Abstract
An enzyme-linked immunosorbent assay (ELISA) was developed to detect organophosphorus pesticides using a phage-borne peptide that was isolated from a cyclic 8-residue peptide phage library. The IC50 values of the phage ELISA ranged from 1.4 to 92.1 μg L−1 for eight organophosphorus pesticides (parathion-methyl, parathion, fenitrothion, cyanophos, EPN, paraoxon-methyl, paraoxon, fenitrooxon). The sensitivity was improved 120- and 2-fold compared to conventional homologous and heterologous ELISA, respectively. The selectivity of the phage ELISA was evaluated by measuring its cross-reactivity with 23 organophosphorus pesticides, among which eight were the main cross-reactants. The spike recoveries were between 66.1% and 101.6% for the detection of single pesticide residues of parathion-methyl, parathion and fenitrothion in Chinese cabbage, apple and greengrocery, and all of the coefficient of variation were less than or equal to 15.9%. Moreover, the phage ELISA results were validated by gas chromatography. The results indicate that isolating phage-borne peptides from phage display libraries is an alternative method for the development of a heterologous immunoassay and that the developed assay has a lower limit of detection than the chemically synthesized competitor assay.
Keywords: Organophosphorus pesticides, Multi-analyte assay, Enzyme-linked immunosorbent assay, Phage-borne peptide
1. Introduction
Organophosphorus (OP) pesticides are widely used in agriculture for sucking and biting insect pest control but are considered hazardous because of their high toxicity to non-target species in the environment.1 The carry-over of OP pesticides to agricultural products increases human exposure. The most effective control measure depends on a rigorous program of monitoring the food-producing chain using sensitive and reliable analytical methods to minimize health risks.
With the advantage of fast detection, low cost and large parallel-processing capacity, immunoassays are practical analytical tools for detecting OP pesticides.2, 3 OP pesticides are low-molecular-weight chemicals that can accommodate binding by only one antibody. Therefore, they are usually detected using a competitive format. In this format, a competitor (a competing hapten conjugated with a carrier protein, enzyme, fluorophore, etc.) that competes with the analyte for binding to the antibody is a necessary reagent.4 Generally, the competing hapten is the same as the immunizing hapten used to elicit the analyte-specific antibodies (homologous assay). However, the sensitivity of the immunoassay can be improved by orders of magnitude if structural variants of the immunizing hapten are used as heterologous competitors (heterologous assay).5–8 In our previous study, the sensitivities of the immunoassays were improved 60-fold compared to homologous immunoassays using a heterologous competitor that was selected from 11 candidates.9 These heterologous assays require considerable effort in chemical synthesis to generate a panel of candidate haptens from which the best competing hapten is selected. Depending on the analyte, the synthesis may be particularly challenging. Additionally, some competing haptens are highly toxic and may pose a hazard to manufacturers and users. Thus, a simple, convenient and nontoxic approach to the development of heterologous immunoassays is important.
Since the phage display of peptides was first reported,10 it has been a powerful tool for a variety of applications, including antibody engineering11, 12 and the isolation of peptide ligands for antibodies and enzymes13–15 and peptide receptors for small molecules16, 17. The principle is that the gene fragment of a polypeptide or protein is inserted into the proper position of a phage coat protein gene without affecting other functions and expressed in progeny phage. The displayed peptide or protein maintains a relatively independent spatial structure and biological activity to facilitate the binding with target. When the displayed peptide is a random sequence, the phage mixture is a phage displayed random peptide library.18 To date, some random peptide libraries have been reported,19–21 and some libraries or phage peptide display cloning systems have also been commercialized (New England Biolabs).
