Abstract
Atomistic descriptions of the μ-opioid receptor (μOR) noncovalently binding with two of its prototypical morphinan agonists, morphine (MOP) and hydromorphone (HMP), are investigated using molecular dynamics (MD) simulations. Subtle differences between the binding modes and hydration properties of MOP and HMP emerge from the calculations. Alchemical free energy perturbation calculations show qualitative agreement with in vitro experiments performed in this work: indeed, the binding free energy difference between MOP and HMP computed by forward and backward alchemical transformation is 1.2±1.1 and 0.8±0.8 kcal/mol, respectively, to be compared with 0.4±0.3 kcal/mol from experiment. Comparison with an MD simulation of μOR covalently bound with the antagonist β-funaltrexamine hints to agonist-induced conformational changes associated with an early event of the receptor’s activation: a shift of the transmembrane helix 6 relative to the transmembrane helix 3 and a consequent loss of the key R165-T279 interhelical hydrogen bond. This finding is consistent with a previous proposal suggesting that the R165-T279 hydrogen bond between these two helices indicates an inactive receptor conformation.
Introduction
Opioid drugs, such as morphine, are widely used in clinics for the treatment of acute, postoperative, and chronic pain. Owing to their exceptional analgesic properties, they are consistently among the most commonly prescribed drugs nowadays [1]. However, although frequently highly effective, opioids consumption in a regular basis leads to the appearance of undesirable side effects, such as constipation or respiratory depression, which limit their clinical applicability. Moreover, their usage often leads to addiction, tolerance and withdrawal [2]. This poses a major problem for the use of the existing opioids in clinics, complicating dosing regimens for patients and strongly restricting the prescription of these drugs [3, 4].
Opioid compounds exert their analgesic and intense rewarding effects by acting upon opioid receptors expressed on the plasma membrane of neuronal cells. These receptors, as members of the class A seven transmembrane (TM) G-protein coupled receptors (GPCRs) superfamily, are specialized in transmitting stimuli from the extracellular environment to the cytoplasm [5]. According to their preferential interaction with endogenous opioid peptides, opioid receptors are divided in four different types: the nociceptin/orphanin FQ peptide receptor, and the classical μ-, δ- and κ-opioid receptors [6–8]. In spite of their relatively high sequence identity [9], they present a very specific functional outcome, each receptor being responsible for distinct pharmacological effects. In particular, μ-opioid receptors (µORs) in the peripheral and central nervous system mediate pain perception. They are the primary target of exogenous analgesics such as morphine and other prototypical opioid drugs [10]. Both beneficial and adverse pharmacodynamic effects of classical morphine-like drugs are attributable to the activation of μORs [11]. They may be separable by using biased ligands [12]. Indeed, it has been demonstrated in a large number of studies that different ligands, while acting on the same receptor, can stimulate and inhibit GPCRs signaling through several intracellular pathways differentially, not simply stimulating or inhibiting all pathways to the same extent. This phenomenon, termed functional selectivity or biased signaling [13, 14], is a key concept in GPCRs signaling, in general, and through opioid receptors, in particular. These receptors can signal selectively through G-protein and β-arrestin pathways in a ligand-specific manner [12, 15, 16].
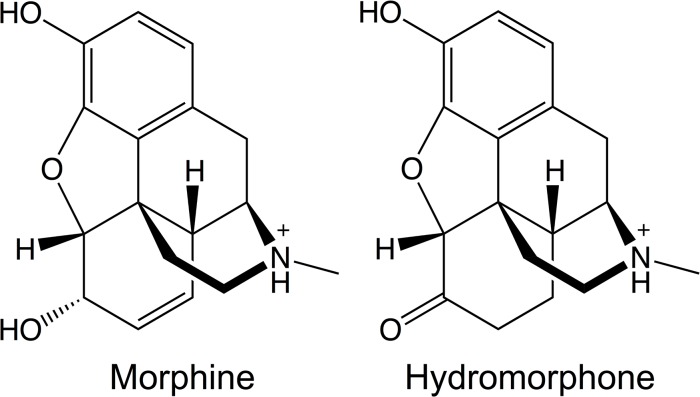
Characterizing opioid-μOR interactions may help to understand how different compounds can trigger distinct downstream responses and cause the selective activation of particular signaling and regulatory pathways. This knowledge can be in turn exploited to develop novel potent analgesics lacking some of the undesirable properties of current opioids by activating specific μOR signaling pathways. Within this perspective, here we investigate the binding determinants and energetics of two prototypical opioid agonists, morphine (MOP) and hydromorphone (HMP), to μOR. The two agonists’ functional activities at the receptor differ. The EC50 values for cAMP (G-protein mediated responses) and for β-arrestin2 (Table 1) point to 3~4-fold increase in the potency of HMP relative to MOP [17]. By contrast, the agonists’ structural features (Fig 1) and binding affinities for μOR are very similar (Table 1). In this paper, we report structural features of the two agonists’ binding modes, along with the agonist-induced conformational changes in μOR. These changes might be present in the early steps toward an active state of the receptor.
Table 1. EC50 (nM) cAMP and β-arrestin2 values for MOP and HMP on human μOR [17], as well as the binding affinities of the two agonists for μOR.
| K i (nM) | EC50 cAMP | EC50 β-arrestin2 | |
|---|---|---|---|
| MOP | 8.0±3.6 | 50 | 501 |
| HMP | 4.0±0.8 | 16 | 126 |
The binding affinities are measured in this study (see “Materials and Methods” for details).
Fig 1. Molecular structures of morphine and hydromorphone.
