2-M abundance was not caused by activation of the LXR signaling pathway.
We have tried to detect SREBP1 protein levels in control and ANA-treated cells by using several commercial anti-SREBP1 antibodies. Unfortunately, these antibodies failed to detect specific signals of SREBP1 by Western blotting in both MPH and HepG2 cells due to the high background and appearance of multiple nonspecific bands within the molecular mass of 50 to 100 kDa. However, qRT-PCR results clearly demonstrated reduced mRNA levels of FASN, a SREBP1 target gene, in ANA treated MPH, which suggested that the active form of SREBP1 was also reduced by ANA.
3.4 Antagonism of ANA to statin in mouse primary hepatocytes
Statins reduce LDL-C in hypercholesterolemic individuals primarily by activation of liver LDLR transcription mediated by SREBP2. Through this process, PCSK9 levels were also induced by statin, particularly at higher statin doses [37,45]. So far our results indicated that CETP inhibitor ANA acts in opposite direction of statin to inhibit SREBP2 activation regardless of CETP expression status, which could lead to a potential antagonism to statin drug. To address this important question, we treated MPH with 10 µM ANA, 5 µM RSV and the combination. At protein levels, RSV treatment elevated SREBP2-M and LDLR protein levels by 58% and 46% as compared with control, which were in clear contrast to the effects of ANA that lowered SREBP2-M and LDLR protein abundances to 56% and 64% of control, respectively. In cells treated with the combination of RSV and ANA, LDLR protein expression was unchanged and SREBP2-M protein levels were higher than control but the induction was less prominent as compared with RSV alone (Fig. 5A & B). Analysis of medium PCSK9 levels showed a similar trend of regulation as LDLR protein (Fig. 5C). Gene expression analysis of qRT-PCR largely confirmed the results of protein analysis and showed the opposite effects of RSV and ANA (Fig. 5D). The RSV-induced increases in LDLR and PCSK9 mRNA levels were still detectable in the presence of ANA but were statistically lower than RSV alone. Altogether, these in vitro study results of HepG2 and MPH consistently demonstrated the inhibitory activity of CETP inhibitor ANA on SREBP2 mediated transcriptional activation of LDLR and PCSK9.
Figure 5. Antagonism to statin-induced activation of SREBP2 by ANA in MPH.
MPH in triplicate wells were treated with 5 µM RSV, 10 µM ANA or RSV+ANA for 24 h. In A, protein levels of LDLR and SREBP2 were examined by Western blotting and specific signals were quantified and presented in B. In C, concentrations of secreted PCSK9 in medium of different treatments were measured by ELISA. Significant differences between control and treatment were assessed by One-way ANOVA with posttest of Bonferroni’s multiple comparison for B & C *p < 0.05 and **p < 0.01 as compared to DMSO vehicle control. In D, total RNA was isolated and mRNA levels of SREBP-target genes were measured and presented relative to control without treatment. Data are mean ± SEM of triplicate RNA samples with duplicate measurements of each cDNA sample. Significant differences between control and treatment were assessed by One-way ANOVA with posttest of Tukey’s multiple comparison, *p < 0.05 and ***p < 0.001 as compared to DMSO control. #p < 0.05 and ##p < 0.01 as compared to RSV treatment alone.
3.5 Reduction of active form of SREBP2 and LDLR protein levels in liver tissue and elevation of serum cholesterol in dyslipidemic mice treated with ANA
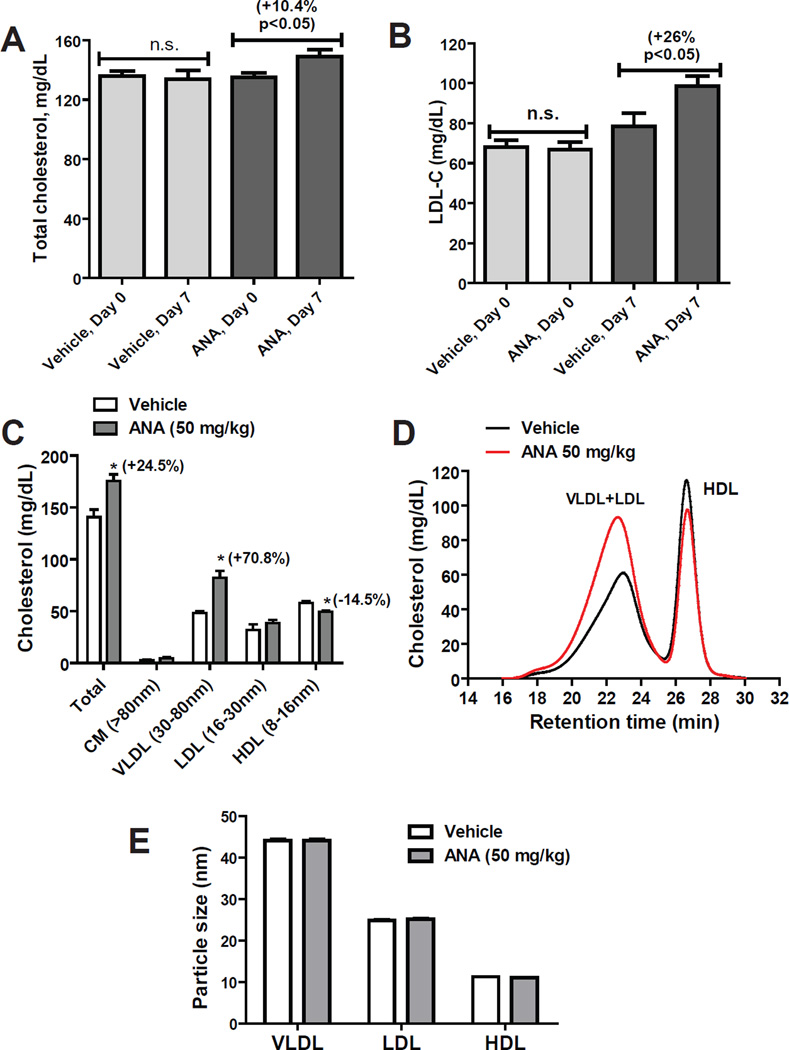
Animal models of hamsters [24–27] and CETP-transgenic mice [28] have been used to demonstrate the activity of CETP inhibitors in raising HDL-C and lowering LDL-C in vivo. Unfortunately, none of those studies have described a consequential impact of plasma CETP inhibition on hepatic LDLR and PCSK9 expression, no matter through indirect or direct actions of CETP inhibitors. To further investigate the potential effect of CETP inhibitors on LDL-C metabolism through their negative effects on SREBP pathway, we employed a dyslipidemic mouse model. Male C57BL/6J mice were fed a HFHC diet for two weeks that elevated serum total cholesterol (TC) by 35% (p<0.0001), HDL-C