The peptides screened from the peptide library are connected on the phage coat protein. Labeled anti-phage antibodies have been commercialized, allowing the phage-borne peptides to be directly used as a competitor without conjugation to detectors such as enzymes or fluorophores. Some investigators have successfully obtained phage-borne peptides that can be used as a competitor for low-molecular-weight compounds from a phage display peptide library,22–25 but few pesticides or their metabolites have been reported. To the best of our knowledge, only atrazine and two metabolites from pyrethroid insecticides have been studied.4, 26 Kim et al. reported a heterologous enzyme-linked immunosorbent assay (ELISA) based on a phage-borne peptide with 100-fold higher sensitivity than the homologous assay.4 These articles demonstrate that phage-borne peptides are an alternative competitor for the development of highly sensitive competitive immunoassays.
In this study, a phage ELISA was developed to detect eight OPs in agricultural products using phage-borne peptides. Compared to the conventional ELISAs reported earlier, the sensitivity was improved 120- and 2-fold for homologous and heterologous ELISAs, respectively.
2. Materials and methods
2.1. Reagents
All reagents were of analytical grade unless specified otherwise. Parathion-methyl, chlorpyrifos-methyl, azinphos-methyl, dimethoate, fenitrooxon, EPN, paraoxon-ethyl, paraoxon-methyl, dicapthon, cyanophos and famphur were all purchased from Dr. Ehrenstorfer (Germany). Other pesticide standards were provided by the Jiangsu Pesticide Research Institute (China). Anti-OP pesticide monoclonal antibody (mAb) C8/D3, immunogenic hapten and synthetic heterologous hapten were produced in our laboratory (Fig. 1).9 Mouse anti-M13 monoclonal antibody-horse radish peroxidase (HRP) conjugate was purchased from GE Healthcare (Piscataway, NJ). Escherichia coli ER2738 was purchased from New England Biolabs (Ipswich, MA). The cyclic 8-amino-acid random peptide library was developed previously.21 Tetramethylbenzidine (TMB), isopropyl-β-D-thiogalactoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-D-galactoside (Xgal) were purchased from Sigma (USA).
Fig. 1.
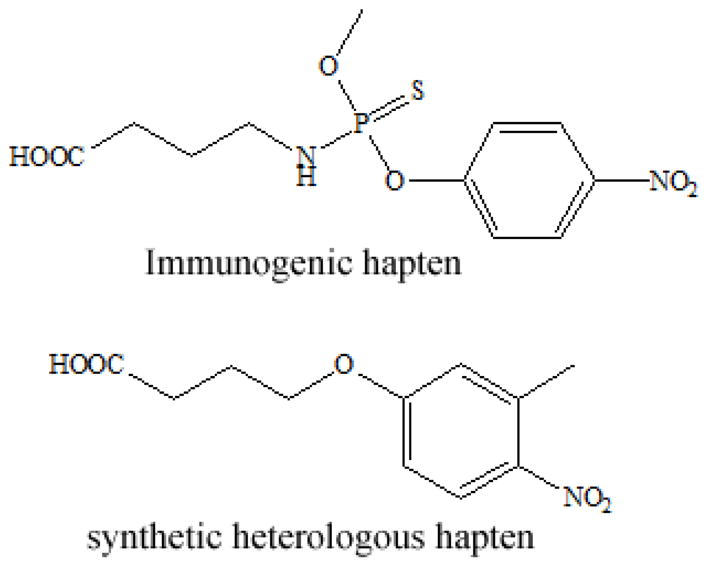
Structures of the immunogenic hapten and synthetic heterologous hapten.
2.2. Preparation of phage-borne peptides
Using methods described by Wang et al.,21 four phage-borne peptides were selected from a cyclic 8-amino-acid random peptide library that selectively bound to the mAb C8/D3 (Table 1). After amplification, the titers of C1-1, C3-3, C5-5 and C11-2 were 1.4×1012, 4.5×1011, 5.7×1011 and 1.7×1011 pfu mL−1, respectively.
Table 1.
Peptide sequences of four competitive clones.
| Clone name | Sequence |
|---|---|
| C1-1 | PPWPARPG |
| C3-3 | PPWPLRPG |
| C5-5 | APWPPRPG |
| C11-2 | SPPWPPRP |
2.3. Phage ELISA
A microtiter plate was coated with C8/D3 mAb and blocked with 300 μL of phosphate-buffered saline (PBS) containing 3% dried skimmed milk. Fifty microliters of phage supernatant in PBS was mixed with 50 μL of 100 μg L−1 parathion-methyl in 10% methanol-PBS or pure dilution buffer. The mixtures were added to the wells, and the preparations were incubated at room temperature for 1 h with shaking. After the wells were washed six times with 0.1% PBST (PBS containing 0.1% Tween 20), 100 μL of anti-M13 phage antibody conjugated with HRP (1:5000 dilution in PBS) was added. After 1 h of incubation and washing six times, the amount of bound enzyme was determined by adding 100 μL of peroxidase substrate, 25 mL of 0.1 M citrate acetate buffer (pH 5.5), 0.4 mL of 6 mgmL−1 TMB in dimethyl sulfoxide (DMSO) and 0.1 mL of 1% H2O2. The absorbance at 450 nm was determined after the reaction was stopped by adding 50 μL of 2 M H2SO4 per well (Fig. 2).
Fig. 2.