Our calculations are based on the X-ray structure of mouse μOR covalently bound with β-funaltrexamine (β-FNA), a semisynthetic opioid antagonist derived from morphine, resolved at 2.8 Å resolution (PDB entry: 4DKL) [18]. Mouse μOR shares 94% sequence identity with human μOR. Therefore, the 3D structure of mouse μOR provides an excellent model for investigating μOR-ligand interactions. We find that MOP and HMP bind to μOR with subtle differences that may account for their different binding affinities, which are consistent with in vitro experiments performed here.
Materials and Methods
Computational details
Structural models of the agonists in complex with μOR
We started from the X-ray structure of mouse μOR [18]. The structure contains a T4 lysozyme replacing the third intracellular loop (IL3), which is part of the putative G-protein binding epitope on class A GPCRs [19]. We constructed the missing IL3 using homology modeling by Modeller 9.9 [20]. In total 20,000 μOR models were generated, from which the one with the lowest Discrete Optimized Protein Energy (DOPE) score [20] was selected. The water molecules in the X-ray structure were preserved during the homology-modeling step. The NQ-Flipper [21] and H++ [22] webservers were used to examine the asparagine and glutamine side-chain rotamers and to predict the histidine side-chain protonation states, respectively. MOP and HMP were built and docked into the orthosteric binding site of μOR using MOE 2012.10 [23]. The protonation state of the side-chain residues forming μOR was estimated by the Protonate3D algorithm [24]. The tertiary amines of the ligands were protonated as they should be at physiological pH (Fig 1) and rotatable bonds were allowed to rotate. Docking was realized using an induced fit procedure that allows taking into account the flexibility of the protein side-chains located in the binding pocket. The electrostatic solvation energy was used to score the resulting binding modes using a generalized born/volume integral method [25]. From the top-ranked 25 modes of each ligand, we selected the mode in best agreement with available experimental data (see S1 File for details).
Embedding in membrane and solvation.
The DOWSER program [26] was used to detect and prefill possible hydrophilic cavities inside the MOP-μOR, HMP-μOR and β-FNA-μOR complexes. The complexes were oriented according to the OPM (Orientations of Proteins in Membranes) database [27] and then embedded in a bilayer of 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), the most abundant phospholipid in animal cell membranes [28]. We used the ‘Stockholm Lipids’ parameters that have been shown to be compatible with AMBER force fields [29], and the POPC bilayer configuration pre-equilibrated in water at 303 K [30]. The embedding was performed using the Inflategro2 program [31]. Each system was then immersed in a periodic 64×67×113 Å3 box, which ensured a distance always larger than 20 Å between adjacent images of the protein during the simulation. The systems were solvated in explicit water and neutralized with 0.15 M NaCl, resulting in ca. 8,430 water molecules, 23 Na+ and 37 Cl- ions. Each system consisted of ca. 42,700 atoms in total.
Molecular Dynamics simulations
Energy minimizations and MD simulations were carried out using Gromacs 4.6 [32]. The TIP3P water model [33], the Åqvist parameters [34], and the Amber99SB_ILDN force field [35] were used for the water molecules, the ions, and the protein, respectively. As for the parameters of the ligands, we calculated their partial atomic charges at the DFT level with the HF/6-31G* basis set using Gaussian 09 [36]. The obtained electrostatic potential was fitted by the RESP program [37] in Amber11 to generate atomic point charges (provided in S2 File). The other parameters were taken from the General Amber force field (GAFF) [38] to build the topology of the ligands (S2 File). The ligand-μOR complexes underwent 1,000 steps of steepest-descent energy minimization with 5,000 kcal·mol-1·Å-2 harmonic positional restraints on the protein-ligand complex, followed by 2,500 steps of steepest-descent and 2,500 steps of conjugate-gradient minimization without restraints. Then, the systems were gradually heated to 300 K in 6 steps of 100-ps MD simulations (from 2 K to 60 K, 120 K, 180K, 240 K and 300 K). The velocities were generated consistent with a Maxwell-Boltzmann distribution at the corresponding temperature. Each system underwent 5 ns equilibration and 0.8-μs production MD simulations in the NPT ensemble (P = 1 bar, T = 300 K) by using the Andersen-Parrinello-Rahman barostat [39, 40] and the Nose-Hoover thermostat [41]. Semi-isotropic pressure coupling was applied to allow the simulation box in the z-axis (perpendicular to the lipid bilayer) to vary independently of the x-y plane. The LINCS algorithm [42] was employed to constrain all the bond lengths. Van der Waals and short-range electrostatic interactions were cut off at 12 Å. Long-range electrostatic interactions were computed using the Particle Mesh Ewald summation (PME) [43] method with a Fourier grid spacing of 1.2 Å. A 2-fs time step was used in the MD calculations. Using the same procedure, 0.5-μs additional MD simulations for each system were obtained starting from different initial velocities.
Free energy calculations
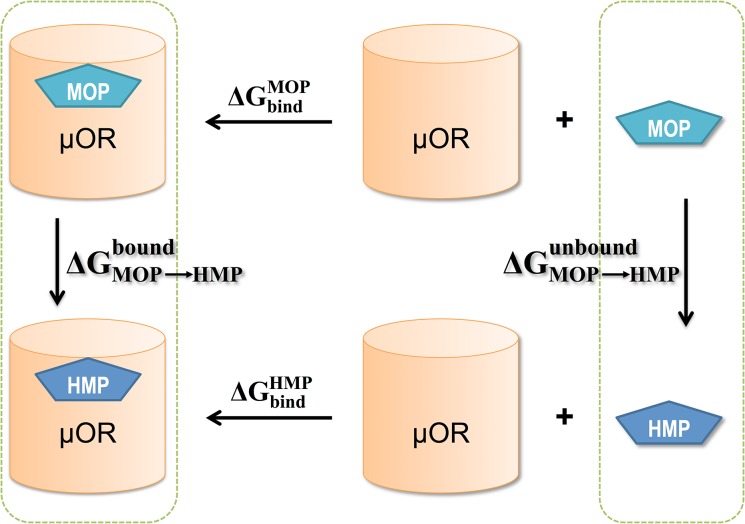
Alchemical free energy perturbation (FEP) methodologies were employed to estimate the difference in the binding free energy to μOR, ∆∆G bind, between MOP and HMP. Using the thermodynamic cycle in Fig 2, ∆∆G bind from MOP to HMP can be calculated as the free energy difference between two alchemical pathways: the transformation of MOP to HMP while bound to μOR, and that of MOP to HMP in solution (unbound). We performed alchemical transformation from MOP to HMP and vice versa.