by 28% (p<0.0001) and LDL-C by 3.1-fold (p<0.0001) (Supplementary Fig. 2). While continued on HFHC diet, mice were randomly divided into two groups. One group was given ANA at a daily dose of 50 mg/kg by oral gavage and the control group received equal volume of vehicle. This dose in mice is extrapolated to a human equivalent dose (HED) of 4 mg/kg [46]. After 1 week of treatment, mice were fasted for 4 h and sacrificed for terminal serum and liver tissue collection. We first measured individual serum lipid levels using enzymatic methods with commercial TC, LDL-C and HDL-C kits. ANA treatment modestly elevated serum total serum cholesterol levels ~10% (p<0.05) (Fig. 6A) and increased serum LDL-C by 26% (p<0.05) as compared to vehicle control (Fig. 6B). No significant changes were detected by HDL-C measurement kit. Next, we performed HPLC analysis of lipoprotein-cholesterol profiles in vehicle and ANA-treated serum samples (Fig. 6C, D). The results showed a 24% (p<0.05) increase in total cholesterol and a prominent increase of 70.8% (p<0.05) in VLDL-associated cholesterol by ANA treatment while the increase in LDL-C (21%) in ANA group did not reach a statistical significance. In addition, the amount of cholesterol in HDL fraction was slightly reduced in ANA group with a statistical significance. Particle sizes of lipoproteins were similar between the two groups (Fig. 6E). Altogether, the analyses of serum lipid levels by two different measurements consistently showed an increase in serum cholesterol level and further confined that increase to VLDL/LDL fractions in ANA-treated dyslipidemic mice.
Figure 6. ANA treatment affected serum cholesterol levels of mice fed a HFHC diet.
(A, B) Male C57BL/6J mice fed a HFHC diet were orally dosed 50 mg/kg/day ANA (n=8) or equal volume of vehicle (0.5% methyl cellulose) as the control group (n=8) for 7 days. Serum TC and LDL-C were measured at day 0 before the drug treatment and day 7 after the last dosing by commercial kits. Data are mean ± SEM.
(C–E) Fifty µl of serum sample from two serum samples of the same treatment group were pooled together and a total of 4 pooled samples from vehicle group and 4 pooled samples from ANA-treated group were individually analyzed for cholesterol levels of each of the major lipoprotein classes including chylomicron (CM), VLDL, LDL, and HDL after HPLC separation. In C, cholesterol levels in individual lipoprotein fractions are presented as mean ± SEM. In D, the mean value of cholesterol in vehicle and ANA-treated samples are plotted. The calculated particle sizes are presented as mean ± SEM. Significant differences between control and ANA treatment were assessed by two-tailed Student’s t-test.
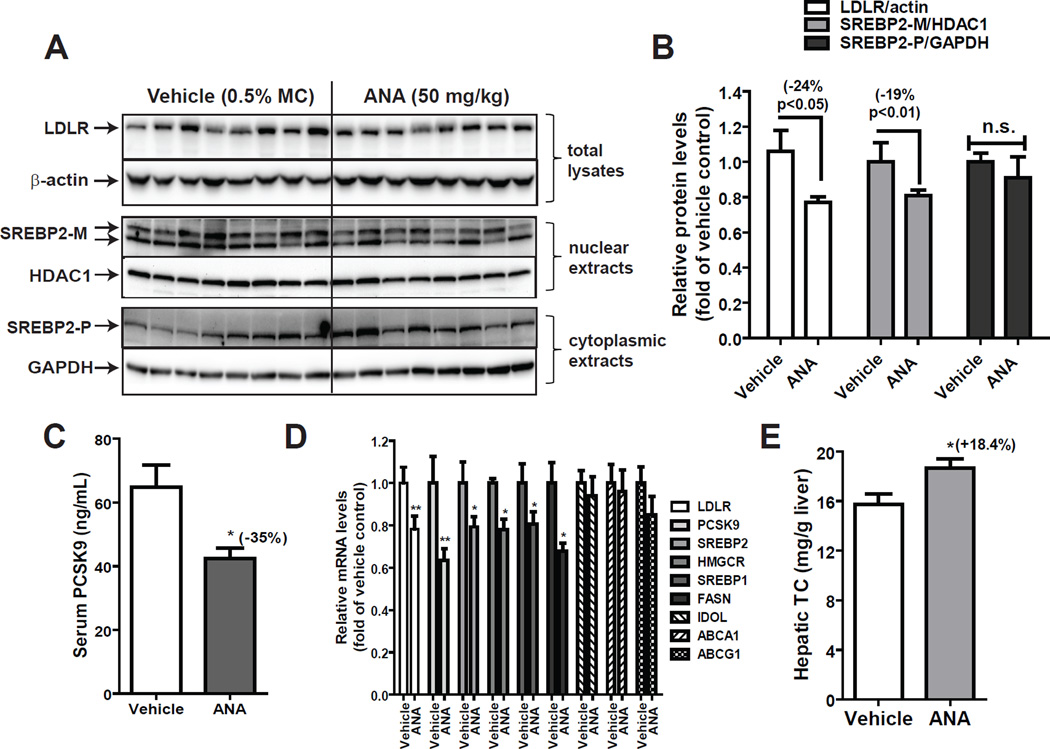
To seek a clear understanding of the effect of ANA on serum cholesterol metabolism in mice in the absence of its target protein CETP, protein levels of LDLR in liver homogenates, SREBP2-M in liver nuclear extracts, and SREBP2-P in cytoplasmic extracts were individually assessed by Western blotting. Fig. 7A shows results of Western blots of individual liver samples. Quantitative analyses of the results are presented in Fig. 7B. Amounts of SREBP2-M in nuclear extracts of liver samples were 19% lower (p<0.01) and LDLR protein levels in whole liver homogenates were 24% lower (p<0.05), respectively in ANA treated group as compared to vehicle group. Similar to HepG2 cells, no significant changes in protein levels of SREBP2-P were observed among liver tissues of control and ANA treated mice. We also determined serum PCSK9 levels in individual mice by ELISA, which indicated a 35% (p<0.05) reduction of serum PCSK9 by ANA treatment (Fig. 7C). qRT-PCR measurements of mRNA levels of LDLR, PCSK9 and other four SREBP target genes in all liver samples revealed that mRNA levels of SREBP2-target genes were reduced in the range of 20–40% in ANA group as compared to vehicle group (Fig. 7D), which were corroborative to the results of protein analyses. In contrast, mRNA levels of three LXR target genes IDOL, ABCA1 and ABCG1 were the same between two groups, which confirmed our in vitro studies in MPH and underscored the negative effect of CETP inhibitor on SREBP pathway.