Schematic diagram of phage ELSIA.
2.4. Selection of the phage-borne peptide
The optimal concentrations of phage-borne peptide and antibody were confirmed using the two-dimensional checkerboard method.27 Selection of the phage-borne peptide was performed by phage ELISA. A series of concentrations (0.01~100 μg L−1) of parathion-methyl, parathion and fenitrothion were prepared in 10% methanol-PBS and tested using the four phage ELISAs. The evaluations of the phage-borne peptides were based on the Amax/IC50 and IC50.28
2.5. Optimization of the phage ELISA
The experimental parameters, including the ionic strength, pH value and organic solvent, were sequentially studied to determine the optimal conditions to achieve the maximum sensitivity. Competitive curves of parathion-methyl were run in PBS solutions containing different concentrations of NaCl (from 0.35 to 3.2 mol/L) and methanol (from 5 to 20%, v/v) as well as solutions with various pH values (from 5 to 9), and the evaluations of the parameters were based on the Amax/IC50 and IC50. The combination of low IC50 and high Amax/IC50 was the most desirable.
2.6. Sensitivity and selectivity of the phage ELISA
A series of concentrations (0.01~50 000 μg L−1) of 23 analytes were prepared in 10% methanol-PBS and tested using the phage ELISA. To evaluate the selectivity of the phage ELISA, the cross-reactivity (CR) was calculated based on the IC50 values using the following formula: CR = [IC50 (parathion-methyl) / IC50 (compound)] × 100%. Here, the CR of parathion-methyl was defined as 100%.
2.7. Accuracy (analysis of spiked agricultural samples)
Three different agricultural samples (Chinese cabbage, apple and greengrocery) were chosen to evaluate the performance of the phage ELISA. Chinese cabbage, apple and greengrocery were purchased from local markets (99 Ranch Market, CA, USA). All of the samples were spiked with known concentrations of parathion-methyl, parathion and fenitrothion in methanol (the final concentrations were 100 and 200 μg L−1).
All samples were cut into pieces and homogenized. The spiked samples were thoroughly mixed and allowed to stand at room temperature for 1 h. The sample pretreatment procedure was as follows. All samples (5 g) were extracted twice by a vortex mixer in 10 mL of methanol for 1 min and centrifuged for 5 min at 4 000 rpm. After centrifugation, the supernatant was transferred into a 25-mL volumetric flask and adjusted to 25 mL with PBS. After appropriate dilution, the solutions were analysed via the phage ELISA. Each pesticide was spiked and analysed in triplicate.
2.8. Gas chromatography (GC) analysis and validation
To validate the developed multi-analyte ELISA, the samples were spiked as described above and analysed by GC. Acetonitrile extraction solvent (10 mL) was added to 10 g of the homogenized sample, and the mixture was oscillated for 1 min with a vortex mixer. Next, 1.5 g of sodium chloride and 4 g of sodium sulfate were added, followed by another 3-min oscillation. After centrifugation (5 min, 4 000 rpm), 5 mL of the supernatant was transferred into a 50-mL flask, and the solvent was evaporated to dryness using a rotary vacuum evaporator with a water bath heated between 45 and 50 °C. The residue was dissolved in 1 mL of acetone. The solution was transferred to a sample vial for analysis by GC (Agilent 6890N-FPD) with a DB-17 fused silica capillary column (30 m×0.32 mm×0.25 μm). The column temperature was initially held at 80 °C for 1 min, after which it was increased to 240 °C at 20 °C/min and held for 5 min. Nitrogen was used as the carrier gas. The matrix-matched standard solutions were prepared by dissolving the residue in 1-mL mixed standard solution for the recovery study.29
3. Results and discussion
3.1. Selection of the phage-borne peptide