Fig 2. The thermodynamic cycle for computing the free energy difference between MOP and HMP upon binding to μOR:
The unbound state requires transformation of the ligands alone in solution, since the receptor is the same in both cases.
The transformations in solution (the unbound state) started from the agonists pre-equilibrated in a periodic 50×50×50 Å3 box containing ca. 4,120 water molecules and a Cl- counterion. The force field parameters and the simulation procedure were the same as those used above. For each agonist, 5 ns pre-equilibration MD was carried out. The transformations of the agonists bound to μOR (the bound state) were started from representative configurations obtained from a cluster analysis of the agonist-μOR trajectories using g_cluster in Gromacs tools [44] and the Gromos method [45].
The transformations from one agonist (A) to the other (B) were performed in the following steps: (i) charge annihilation by scaling the electrostatic potential energy of A, in 30 steps (∆λ = 0.025 for the λ values from 0 to 0.25 and from 0.8 to 1, and ∆λ = 0.05 for the λ values from 0.25 to 0.8); (ii) decoupling the van der Waals interactions and transforming A into B in 25 steps (∆λ = 0.01 for the λ values from 0 to 0.05, and ∆λ = 0.05 for the λ values from 0.05 to 1); and (iii) charge generation for B by scaling its electrostatic potential energy in 30 steps (using the same λ values as those in (i)). The simulations in the unbound state at each λ value included 5,000 steps of steepest descent energy minimization and 5,000 steps L-BFGS energy minimization, 200 ps NVT (T = 300 K) equilibration, 1 ns NPT (P = 1 bar, T = 300 K) equilibration and 10 ns NPT (P = 1 bar, T = 300 K) production run with dH/dλ collection every 10 steps. The temperature was controlled via Langevin dynamics and the pressure, Andersen-Parrinello-Rahman barostat [39, 40]. The simulations in the bound state were performed in the same manner except that the timescales in this case were: 500 ps NVT equilibration, 5 ns NPT equilibration and 10 ns NPT production run.
The free energies of the steps involved in the transformations were calculated using the Bennett’s acceptance ratio method (BAR) [46] implemented in Gromacs 4.6. BAR combines the information normally used for forward and reverse free energy perturbations. The free energy difference of a transformation is a sum over those of n successive λ values, , in which δG i is computed iteratively according to
where U is the potential energy as a function of the system’s configuration, x.
Experimental details
Human μOR binding assay. The human μOR binding assay was performed as homogeneous SPA-assay (scintillation proximity assay) using the assay buffer 50 mM TRIS-HCl (pH 7.4) supplemented with 0.052 mg/ml bovine serum albumin (Sigma-Aldrich Co., St. Louis, MO). The final assay volume (250 μl/well) included 1 nM of [N-allyl-2,3-3H]naloxone as ligand (PerkinElmer Life Sciences, Inc. Boston, MA, USA), and either test compound in dilution series or 25 μM unlabeled naloxone for determination of unspecific binding. The test compound was diluted with 25% DMSO in H2O to yield a final 0.5% DMSO concentration, which also served as a respective vehicle control. The assay was started by adding wheat germ agglutinin coated SPA beads (GE Healthcare UK Ltd., Buckinghamshire, UK), which had been preloaded with human μOR membranes (PerkinElmer Life Sciences, Inc. Boston, MA, USA). After incubation for 90 minutes at RT and centrifugation for 20 minutes at 500 rpm the signal rate was measured by means of a 1450 Microbeta Trilux β-counter (PerkinElmer Life Sciences/Wallac, Turku, Finland). Half-maximal inhibitory concentration (IC50) values reflecting 50% displacement of [3H]naloxone-specific receptor binding were calculated by nonlinear regression analysis and Ki values were calculated by using the Cheng-Prusoff equation [47].
Results and Discussion
The initial binding modes of MOP and HMP obtained from docking are very similar (see S1 File for details). During 0.8 μs MD simulations, the transmembrane domain of μOR remain relatively rigid (Cα RMSF ranging from 0.4 Å to 1.5 Å) while, as expected, the loop regions of μOR display larger fluctuations (Cα RMSF ranging from 1.0 to 7.1 Å) (S1 Fig). The largest fluctuations occur in the modeled IL3, the region where a T4-lysozyme is located in the X-ray structure of the receptor, for the case of the HMP-μOR complex (S1 Fig). The Cα RMSD of μOR transmembrane domain are 1.7±0.3 Å, 1.3±0.2 Å and 1.2±0.1 Å in the MOP-μOR, HMP-μOR and β-FNA-μOR complexes, respectively, well below the resolution of the X-ray structure (S2 Fig). MOP and HMP mostly maintain their initial docking mode, while β-FNA well preserves its binding mode in the X-ray structure, the salt-bridge with D147 being preserved during the timescale of all the MD simulations. The RMSD of the ligand non-hydrogen atoms are 3.3±0.4 Å, 2.0±0.4 Å and 1.6±0.2 Å for MOP, HMP and β-FNA, respectively (S2 Fig).