Figure 7. ANA treatment reduced the mature form of SREBP2 and LDLR protein levels in liver tissue and decreased serum PCSK9 levels in dyslipidemic mice.
(A) Individual liver protein extracts were prepared and protein concentrations were determined. 50 µg of homogenate proteins of individual liver samples were resolved by SDS-PAGE and LDLR protein was detected by immunoblotting using anti-LDLR antibody. The membrane was reprobed with anti-β-actin antibody.
Nuclear fraction and cytoplasmic fraction of individual liver homogenate from the ANA and vehicle groups were analyzed for SREBP2-M and SREBP2-P protein levels by Western blotting. The membranes were reprobed with anti-HDAC1 antibody as a control of equal nuclear protein loading or GAPDH as a control of equal cytoplasmic protein loading.
(B) The protein abundance of LDLR was quantified with the Alpha View Software with normalization by signals of β-actin. Values are mean ± SEM of 7 samples per group. * p < 0.05 compared to the vehicle group. The protein abundance of SREBP2-M in nuclear extracts was quantified with normalization by signals of HDAC1. Values are mean ± SEM of 8 samples per group. ** p < 0.01 compared to the vehicle group. The protein abundance of SREBP2-P in cytoplasmic extracts was quantified with normalization by signals of GAPDH. Values are mean ± SEM of 8 samples per group. n.s., not statistically significant as compared to the vehicle group.
(C) Individual serum PCSK9 levels were quantified by ELISA. Values are mean ± SEM of 8 mice per group.
(D) qPCR analysis of liver mRNA levels of LDLR and PCSK9 along with 4 additional SREBP-target genes and 3 LXR regulated genes in ANA-treated and vehicle-treated control mice. Values are mean ± SEM of 8 mice per group
(E) Cholesterol levels in vehicle and ANA-treated liver samples were measured. Values are mean ± SEM of 8 mice per group.
In an attempt to identify the underlying cause of attenuated activation of SREBP2, we measured cholesterol levels of all liver samples and observed an 18.4% increase (p<0.05) by ANA treatment (Fig. 7E), which could be responsible for the reduction of SREBP2-M in ANA-treated liver.
Next, we examined the effect of ANA in normolipidemic mice. C57BL/6J mice fed a normal chow diet was given ANA at the same daily dose of 50 mg/kg for 10 days. The results are presented in Supplementary Figure 3A–F. ANA treatment increased serum LDL-C by 33% (p<0.05), which was consistent to the observation made in hypercholesterolemic mice. Hepatic gene expression analysis showed that LDLR mRNA levels were reduced by 20% (p<0.01) and PCSK9 mRNA levels were reduced by 35% (p<0.01) by ANA treatment. Serum PCSK9 levels were 24% (p<0.05) lower in ANA-treated group. Western blot analysis of individual liver homogenate revealed a 35% reduction of SREBP2-M (p<0.05) upon ANA treatment while LDLR protein levels did not significantly differ between the two groups. Importantly, similar to what we have observed in the HFHC diet study, the reduction of the mature form of SREBP2 was accompanied by a 43.3% (p<0.01) increase in hepatic cholesterol mass in ANA-treated liver samples as compared to control.