The Amax/IC50 and IC50 values of the four phage ELISAs for parathion-methyl, parathion and fenitrothion are shown in Table 2. The sensitivity of the four phage ELISAs decreased in the following order: C11-2 > C3-3 > C1-1 > C5-5. C11-2 was superior to the other phage-borne peptides in terms of sensitivity when used as a competitor. Because the phage-borne peptide C11-2 yielded the most sensitive assays, it was used for the next study. Because the Amax/IC50 and IC50 values for parathion-methyl, parathion, and fenitrothion had the same sensitivity ranking in the four phage ELISAs, only parathion-methyl was utilised in the next optimization.
Table 2.
Average IC50 and Amax/IC50 values of the four phage ELISAs.
| Peptide | Parathion-methyl (μg L−1) | Parathion (μg L−1) | Fenitrothion (μg L−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Amax | IC50 | Amax/IC50 | Amax | IC50 | Amax/IC50 | Amax | IC50 | Amax/IC50 | |
| C1-1 | 1.284 | 2.3 | 0.558 | 1.189 | 5.3 | 0.224 | 1.247 | 3.7 | 0.337 |
| C3-3 | 1.532 | 1.7 | 0.901 | 1.539 | 5.9 | 0.261 | 1.525 | 3.0 | 0.508 |
| C5-5 | 0.878 | 2.8 | 0.314 | 0.924 | 11.4 | 0.081 | 0.852 | 6.4 | 0.133 |
| C11-2 | 1.777 | 1.3 | 1.367 | 1.765 | 4.7 | 0.376 | 1.718 | 2.4 | 0.716 |
3.2. Optimization of the phage ELISA
Phage ELISA is based on antigen-antibody interactions, and the ionic strength and pH value are additional factors influencing the equilibrium constant.30 Therefore, the ionic strength and pH value of the working solution were optimized. The 0.14 M NaCl, pH 7.4 PBS buffer was selected for the phage ELISA because it exhibited superior sensitivity (Table 3 and 4). Organic solvent can not only improve the solubility of the analyte but also extract the analyte in the pretreatment. Based on its weaker effect on antigen antibody binding, methanol is commonly selected as an organic co-solvent in immunoassays. When the final concentration of methanol was less than or equal to 5% (50 μL of phage supernatant in PBS was mixed with 50 μL of analyte in 10% methanol-PBS), the effect of methanol on the phage ELISA was negligible (Fig. 3). In conclusion, the optimized working solution was 0.14 M NaCl and pH 7.4 PBS buffer containing 5% methanol.
Table 3.
Average IC50 and Amax/IC50 values of the phage ELISA in PBS solutions containing different concentrations of NaCl.
| Ionic strength (M) | Parathion-methyl
|
||
|---|---|---|---|
| Amax | IC50 (μg L−1) | Amax/IC50 | |
| 0.35 | 0.313 | 0.7 | 0.465 |
| 0.07 | 1.063 | 0.8 | 1.274 |
| 0.14 | 1.682 | 1.2 | 1.430 |
| 0.2 | 1.909 | 1.5 | 1.270 |
| 0.4 | 1.888 | 1.6 | 1.176 |
| 0.8 | 2.008 | 1.8 | 1.091 |
| 1.6 | 2.111 | 2.1 | 0.989 |
| 3.2 | 2.117 | 4.5 | 0.467 |
Table 4.
Average IC50 and Amax/IC50 values of the phage ELISA in PBS solutions of various pH.
| pH value | Parathion-methyl
|
||
|---|---|---|---|
| Amax | IC50 (μg L−1) | Amax/IC50 | |
| 5 | 1.949 | 5.9 | 0.332 |
| 6 | 1.837 | 2.5 | 0.733 |
| 6.5 | 1.973 | 1.7 | 1.146 |
| 7 | 1.993 | 1.7 | 1.160 |
| 7.4 | 1.824 | 1.5 | 1.188 |
| 8 | 1.760 | 1.5 | 1.131 |
| 9 | 0.726 | 1.2 | 0.597 |
Fig. 3.
Effect of methanol on the phage ELISA. Parathion-methyl was run in PBS solutions containing different concentrations of methanol. Each point is the mean of three wells.
3.3. Sensitivity of the phage ELISA
As shown in Table 5, 23 OP pesticides were evaluated using the phage ELISA. The IC50 ranged from 1.4 to 92.1 μg L−1 for the eight OP pesticides (parathion-methyl, parathion, fenitrothion, EPN, cyanophos, paraoxon-methyl, paraoxon-ethyl and fenitrooxon), and the limits of detection (LOD, IC10) ranged from 0.6 to 29.2 μg L−1. For the same mAb,9 the sensitivity of the phage ELISA was improved 2-fold and 120-fold compared to the heterologous ELISA (with the competitor prepared by chemical synthesis) and homologous ELISA, respectively. This result indicated that the phage-borne peptide was an alternative competitor for the development of a heterologous ELISA, and the assay had a lower limit of detection compared to the chemically synthesized competitor assay.
Table 5.
IC10 and IC50 values and cross-reactivity of a set of analogues structurally related to parathion-methyl by phage ELISA.
| Compound | Structure | IC10 (μg L−1) | IC50 (μg L−1) | CR (%) |
|---|---|---|---|---|
| Parathion-methyl |