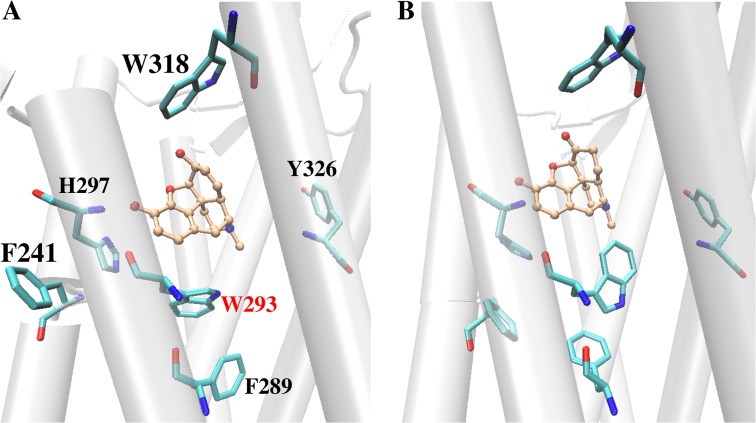
The two agonists (MOP and HMP) accommodate into the active site cavity in a slightly different manner, forming different H-bond patterns with residue H297 in TM6. In particular, MOP establishes a direct H-bond with H297 side chain, whereas HMP maintains water-mediated H-bonds with H297 side chain and K233 backbone mostly via one bridging water molecule (Fig 3 and S3 Fig). The presence of a cyclohexanone moiety within the HMP skeleton as opposed to a cyclohexenol ring, which is its cyclic counterpart in MOP, determines this different interaction pattern as it alters the drug surface available for hydrophobic interactions with protein residues in this region and slightly modifies the conformation of the polycyclic structure. MOP fits into the active site cavity establishing additional van der Waals contacts with G325, I296 and M151 while disrupting the initial interaction between D147 and the Y326 hydroxyl group. This disruption occurs at ~220 ns when Y326 moves away from D147 and eventually forms a very stable H-bond with T120. On the contrary, HMP binding causes the flip of the W293 side chain during the first 225 ns and gives rise to a stable interaction between the W293 and Y326 side chains (Fig 4). As a consequence, Y326 remains in close proximity to the ligand and preserves a H-bond with D147 (Fig 4B). The flip of the W293 side chain also leads to a rearrangement of the nearby aromatic residues, which establish different van der Waals contacts than those in MOP-μOR (Fig 4). The W293 reorientation separates HMP from G325 and places the ligand closer to I322, resulting in slightly different HMP-μOR hydrophobic contacts from those between MOP and the receptor (see Table 2). In addition, the flip of the W293 side chain allows more water molecules to enter the binding cavity below HMP (i.e. opposite to the extracellular side). After W293 flips, 15±2 water molecules are present inside the cavity of HMP-µOR, to be compared with 10±2 in MOP-μOR (S3 File). The different degrees of hydration are statistically significant, as indicated by normalized histograms and Welch’s t-test on the data (S3 File). Additional 0.5-μs MD simulations of MOP-μOR and HMP-μOR, starting from different initial velocities, reproduce the above-observed differences of the H-bond pattern, the W293 side chain orientation, the van der Waals contacts and the hydration in the binding cavity (S3 File).
Fig 3. Representative structure of the MD simulations for (A) the MOP-μOR and (B) the HMP-μOR complexes obtained from clustering analysis.
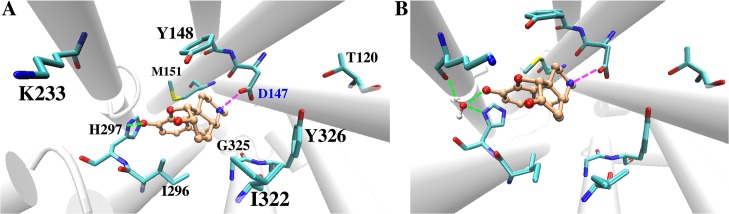
The ligand carbon atoms are in orange. H-bonds and salt-bridges are shown in green and magenta dashed lines, respectively. For clarity hydrogen atoms of the ligands and the μOR residues are not shown. H297 is monoprotonated at the Nε atom.
Fig 4. Arrangements of aromatic residues at the μOR orthosteric binding site upon binding with (A) MOP and with (B) HMP.
Table 2. Minimum distances between both morphinan drugs (MOP and HMP) and selected protein residues.
| I296 | M151 | G325 | I322 | |
|---|---|---|---|---|
| MOP | 2.4 ± 0.2 | 2.2 ± 0.2 | 2.4 ± 0.3 | 3.3 ± 0.5 |
| HMP | 2.4 ± 0.2 | 2.3 ± 0.2 | 5.0 ± 0.3 | 2.7 ± 0.2 |
Distances are given in Å and averaged over the finite temperature MD trajectories.
In addition, our MD simulations show the impact of agonist binding on the flexibility of the extracellular loop 3 (EL3). MOP binding is associated with conformational changes in this loop (Fig 5). Notably, the presence of MOP in the active site pocket causes W318 side chain to move away from the ligand after 470 ns, which allows E310 in EL3 to form salt-bridge interactions with K233 that reduces EL3 mobility (Fig 5).
Fig 5. Conformational change of μOR EL3 observed in the case of MOP binding.
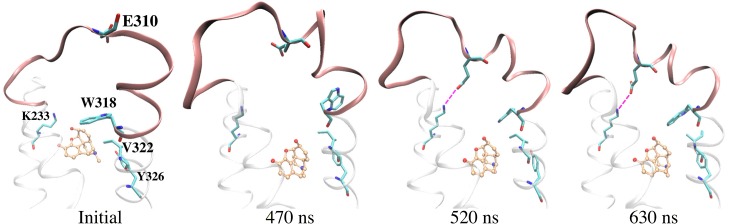
E310 in EL3 forms a salt-bridge with K233 (magenta dashed lines), which remains until the end of the simulation.