3.6 Downregulation of SREBP2-mediated LDLR and PCSK9 expression by EVA and TOR in HepG2 cells
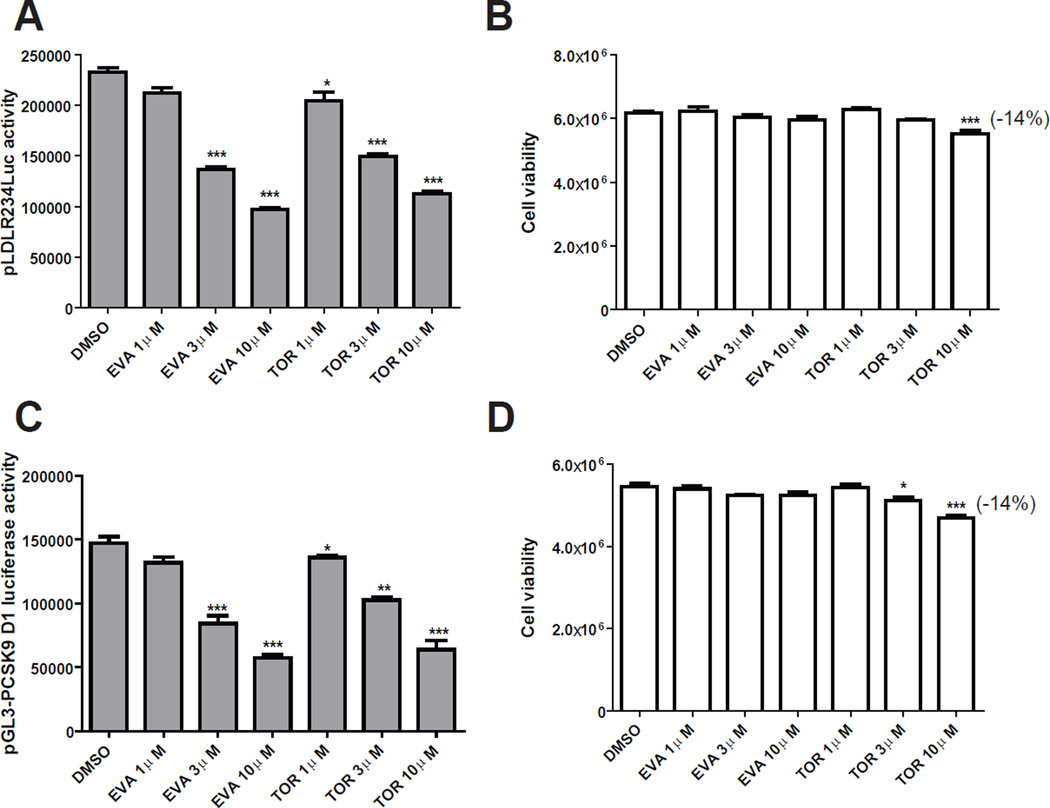
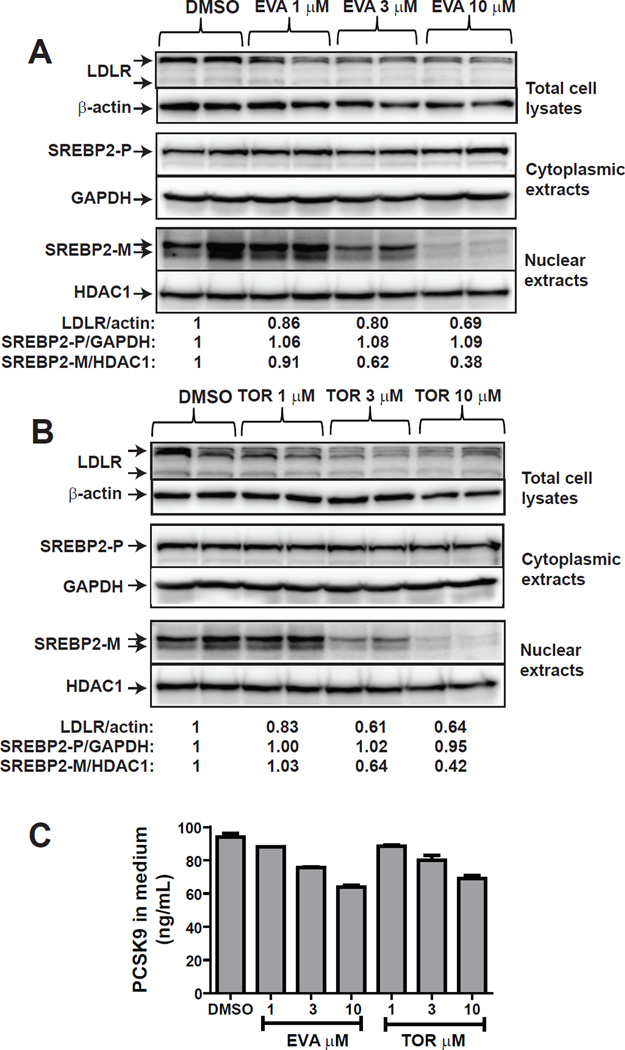
We wanted to know if the negative effect of ANA on SREBP pathway is shared by other strong CETP inhibitors. We tested dose-dependent effects of EVA and TOR on PCSK9 and LDLR promoter activities in CL26 and B11 cells treated with 1–10 µM of each of these inhibitors. Both inhibitors exerted a strong inhibitory effect on LDLR (Fig. 8A) and PCSK9 (Fig. 8C) promoter activities at 3 µM concentration with no effect (EVA) or small effect (TOR) (Fig. 8B & D) on cell viability. Western blot analysis of protein levels of LDLR in total cell lysates, SREBP2-P in cytoplasmic extracts and SREBP2-M in nuclear extracts (Fig. 9A & B) revealed dose-dependent reductions of LDLR and SREBP2-M by EVA and TOR. Likewise, serum PCSK9 levels were reduced by these strong CETP inhibitors in the same concentration range (Fig. 9C).
Figure 8. EVA and TOR inhibited LDLR and PCSK9 promoter activity.
HepG2-B11 cells (A, B) or HepG2-CL26 (C, D) cells were incubated with EVA or TOR at indicated concentrations for 24 h. Luciferase activities were measured to determine LDLR promoter activity in B11 cells and PCSK9 promoter activity in CL26 cells. Cell viability was determined as described in methods. Significant differences between control and treatments were assessed by One-way ANOVA with posttest of Dunnett’s Multiple Comparison Test.
Figure 9. Downregulation of SREBP2-M by EVA and TOR reduced LDLR and PCSK9 expression.
HepG2 cells were treated with EVA (A) or TOR (B) at indicated concentrations for 24 h. Total cell lysates (30 µg) were analyzed for the protein abundance of LDLR and β-actin. Nuclear and cytoplasmic extracts were isolated from each sample. Nuclear extracts of 30 µg protein per sample were used to detect SREBP2-M. The membrane was reprobed with anti-HDAC1. Cytoplasmic extracts of 30 µg protein per sample were used to detect SREBP2-P. The membrane was reprobed with anti-GAPDH.
The amount of secreted PCSK9 in culture medium was determined by a human PCSK9 ELISA kit (C).