|
0.6 | 1.4 | 100 |
| Parathion |

|
1.6 | 4.2 | 33.3 |
| Fenitrothion |

|
1.1 | 2.7 | 51.9 |
| Cyanophos |

|
3.4 | 10.3 | 13.6 |
| EPN |

|
5.9 | 21.1 | 6.6 |
| Paraoxon-methyl |

|
29.2 | 92.1 | 1.5 |
| Paraoxon |

|
29.0 | 85.7 | 1.6 |
| Fenitrooxon |

|
28.9 | 88.7 | 1.6 |
| Dicapthon |

|
59.3 | 199.6 | 0.7 |
| Famphur |

|
5850.1 | 17142.1 | 0.008 |
| Isocarbophos |

|
>50000 | >50000 | <0.003 |
| Fenthion |

|
>50000 | >50000 | <0.003 |
| Triazophos |

|
>50000 | >50000 | <0.003 |
| Chlorpyrifos |

|
>50000 | >50000 | <0.003 |
| Chlorpyrifos-methyl |

|
>50000 | >50000 | <0.003 |
| Phoxim |
|
>50000 | >50000 | <0.003 |
| Malathion |

|
>50000 | >50000 | <0.003 |
| Phorate |

|
>50000 | >50000 | <0.003 |
| Dimethoate |

|
>50000 | >50000 | <0.003 |
| Acephate |

|
>50000 | >50000 | <0.003 |
| Dichlorvos |

|
>50000 | >50000 | <0.003 |
| Tolclofos-methyl |

|
>50000 | >50000 | <0.003 |
| Azinphos-methyl |

|
>50000 | >50000 | <0.003 |
3.4. Selectivity of the phage ELISA
The selectivity of the phage ELISA was evaluated by measuring the cross-reactivities (CRs) with the 23 OP pesticides, and the results are summarized in Table 5. Parathion-methyl, parathion, fenitrothion, EPN, cyanophos, paraoxon-methyl, paraoxon-ethyl and fenitrooxon were the main cross-reactants among the 23 OP pesticides, and the CRs for the phage ELISA were similar to those from both the conventional ELISA and immunochromatographic assay.9 Therefore, the developed phage ELISA was selective for the eight OP pesticides.
3.5. Analysis of spiked samples
Matrix interference is one of the most common challenges in developing an immunoassay. It can be reduced in a number of ways, and dilution with buffer is a commonly used procedure. Based on the optimized methanol concentration for the phage ELISA, the final extract of the Chinese cabbage, apple and greengrocery samples (containing approximately 80% methanol) was diluted 8-fold to remove the methanol effect. Using this dilution, the absorbance values of the standard curves prepared during the matrix dilution were similar to those in the matrix-free buffer (Fig. 4). This result indicates that 8-fold dilution (the total dilution was 80-fold: 2-fold by mixing with phage in the phage ELISA procedure, 5-fold during extraction and 8-fold to remove the methanol effect) was adequate to remove the matrix interference from the samples. Therefore, the final extract of the spiked samples was diluted 8-fold before the phage ELISA analysis.
Fig. 4.
Standard inhibition curves of parathion-methyl in PBS and different matrices by phage ELISA (n = 3). The extracts of Chinese cabbage, apple and greengrocery samples were diluted 8-fold.