Therefore, our MD simulations confirm the key role of D147 and H297 in binding opioid ligands [48–50] and illustrate subtle differences in the binding mode of MOP and HMP, which may account for the slightly difference in their binding affinity. Indeed, HMP accommodates in the binding pocket slightly differently from MOP, maintaining a larger propensity of water-mediated H-bonds with H297 and K233 rather than a direct H-bond with H297. HMP also leads to slightly higher hydration in the binding pocket than MOP.
Next, we calculate the relative binding affinities of MOP and HMP to μOR using alchemical free energy perturbation. This approach is highly suitable for these complexes as the ligands involved in the alchemical transformation are structurally and chemically similar (Fig 1) and, therefore, one ligand can smoothly and gradually be transformed into the other. Moreover, the salt-bridge between the positively charged basic nitrogen atom of the morphinan drugs and the negatively charged D147 is preserved during our simulations. It acts as an anchor, which limits ligand diffusion and allows adequately sampling of all the relevant drug conformations inside the active site cavity. This fact significantly reduces the statistical error associated to this kind of calculations. We critically evaluate the convergence of the computed transformation by examining the hysteresis resulting from the FEP calculations of both the forward (MOP to HMP) and the reverse (HMP to MOP) transformations. This turns out to be very satisfactory (Table 3). Notably, the accommodation of both agonists inside the μOR active site is fully consistent with that found during the standard MD simulations of the MOP-μOR and HMP-μOR complexes. In particular, the H-bond pattern emerged from both forward and reverse alchemical transformations is consistent with that in the standard MD simulations, i.e. water-mediated H-bonds between HMP and H297, and direct H-bond between MOP and H297 (S3 Fig). As in the MD, the W293 side chain orientation differs in the complexes with HMP and with MOP during the alchemical free energy perturbation (S3 File). Therefore, the forward and backward alchemical transformations further corroborate the findings emerging from the MD that the differences in the H-bond patterns, the van der Waals contacts and the W293 side chain orientations are significant and are not statistical fluctuations. Instead, the aforementioned EL3 conformational changes observed in the MD simulations of MOP-μOR are not present in the alchemical transformations. Thus, this feature is possibly due to statistical fluctuations and may contribute insignificantly to the ligand binding affinity.
Table 3. Free energy differences (in kcal/mol) for the described transformations.
| Bound state | Unbound state (in solution) | ∆∆G bind | Exp. a | |
|---|---|---|---|---|
| MOP→HMP | 16.7 ± 0.6 | -17.9 ± 0.5 | -1.2 ± 1.1 | -0.4±0.3 |
| HMP→MOP | -17.5 ± 0.6 | 18.3 ± 0.2 | 0.8 ± 0.8 | 0.4±0.3 |
Error estimates are included.
a Calculated as
Despite the relatively large standard deviations of our free energy calculations, the calculated and measured differences in binding affinity are in qualitative agreement. However, because these differences are very small and are close to being non-significant, these results must be interpreted with caution. Exhaustive computational studies on multiple ligands, such as those in refs. [51, 52] for the glutamate receptor, would be required to establish quantitative conclusions on ligand-μOR energetics.
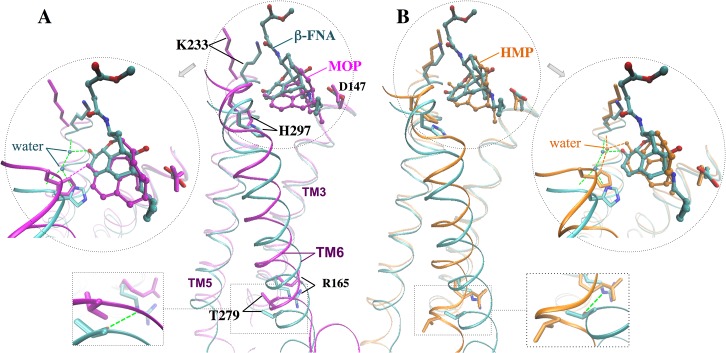
Finally, we compare the two agonists with β-FNA. The three ligands feature very similar polycyclic ring skeletons and a common salt bridge with D147. However, β-FNA shows different H-bond pattern: two water molecules mediate the ligand’s H-bonds with H297 and K233, which are preserved throughout the two independent MD simulations. Such pattern is not retained in either of the two agonists (Fig 6). This is likely due to the covalent bond between β-FNA and K233, which constraints the antagonist’s location and mobility in the binding pocket.
Fig 6. Superimposition of representative MD structures obtained by cluster analysis (see Materials and Methods) for (A) MOP-μOR and (B) HMP-μOR onto the X-ray structure of β-FNA-μOR [18].
Dashed lines indicate H-bonds.