4. Discussion
It is well documented that the expression level of hepatic LDLR and its function in removing LDL-C from circulation are dictated by two critical factors: intracellular cholesterol levels and serum PCSK9 concentrations. Sterols inhibit LDLR gene transcription by suppression of the processing and the release of the mature form of SREBP2 to prevent its entry into the nucleus to bind to the SRE-1 site of LDLR promoter as well as PCSK9 promoter. The serum PCSK9 reduces hepatic LDLR protein levels by promoting its lysosomal degradation. Thus, any modulators affect PCSK9 expression will ultimately change liver LDLR density and influence its capacity in removing LDL-C from the circulation.
We set out to understand the potential impact of CETP-inhibitor mediated suppression of PCSK9 expression on hepatic LDLR protein level and the consequential impact on serum LDL-C metabolism, following up an initial report that the CETP inhibitor K-312 repressed PCSK9 expression in HepG2 cells through a SRE-1 dependent mechanism [34,35]. We were interested in learning whether other CETP inhibitors share similar functions. This investigation led us to uncover a previously unrecognized effect of ANA as well as EVA and TOR that inhibited LDLR and PCSK9 transcription via reducing the active mature form of SREBP2 in hepatic cells. This shared property of CETP inhibitors is independent of their actions to inhibit CETP enzymatic activity and is effective within a concentration range of 1–10 µM when added to HepG2 cells. Our conclusion was built upon several different lines of investigations.
Our initial studies of the dose-dependent effect of ANA and DAL on promoter activity, mRNA level and protein abundance of PCSK9 and LDLR showed a coordinate suppression of PCSK9 and LDLR gene expression by ANA but not by DAL. Since ANA is a stronger CETP inhibitor than DAL [9, 47], at first, we considered the possible involvement of CETP activity in ANA mediated inhibition of LDLR/PCSK9 expression. By transfection of specific siRNAs to knockdown CETP mRNA expression in HepG2 cells, we observed that ANA produced a similar inhibitory effect on LDLR mRNA expression in si-CETP transfected cells and cells transfected with a nonspecific siRNA control oligonucleotides, thereby suggesting that CETP activity is not required for the observed downregulation of LDLR and PCSK9.
We also considered the nature of HepG2 cells that are derived from a human hepatoma which might not truthfully represent the response of normal liver cells exposed to CETP inhibitors. Thus, we tested primary human hepatocytes derived from three individual donors. The results of Western blotting showed a clear decrease in LDLR and PCSK9 protein level in ANA-treated hepatocytes at a concentration of 3 µM, which was not further reduced by increasing the drug concentration to 10 µM, implying that human normal liver cells could be more sensitive to this CETP inhibitor than HepG2 cells.
The inhibitory effect of ANA on PCSK9 and LDLR promoter activity was abolished by mutations of SRE-1 sequences on both promoters. This observation promoted us to examine the SREBP processing in control and drug treated cells. By analyzing total cell lysates as well as cytoplasmic and nuclear extractions, we found that ANA treatment did not notably reduce the amount of precursor form of SREBP2 but it dose-dependently lowered abundance of mature form of SREBP2. Because SREBP2 precursor form is highly abundant and the level of SREBP2-M is extremely low in HepH2 cells, it is highly possible that the method of Western blotting is not sensitive enough to detect the small changes in SREBP2-P density after ANA treatment. One piece of supporting evidence is that RSV treatment increased SREBP2-M in mouse primary hepatocytes without altering SREBP2 precursor levels (Fig. 5A). We could not detect changes in SREBP1 protein levels due to the lack of a specific anti-SREBP1 antibody. Therefore, we measured mRNA levels of several genes that are target of SREBP2 and SREBP1 and demonstrated that these mRNA expressions were all decreased after ANA treatment, which indicated the reduced transactiviting activities of both SREBP2 and SREBP1. It is worthy to note that ANA did not change mRNA levels of SCAP, S1P and INSIG2 whose gene products are upstream modulators of the processing pathway of SREBPs [48]. Furthermore, HNF1α mRNA levels were not altered by ANA treatment that provided additional evidence against the involvement of HNF1α in PCSK9 suppression by this CETP inhibitor.
Mice and MPH have not been previously described in functional studies of CETP inhibitors due to the natural absence of CETP in this species. However, in this study MPH provided us a clean system to investigate the CETP-independent effect of this class of compounds on LDLR/PCSK9 pathway. By treating MPH with ANA at different doses, we demonstrated the same inhibitory effect of this compound on reduction of SREBP2-M cellular levels and the consequential decrease in cellular protein levels of LDLR and PCSK9 as we observed in human liver cells. The reduction of PCSK9 was further confirmed by quantitative measurement of medium PCSK9 of cultured MPH. We noticed that in mouse system, inhibition of Ldlr gene transcription appeared to exert a more dominant role in determination of the LDLR protein levels than the reduction of serum PCSK9. This is evident by the observation that PCSK9 serum levels were reduced by ANA due to the reduced SREBP2-mediated transcription but the LDLR protein levels were still lower in ANA treated MPH than control.