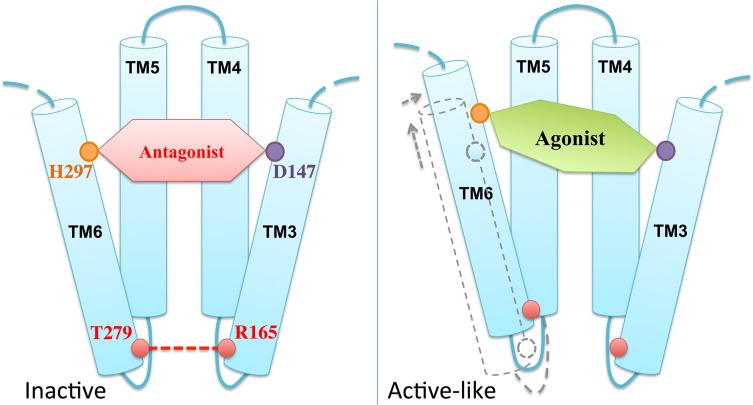
We close this section by comparing the available experimental structural information on the antagonist-bound receptor with the structural predictions emerging from our simulations. In the X-ray structure of the antagonist β-FNA covalently bound to μOR [18]–so far the only structure for this receptor–R165 in TM3 forms a H-bond with T279 in TM6 (Fig 6) [18], which turns out to be very stable during the MD simulation (lifetime 89%). By contrast, the presence of the two agonists in our simulations cause a loss of this key H-bond (lifetime < 2% in both cases) along with a translational shift of TM6 with respect to TM3 (Fig 6). TM6 moves toward TM5 and, more, to the extracellular side. The Cα RMSDs of TM6 with respect to the X-ray structure are 3.0±0.5 Å, 2.3±0.5 Å and 1.2±0.3 Å for MOP-μOR, HMP-μOR and β-FNA-μOR, respectively; whereas those of TM3 are 0.6±0.1 Å, 0.6±0.1 Å and 0.5±0.1 Å, respectively (S4 Fig). Hence, we suggest that this shift, along with the loss of the R165-T279 H-bond, is a specific feature of an active-like state. Our hypothesis is fully consistent with the following facts: (i) the T279K mutation gave rise to a constitutively active µOR variant [53]; (ii) R165 is conserved in 98% of class A GPCRs and is part of the D(E)RY motif found at the intracellular end of TM3, conserved across ~70% of class A GPCRs [54]; (iii) the R165-T279 H-bond may stabilize the receptor in an inactive state [18, 53], similar to the role of the so-called ‘ionic lock’–a salt bridge in the equivalent position between TM3 and TM6 in rhodopsin [55]; (iv) previous structural studies have suggested an activation mechanism across GPCRs based on a global toggle switch mechanism involving similar movements of TM6: this helix would move toward TM5 while increasing its separation from TM3 [56]. A common activation mechanism has been proposed for class A GPCRs based on a number of X-ray structures of active and inactive GPCRs [57]. The mechanism involves a significant outward movement of the intracellular end of TM6. Hence, the relatively small movement between this helix and TM3 observed here might be an early event toward the activation.
Comparison of the structural determinants at the active site suggests that H297 at TM6 plays an important role in the agonist-induced shift of TM6. H297 forms direct or water-mediated H-bonds as well as van der Waals interactions with the two agonists, consistent with its role as an “anchor” for opioids binding to μOR [50]. However, the agonists stabilize the residue in a position that is rather shifted relative to that in the β-FNA-μOR complex (Fig 6). Hence, we suggest that the ligand-H297 interaction is a key factor for the displacement of TM6 relative to TM3 (Fig 7).
Fig 7. Schematic model of the agonist-induced μOR conformational change into an active-like state.
Conclusions
In the present work, we investigated the binding characteristics of two morphinan agonists (MOP and HMP) to μOR by molecular simulations. Comparison is made with a simulation of the antagonist β-FNA covalently bound to μOR. An atomistic description of the ligand-receptor interactions was obtained using 3.9-μs MD simulations and 1.7 μs alchemical free energy perturbation calculations. The MOP binding mode obtained here is consistent with previous 5.6 μs long MD simulations [58]. Our simulations point to the crucial role of D147 and H297, which anchor the agonists via highly stable salt-bridge and H-bond interactions, respectively. MOP forms a direct H-bond with H297 whereas HMP forms water-mediated H-bonds with H297, both of which differ from the H-bond pattern of β-FNA. HMP also forms water-mediated H-bonds with K233. The two agonists form slightly different van der Waals contacts with residues W293, I322 and G325. W293 side chain adopts distinct orientations upon MOP and HMP binding. The HMP-bound pocket is slightly more hydrated than in the MOP-bound one. These subtle differences in the agonists’ binding mode likely account for the small difference in their binding affinity, which is here measured in vitro and calculated with alchemical free energy perturbation. Both agonists alter the relative position between TM3 and TM6 of the receptor, likely via their interactions with H297 in TM6. The alteration disrupts the R165-T279 H-bond between TM3 and TM6 that has been suggested to stabilize the receptor in an inactive conformation in the X-ray structure [18]. By contrast, in the simulation of β-FNA-μOR the relative position between TM3 and TM6 as well as the R165-T279 H-bond remains similar to those in the X-ray structure [18].
In conclusion, this study characterizes the structural features accounting for the two agonists’ different affinities, and more importantly identifies the key agonist-receptor interactions that likely promote receptor activation.
Supporting Information
(TIFF)
(TIFF)
(A) The dominant H-bond patterns during the MD simulations for MOP-μOR and HMP-μOR. The number of bridging water molecules in these H-bonds is plotted in (B, C) as a function of simulation time in the two independent MD simulations (lasting 0.8 μs and 0.5 μs, respectively), and in (D) during the course of the forward and backward alchemical transformations.
(TIFF)
(TIFF)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors acknowledge the Jülich-Aachen Research Alliance (JARA, Germany) and the Jülich Supercomputing Center (Jülich, Germany) for computing recourses.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been funded by the European Union Seventh Framework Programme (FP7/2007-2013, http://ec.europa.eu/research/fp7) under Grant Agreement N. 604102 (the Human Brain Project) to P. Carloni. The in vitro data were provided by Grünenthal GmbH through research collaboration without additional financial support. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Grünenthal GmbH provided support in the form of salaries for authors [AK, IS, MW and TK] but did not have any additional role in the study design, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the 'author contributions' section.