Since we have observed a CETP-independent inhibitory effect of ANA on LDLR/PCSK9 pathway in human and mouse liver cells in vitro, it is important to know whether this aspect of ANA action alone could affect serum cholesterol metabolism. First, we treated dyslipidemic mice with ANA at 50 mg/kg, which is translated into 4 mg/kg of HED and equates to a 240 mg dose of ANA for a 60 kg person [46]. In a clinical study to examine the efficacy, safety and tolerability of ANA, up to a daily dose of 300 mg ANA was applied to patients with dyslipidemia [22]. Thus, treating mice with 50 mg/kg/day was within its clinical dosing range. By using conventional enzymatic reagents to measure serum lipid levels as well as HPLC method to comprehensively profile cholesterol levels in different lipoprotein fractions, we provided consistent results that ANA treatment significantly increased VLDL-C/LDL-C levels in dyslipidemic mice. Analysis of live tissues of the two groups by Western blotting confirmed the reductions of SREBP2-M in nuclear extracts of drug treated liver versus control liver. Despite the reduced serum PCSK9 levels, liver LDLR protein amounts were lower in ANA treated mice as compared to control mice fed the HFHC diet. These results suggest that in the absence of CETP inhibition, the negative effect of ANA on SREBP2 activation manifested and produced an unfavorable impact on serum cholesterol metabolism. The purpose of our second in vivo study was to examine the effect of ANA on LDLR/PCSK9 pathway in mice under a normal diet. The overall results of lipid analysis and hepatic gene expression analysis largely confirmed the findings made in hyperlipidemic mice. Importantly, by measuring hepatic cholesterol contents in all liver samples, we demonstrate that ANA treatment was associated with a significant increase in liver cholesterol levels in normolipidemic as well as hyperlipidemic mice. Since hepatic cholesterol is the most effective regulator of SREBP2 processing, the raise in cellular cholesterol might be the causing factor for the fall in SREBP2-M abundance by ANA treatment in absence of CETP. It was reported that two week treatment of dyslipidemic hamsters with ANA at a daily dose of 60 mg/kg lowered liver cholesterol mass [24], presumably through the accelerated hepatic excretion of cholesterol upon ANA treatment in the CETP-positive rodent model.
Our further in vitro studies in HepG2 cells showed that two other strong CETP inhibitors EVA and TOR behaved quite similarly to ANA with regards to lowering SREBP2-M/PCSK9/LDLR in a similar concentration range of 1–10 µM, which are substantially higher than their reported IC50 in CETP activity inhibition. It is unclear how this class of compounds reduces the mature form of SREBP2. It has been reported that certain oxysterol ligands of LXR inhibit the processing of SREBP and suppress SREBP-target genes while activating LXR target genes [49]. We considered a possibility that the inhibition of SREBP2 processing by CETP inhibitors was mediated through a similar mechanism of LXR agonists. However, examination of mRNA expressions of three authentic LXR regulated genes ABCA1, ABCG1 and IDOL in MPH and in mouse liver tissues did not reveal any changes by ANA treatment. Clearly, further investigations are required to fully understand this off-target action of CETP inhibitors.
5. Conclusion
We have demonstrated that treatments of liver cells with ANA as well as EVA and TOR reduce the mature form of SREBP2 which leads to an attenuated transcription of LDLR and its degrader PCSK9. This negative effect on LDLR/PCSK9 pathway manifested in mice by ANA in the absence of CETP inhibition and modestly impacted serum cholesterol metabolism.
Importantly, one must be cautious in interpreting our results and in relating these findings with human clinical studies of ANA or other CETP inhibitors. Clinical studies of ANA have demonstrated its strong efficacies in raising HDL-C and lowering LDL-C by itself and with a statin [22]. It is possible that the inhibition of CETP activity in humans naturally lowered LDL-C which is dominant over the modest inhibitory activity of this compound in reducing LDLR transcription. Furthermore, the concentration required to impact SREBP pathway in our cell culture studies are significantly higher than doses of these inhibitors in producing complete inhibitions of CETP activity in cell free system and in plasma. Nevertheless, considering the critical roles of LDLR and PCSK9 in control of plasma LDL-C levels and their relations to CVD risk, further in vivo studies in CETP-positive animal models to detect changes in serum PCSK9 and liver LDLR expression by CETP inhibitors will provide additional critical information for a better understanding of the impact of CETP inhibition on plasma LDL-C metabolism mediated through the LDLR/PCSK9 pathway, a major route for removing circulating atherogenic LDL-cholesterol.
Supplementary Material
Highlights.
CETP inhibitors reduce the mature form of SREBP2 leading to attenuated expressions of LDLR and PCSK9
Effects of CETP inhibitors on SREBP pathway are off-target actions
Anacetrapib treatment reduced serum PCSK9 and lowered liver LDLR in mice
CETP inhibitors may affect serum LDL-C metabolism by modulating LDLR/PCSK9 pathway
Acknowledgement
This study was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service) and by grants (1R01 AT002543-01A1 and 1R01AT006336-01A1) from National Center of Complementary and Alternative Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Statement of originality
All results presented in this manuscript are original and unpublished data. This manuscript has not been submitted to any other journals.