References
- 1. Kuehn BM. Prescription drug abuse rises globally. Jama-Journal of the American Medical Association. 2007;297(12):1306–. 10.1001/jama.297.12.1306 . [DOI] [PubMed] [Google Scholar]
- 2. Christie MJ. Cellular neuroadaptations to chronic opioids: tolerance, withdrawal and addiction. British Journal of Pharmacology. 2008;154(2):384–96. 10.1038/bjp.2008.100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ross S, Peselow E. The Neurobiology of Addictive Disorders. Clinical Neuropharmacology. 2009;32(5):269–76. 10.1097/WNF.0b013e3181a9163c . [DOI] [PubMed] [Google Scholar]
- 4. Bailey CP, Connor M. Opioids: cellular mechanisms of tolerance and physical dependence. Current Opinion in Pharmacology. 2005;5(1):60–8. 10.1016/j.coph.2004.08.012 . [DOI] [PubMed] [Google Scholar]
- 5. Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, Rosen H, et al. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nature Reviews Drug Discovery. 2013;12(1):25–34. 10.1038/nrd3859 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lord JA, Waterfield AA, Hughes J, Kosterlitz HW. Endogenous opioid peptides: multiple agonists and receptors. Nature. 1977;267(5611):495–9. Epub 1977/06/09. . [DOI] [PubMed] [Google Scholar]
- 7. Brownstein MJ. A brief history of opiates, opioid peptides, and opioid receptors. Proc Natl Acad Sci U S A. 1993;90(12):5391–3. Epub 1993/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mollereau C, Parmentier M, Mailleux P, Butour JL, Moisand C, Chalon P, et al. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS Lett. 1994;341(1):33–8. Epub 1994/03/14. . [DOI] [PubMed] [Google Scholar]
- 9. Dreborg S, Sundstrom G, Larsson TA, Larhammar D. Evolution of vertebrate opioid receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15487–92. 10.1073/pnas.0805590105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spetea M, Asim MF, Wolber G, Schmidhammer H. The mu Opioid Receptor and Ligands Acting at the mu Opioid Receptor, as Therapeutics and Potential Therapeutics. Current Pharmaceutical Design. 2013;19(42):7415–34. . [DOI] [PubMed] [Google Scholar]
- 11. Matthes HWD, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383(6603):819–23. 10.1038/383819a0 . [DOI] [PubMed] [Google Scholar]
- 12. Law PY, Reggio PH, Loh HH. Opioid receptors: toward separation of analgesic from undesirable effects. Trends in biochemical sciences. 2013;38(6):275–82. Epub 2013/04/20. 10.1016/j.tibs.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou L, Bohn LM. Functional selectivity of GPCR signaling in animals. Current Opinion in Cell Biology. 2014;27:102–8. 10.1016/j.ceb.2013.11.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rominger DH, Cowan CL, Gowen-MacDonald W, Violin JD. Biased ligands: pathway validation for novel GPCR therapeutics. Current Opinion in Pharmacology. 2014;16:108–15. 10.1016/j.coph.2014.04.002 . [DOI] [PubMed] [Google Scholar]
- 15. Kenakin T. Functional Selectivity and Biased Receptor Signaling. Journal of Pharmacology and Experimental Therapeutics. 2011;336(2):296–302. 10.1124/jpet.110.173948 . [DOI] [PubMed] [Google Scholar]
- 16. Lamberts JT, Traynor JR. Opioid Receptor Interacting Proteins and the Control of Opioid Signaling. Current Pharmaceutical Design. 2013;19(42):7333–47. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeWire SM, Yamashita DS, Rominger DH, Liu GD, Cowan CL, Graczyk TM, et al. A G Protein-Biased Ligand at the mu-Opioid Receptor Is Potently Analgesic with Reduced Gastrointestinal and Respiratory Dysfunction Compared with Morphines. Journal of Pharmacology and Experimental Therapeutics. 2013;344(3):708–17. 10.1124/Jpet.112.201616 . [DOI] [PubMed] [Google Scholar]
- 18. Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485(7398):321–6. Epub 2012/03/23. 10.1038/nature10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rasmussen SGF, DeVree BT, Zou YZ, Kruse AC, Chung KY, Kobilka TS, et al. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–U311. 10.1038/Nature10361 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5:Unit 5 6. Epub 2008/04/23. 10.1002/0471250953.bi0506s15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weichenberger CX, Sippl MJ. NQ-Flipper: recognition and correction of erroneous asparagine and glutamine side-chain rotamers in protein structures. Nucleic Acids Research. 2007;35:W403–W6. 10.1093/nar/gkm263 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Research. 2005;33:W368–W71. 10.1093/nar/gki464 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molecular Operating Environment (MOE) 2012.10. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: Chemical Computing Group Inc.; 2013.
- 24. Labute P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009;75(1):187–205. 10.1002/Prot.22234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Labute P. The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem. 2008;29(10):1693–8. 10.1002/Jcc.20933 . [DOI] [PubMed] [Google Scholar]
- 26. Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins-Structure Function and Genetics. 1996;24(4):433–8. . [DOI] [PubMed] [Google Scholar]
- 27. Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623–5. 10.1093/Bioinformatics/Btk023 . [DOI] [PubMed] [Google Scholar]
- 28. Tattrie NH, Bennett JR, Cyr R. Maximum and Minimum Values for Lecithin Classes from Various Biological Sources. Can J Biochem Cell B. 1968;46(8):819–&. . [DOI] [PubMed] [Google Scholar]
- 29. Jambeck JPM, Lyubartsev AP. An Extension and Further Validation of an All-Atomistic Force Field for Biological Membranes. J Chem Theory Comput. 2012;8(8):2938–48. 10.1021/Ct300342n . [DOI] [PubMed] [Google Scholar]
- 30. Jambeck JPM, Lyubartsev AP. Derivation and Systematic Validation of a Refined All-Atom Force Field for Phosphatidylcholine Lipids. J Phys Chem B. 2012;116(10):3164–79. 10.1021/Jp212503e . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt TH, Kandt C. LAMBADA and InflateGRO2: Efficient Membrane Alignment and Insertion of Membrane Proteins for Molecular Dynamics Simulations. J Chem Inf Model. 2012;52(10):2657–69. 10.1021/Ci3000453 . [DOI] [PubMed] [Google Scholar]
- 32. Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–18. 10.1002/Jcc.20291 . [DOI] [PubMed] [Google Scholar]
- 33. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79(2):926–35. 10.1063/1.445869 . [DOI] [Google Scholar]
- 34. Aqvist J. Ion Water Interaction Potentials Derived from Free-Energy Perturbation Simulations. J Phys Chem-Us. 1990;94(21):8021–4. . [Google Scholar]
- 35. Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–8. 10.1002/Prot.22711 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09, Revision A.02. Wallingford CT2009.