REFERENCES
- 1.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. J Am Coll Cardiol. 2004;44:720–732. doi: 10.1016/j.jacc.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Spady DK. Hepatic clearance of plasma low density lipoproteins. Semin Liver Dis. 1996;12:373–385. doi: 10.1055/s-2008-1040407. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 5.Grundy SM. Statin trials and goals of cholesterol-lowering therapy. Circulation. 1998;97:1436–1439. doi: 10.1161/01.cir.97.15.1436. [DOI] [PubMed] [Google Scholar]
- 6.Clark RW. Raising high-density lipoprotein with cholesteryl ester transfer protein inhibitors. Curr Opin Pharmacol. 2006;6:162–168. doi: 10.1016/j.coph.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Clark RW, Ruggeri RB, Cunningham D, Bamberger MJ. Description of the torcetrapib series of cholesteryl ester transfer protein inhibitors, including mechanism of action. J Lipid Res. 2006;47:537–552. doi: 10.1194/jlr.M500349-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Chapman MJ, Le GW, Guerin M, Kontush A. Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitors. Eur Heart J. 2010;31:149–164. doi: 10.1093/eurheartj/ehp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyares MA, Davis K. Patient considerations and clinical impact of cholesteryl ester transfer protein inhibitors in the management of dyslipidemia: focus on anacetrapib. Vasc Health Risk Manag. 2012;8:483–493. doi: 10.2147/VHRM.S29010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballantyne CM, Miller M, Niesor EJ, Burgess T, Kallend D, Stein EA. Effect of dalcetrapib plus pravastatin on lipoprotein metabolism and high-density lipoprotein composition and function in dyslipidemic patients: results of a phase IIb dose-ranging study. Am Heart J. 2012;163:515–521. doi: 10.1016/j.ahj.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 11.de Grooth GJ, Kuivenhoven JA, Stalenhoef AF, de GJ, Zwinderman AH, Posma JL, van TA, Kastelein JJ. Efficacy and safety of a novel cholesteryl ester transfer protein inhibitor, JTT-705, in humans: a randomized phase II dose-response study. Circulation. 2002;105:2159–2165. doi: 10.1161/01.cir.0000015857.31889.7b. [DOI] [PubMed] [Google Scholar]
- 12.Stroes ES, Kastelein JJ, Benardeau A, Kuhlmann O, Blum D, Campos LA, Clerc RG, Niesor EJ. Dalcetrapib: no off-target toxicity on blood pressure or on genes related to the renin-angiotensin-aldosterone system in rats. Br J Pharmacol. 2009;158:1763–1770. doi: 10.1111/j.1476-5381.2009.00460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein EA, Stroes ES, Steiner G, Buckley BM, Capponi AM, Burgess T, Niesor EJ, Kallend D, Kastelein JJ. Safety and tolerability of dalcetrapib. Am J Cardiol. 2009;104:82–91. doi: 10.1016/j.amjcard.2009.02.061. [DOI] [PubMed] [Google Scholar]
- 14.Gutstein DE, Krishna R, Johns D, Surks HK, Dansky HM, Shah S, Mitchel YB, Arena J, Wagner JA. Anacetrapib, a novel CETP inhibitor: pursuing a new approach to cardiovascular risk reduction. Clin Pharmacol Ther. 2012;91:109–122. doi: 10.1038/clpt.2011.271. [DOI] [PubMed] [Google Scholar]
- 15.Krishna R, Bergman AJ, Green M, Dockendorf MF, Wagner JA, Dykstra K. Model-based development of anacetrapib, a novel cholesteryl ester transfer protein inhibitor. AAPS J. 2011;13:179–190. doi: 10.1208/s12248-011-9254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishna R, Bergman AJ, Jin B, Fallon M, Cote J, Van HP, Laethem T, Gendrano IN, III, Van DK, Hilliard D, Laterza O, Snyder K, Chavez-Eng C, Lutz R, Chen J, Bloomfield DM, De SM, Van Bortel LM, Gutierrez M, Al-Huniti N, Dykstra K, Gottesdiener KM, Wagner JA. Multiple-dose pharmacodynamics and pharmacokinetics of anacetrapib, a potent cholesteryl ester transfer protein (CETP) inhibitor, in healthy subjects. Clin Pharmacol Ther. 2008;84:679–683. doi: 10.1038/clpt.2008.109. [DOI] [PubMed] [Google Scholar]
- 17.Krishna R, Anderson MS, Bergman AJ, Jin B, Fallon M, Cote J, Rosko K, Chavez-Eng C, Lutz R, Bloomfield DM, Gutierrez M, Doherty J, Bieberdorf F, Chodakewitz J, Gottesdiener KM, Wagner JA. Effect of the cholesteryl ester transfer protein inhibitor, anacetrapib, on lipoproteins in patients with dyslipidaemia and on 24-h ambulatory blood pressure in healthy individuals: two double-blind, randomised placebo-controlled phase I studies. Lancet. 2007;370:1907–1914. doi: 10.1016/S0140-6736(07)61813-3. [DOI] [PubMed] [Google Scholar]
- 18.Cao G, Beyer TP, Zhang Y, Schmidt RJ, Chen YQ, Cockerham SL, Zimmerman KM, Karathanasis SK, Cannady EA, Fields T, Mantlo NB. Evacetrapib is a novel, potent, and selective inhibitor of cholesteryl ester transfer protein that elevates HDL cholesterol without inducing aldosterone or increasing blood pressure. J Lipid Res. 2011;52:2169–2176. doi: 10.1194/jlr.M018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJ, Mundl H, Nicholls SJ, Shah PK, Tardif JC, Wright RS. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;29(367):2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz GG, Olsson AG, Ballantyne CM, Barter PJ, Holme IM, Kallend D, Leiter LA, Leitersdorf E, McMurray JJ, Shah PK, Tardif JC, Chaitman BR, Duttlinger-Maddux R, Mathieson J. Rationale and design of the dal-OUTCOMES trial: efficacy and safety of dalcetrapib in patients with recent acute coronary syndrome. Am Heart J. 2009;158:896–901. doi: 10.1016/j.ahj.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Bloomfield D, Carlson GL, Sapre A, Tribble D, McKenney JM, Littlejohn TW, III, Sisk CM, Mitchel Y, Pasternak RC. Efficacy and safety of the cholesteryl ester transfer protein inhibitor anacetrapib as monotherapy and coadministered with atorvastatin in dyslipidemic patients. Am Heart J. 2009;157:352–360. doi: 10.1016/j.ahj.