- 37. Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21(12):1049–74. . [DOI] [Google Scholar]
- 38. Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–74. 10.1002/Jcc.20035 . [DOI] [PubMed] [Google Scholar]
- 39. Andersen HC. Molecular-Dynamics Simulations at Constant Pressure and-or Temperature. J Chem Phys. 1980;72(4):2384–93. . [Google Scholar]
- 40. Parrinello M, Rahman A. Polymorphic Transitions in Single-Crystals—a New Molecular-Dynamics Method. J Appl Phys. 1981;52(12):7182–90. . [Google Scholar]
- 41. Nose S, Klein ML. Constant Pressure Molecular-Dynamics for Molecular-Systems. Mol Phys. 1983;50(5):1055–76. 10.1080/00268978300102851 . [DOI] [Google Scholar]
- 42. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–72. . [DOI] [Google Scholar]
- 43. Darden T, Perera L, Li LP, Pedersen L. New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Struct Fold Des. 1999;7(3):R55–R60. 10.1016/S0969-2126(99)80033-1 . [DOI] [PubMed] [Google Scholar]
- 44. Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–47. 10.1021/Ct700301q . [DOI] [PubMed] [Google Scholar]
- 45. Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide folding: When simulation meets experiment. Angew Chem Int Edit. 1999;38(1–2):236–40. 10.1002/(Sici)1521-3773(19990115)38:1/2<236::Aid-Anie236>3.0.Co;2-M. . [Google Scholar]
- 46. Kirkwood JG. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3(5):300–13. 10.1063/1.1749657 . [DOI] [Google Scholar]
- 47. Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22(23):3099–108. Epub 1973/12/01. . [DOI] [PubMed] [Google Scholar]
- 48. Surratt CK, Johnson PS, Moriwaki A, Seidleck BK, Blaschak CJ, Wang JB, et al. -Mu Opiate Receptor—Charged Transmembrane Domain Amino-Acids Are Critical for Agonist Recognition and Intrinsic Activity. Journal of Biological Chemistry. 1994;269(32):20548–53. . [PubMed] [Google Scholar]
- 49. Serohijos AWR, Yin SY, Ding F, Gauthier J, Gibson DG, Maixner W, et al. Structural Basis for mu-Opioid Receptor Binding and Activation. Structure. 2011;19(11):1683–90. 10.1016/J.Str.2011.08.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mansour A, Taylor LP, Fine JL, Thompson RC, Hoversten MT, Mosberg HI, et al. Key residues defining the mu-opioid receptor binding pocket: A site-directed mutagenesis study. J Neurochem. 1997;68(1):344–53. . [DOI] [PubMed] [Google Scholar]
- 51. Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nat Struct Mol Biol. 2011;18(3):283–U62. 10.1038/Nsmb.2010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15(10):1203–14. 10.1016/J.Str.2007.07.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang P, Li J, Chen CG, Visiers I, Weinstein H, Liu-Chen LY. Functional role of a conserved motif in TM6 of the rat mu opioid receptor: Constitutively active and inactive receptors result from substitutions of Thr6.34(279) with Lys and Asp. Biochemistry-Us. 2001;40(45):13501–9. 10.1021/Bi010917q . [DOI] [PubMed] [Google Scholar]
- 54. Suwa M, Sugihara M, Ono Y. Functional and Structural Overview of G-Protein-Coupled Receptors Comprehensively Obtained from Genome Sequences. Pharmaceuticals. 2011;4(4):652–64. 10.3390/ph4040652 . [DOI] [Google Scholar]
- 55. Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends in biochemical sciences. 2009;34(11):540–52. 10.1016/J.Tibs.2009.07.005 . [DOI] [PubMed] [Google Scholar]
- 56. Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Molecular mechanism of 7TM receptor activation—A global toggle switch model. Annual Review of Pharmacology and Toxicology. 2006;46:481–519. 10.1146/Annurev.Pharmtox.46.120604.141218 . [DOI] [PubMed] [Google Scholar]
- 57. Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS. Unifying Family A GPCR Theories of Activation. Pharmacol Therapeut. 2014;143(1):51–60. 10.1016/J.Pharmthera.2014.02.004 . [DOI] [PubMed] [Google Scholar]
- 58. Yuan SG, Vogel H, Filipek S. The Role of Water and Sodium Ions in the Activation of the mu-Opioid Receptor. Angew Chem Int Edit. 2013;52(38):10112–5. 10.1002/Anie.201302244 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(A) The dominant H-bond patterns during the MD simulations for MOP-μOR and HMP-μOR. The number of bridging water molecules in these H-bonds is plotted in (B, C) as a function of simulation time in the two independent MD simulations (lasting 0.8 μs and 0.5 μs, respectively), and in (D) during the course of the forward and backward alchemical transformations.
(TIFF)
(TIFF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.