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 23.Davidson M, Liu SX, Barter P, Brinton EA, Cannon CP, Gotto AM, Jr, Leary ET, Shah S, Stepanavage M, Mitchel Y, Dansky HM. Measurement of LDL-C after treatment with the CETP inhibitor anacetrapib. J Lipid Res. 2013;54:467–472. doi: 10.1194/jlr.M032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro-Perez J, Briand F, Gagen K, Wang SP, Chen Y, McLaren DG, Shah V, Vreeken RJ, Hankemeier T, Sulpice T, Roddy TP, Hubbard BK, Johns DG. Anacetrapib promotes reverse cholesterol transport and bulk cholesterol excretion in Syrian golden hamsters. J Lipid Res. 2011;52:1965–1973. doi: 10.1194/jlr.M016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SP, Daniels E, Chen Y, Castro-Perez J, Zhou H, Akinsanya KO, Previs SF, Roddy TP, Johns DG. In vivo effects of anacetrapib on prebeta HDL: improvement in HDL remodeling without effects on cholesterol absorption. J Lipid Res. 2013;54:2858–2865. doi: 10.1194/jlr.M041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briand F, Thieblemont Q, Muzotte E, Sulpice T. Upregulating reverse cholesterol transport with cholesteryl ester transfer protein inhibition requires combination with the LDL-lowering drug berberine in dyslipidemic hamsters. Arterioscler Thromb Vasc Biol. 2013;33:13–23. doi: 10.1161/ATVBAHA.112.252932. [DOI] [PubMed] [Google Scholar]
- 27.Niesor EJ, Magg C, Ogawa N, Okamoto H, von der ME, Matile H, Schmid G, Clerc RG, Chaput E, Blum-Kaelin D, Huber W, Thoma R, Pflieger P, Kakutani M, Takahashi D, Dernick G, Maugeais C. Modulating cholesteryl ester transfer protein activity maintains efficient pre-beta-HDL formation and increases reverse cholesterol transport. J Lipid Res. 2010;51:3443–3454. doi: 10.1194/jlr.M008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell TA, III, Graham MJ, Lee RG, Mullick AE, Fu W, Norris D, Crooke RM. Antisense oligonucleotide inhibition of cholesteryl ester transfer protein enhances RCT in hyperlipidemic, CETP transgenic, LDLr−/− mice. J Lipid Res. 2013;54:2647–2657. doi: 10.1194/jlr.M036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidah NG. PCSK9 as a therapeutic target of dyslipidemia. Expert Opin Ther Targets. 2009;13:19–28. doi: 10.1517/14728220802600715. [DOI] [PubMed] [Google Scholar]
- 30.Lambert G, Sjouke B, Choque B, Kastelein JJ, Hovingh GK. The PCSK9 decade. J Lipid Res. 2012;53:2515–2524. doi: 10.1194/jlr.R026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenney JM, Koren MJ, Kereiakes DJ, Hanotin C, Ferrand AC, Stein EA. Safety and efficacy of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 serine protease, SAR236553/REGN727, in patients with primary hypercholesterolemia receiving ongoing stable atorvastatin therapy. J Am Coll Cardiol. 2012;59:2344–2353. doi: 10.1016/j.jacc.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Graham MJ, Lemonidis KM, Whipple CP, Subramaniam A, Monia BP, Crooke ST, Crooke RM. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J Lipid Res. 2007;48:763–767. doi: 10.1194/jlr.C600025-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Frank-Kamenetsky M, Grefhouse A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, Gamba-Vitalo C, Hadwiger P, Jayaraman M, John M, Jayaprakash N, Marier M, Nechev L, Rajeev KG, Read T, Röhl I, Soutschek J, Tan P, Wong J, Wang G, Zimmermann T, Fougerolles AD, Vornlocher H-P, Langer R, Anderson DG, Manoharan M, Koteliansky V, Horton JD, Fitzgerald K. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata H, Murakami K, Murakami T, Miyosawa K, Iwashita M, Yamazaki H, Ohgiya T, Haruki Yamazaki K, Kimiyuki Shibuya K, Akimune Asanuma A, Tanabe S. A Novel and Potent CETP Inhibitor K-312 with PCSK9 Inhibitory Property, Exerts Strong Reduction of LDL-C and Anti-atherosclerotic Effects. Circulation. 2012;126:A11879–a11880. [Google Scholar]
- 35.Miyosawa K, Watanabe Y, Murakami TDesai D, Jiro Matsumoto J, Tanabe S, Aikawa M. A Novel CETP Inhibitor, K-312, Suppresses PCSK9 Expression Through the Modulation of Its Promoter Activity. Circulation. 2012;126:A13436–A13437. [Google Scholar]
- 36.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 38.Jeong HJ, Lee H-S, Kim K-S, Kim Y-K, Yoon D, Park SW. Sterol-dependent regulation of proprotein convertase subtilisin/kexin type 9 expression by sterol-regulatory element binding protein-2. J Lipid Res. 2008;49:399–409. doi: 10.1194/jlr.M700443-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Costet P, Cariou B, Lambert G, Lalanne F, Lardeux B, Jarnoux A-L, Grefhorst A, Staels B, Krempf M. Hepatic PCSK9 expression is regulated by nutritional status via insulin and sterol regulatory element-binding protein 1c. J Biol Chem. 2006;281:6211–6218. doi: 10.1074/jbc.M508582200. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Dong B, Park SW, Lee H-S, Chen W, Liu J. Hepatocyte nuclear factor 1a plays a critical role in PCSK9 gene transcription and regulation by the natural hypocholesterolemic compound berberine. J.Biol.Chem. 2009;284:28885–28895. doi: 10.1074/jbc.M109.052407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, Fenollar-Ferrer C, Cao A, Anselmi C, Carloni P, Liu J. Molecular dissection of human oncostatin M-mediated signal transductions through site-directed mutagenesis. Int J Mol Med. 2009;23:161–172. [PubMed] [Google Scholar]
- 43.Liu J, Streiff R, Zhang YL, Vestal RE, Spence MJ, Briggs MR. Novel mechanism of transcriptional activation of hepatic LDL receptor by oncostatin M. J Lipid Res. 1997;38:2035–2048. [PubMed] [Google Scholar]
- 44.Dong B, Kan CF, Singh AB, Liu J. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J Lipid Res. 2013;54:1241–1254. doi: 10.1194/jlr.M032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong B, Wu M, Li H, Kraemer FB, Adeli K, Seidah NG, Park SW, Liu J. Strong induction of PCSK9 gene expression through HNF1a and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J Lipid Res. 2010;51:1486–1495. doi: 10.1194/jlr.M003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reagan-Shaw S, Nihal M, Ahmad S. Dose translation from animal to human studies revisited. The FASEB Journal. 2007;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 47.Miyares MA. ANA and DAL: two novel cholesteryl ester transfer protein inhibitors. Ann Pharmacother. 2011;45:84–94. doi: 10.1345/aph.1P446. [DOI] [PubMed] [Google Scholar]
- 48.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, Covey DF, Mangelsdorf DJ, Cohen JC, Hobbs HH. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.