The measured recoveries from the spiked samples are summarized in Table 6. The average recoveries ranged from 66.1% to 101.1% for parathion-methyl, parathion and fenitrothion. The coefficients of variation (CV) were less than or equal to 15.9%. According to the maximum residue limit (MRL) of Codex Alimentarious Commission (CAC), the MRL of parathion-methyl, parathion and fenitrothion in most fruits and vegetables, including apple, is greater than or equal to 0.1 mg kg−1. Thus, the phage ELISA met the requirement for detecting OP pesticide residues in most samples.
Table 6.
Average recoveries of samples spiked with parathion-methyl, parathion and fenitrothion by phage ELISA and GC (n=3).
| Phage ELISA | GC | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Compound | Sample | Spiked (ng g−1) | Measured ±SD (ng g−1) | Recovery (%) | CV (%) | Measured ±SD (ng g−1) | Recovery (%) | CV (%) |
| Parathion-methyl | Chinese cabbage | 100 | 80.1±6.5 | 80.1 | 8.1 | 96.5±0.8 | 96.5 | 0.8 |
| 200 | 158.2±5.4 | 79.1 | 3.4 | 216.9±14.5 | 108.5 | 6.7 | ||
| Apple | 100 | 89.0±8.1 | 89.1 | 9.1 | 98.8±3.0 | 98.8 | 3.1 | |
| 200 | 236.3±7.3 | 118.1 | 3.1 | 206.9±19.4 | 103.5 | 9.4 | ||
| Greengrocery | 100 | 105.2±10.7 | 105.2 | 10.1 | 87.1±6.0 | 87.1 | 6.9 | |
| 200 | 215.2±21.9 | 107.6 | 10.2 | 216.8±17.4 | 108.4 | 8.0 | ||
| Parathion | Chinese cabbage | 100 | 86.1±13.7 | 86.1 | 15.9 | 96.0±2.0 | 96.0 | 2.0 |
| 200 | 164.7±5.9 | 82.4 | 3.6 | 199.8±11.2 | 99.9 | 5.6 | ||
| Apple | 100 | 89.0±8.9 | 89.0 | 10.1 | 96.9±2.4 | 96.9 | 2.5 | |
| 200 | 177.4±9.6 | 88.7 | 5.4 | 202.0±7.1 | 101.0 | 3.5 | ||
| Greengrocery | 100 | 112.2±23.3 | 112.2 | 20.8 | 88.1±10.2 | 88.1 | 11.2 | |
| 200 | 221.2±14.7 | 110.0 | 7.4 | 184.2±9.3 | 92.1 | 5.1 | ||
| Fenitrothion | Chinese cabbage | 100 | 90.7±9.7 | 90.7 | 10.7 | 97.8±0.4 | 97.8 | 0.4 |
| 200 | 172.5±10.9 | 86.2 | 6.3 | 216.1±7.4 | 108.1 | 3.4 | ||
| Apple | 100 | 78.9±6.7 | 78.9 | 8.5 | 98.8±2.5 | 98.8 | 2.6 | |
| 200 | 175.4±12.1 | 87.7 | 6.9 | 215.1±10.9 | 107.5 | 5.1 | ||
| Greengrocery | 100 | 89.6±13.9 | 89.6 | 15.6 | 102.7±4.7 | 102.7 | 4.6 | |
| 200 | 217.6±11.1 | 108.8 | 5.1 | 180.8±13.9 | 90.4 | 7.7 | ||
3.6. Validation of the phage ELISA
To confirm the accuracy and applicability of the phage ELISA, the spiked samples were analysed by GC. The average recoveries by GC for all samples ranged from 87.1% to 108.5%, and the CVs were less than or equal to 11.7% (Table 6). The phage ELISA and GC techniques yielded comparable results, indicating that the developed phage ELISA was reliable and accurate.
4. Conclusions
In this study, a phage ELISA was developed to detect eight OP pesticides in agricultural samples using a phage-borne peptide. Using the same mAb, the sensitivity of the phage ELISA was improved 2- and 120-fold compared to the heterologous ELISA (where the competitor was prepared by chemical synthesis) and homologous ELISA, respectively. The accuracy and precision of the assay were validated by GC, and the agreement between the phage ELISA and GC was high. Therefore, the phage ELISA presented in this study is suitable as a convenient quantitative tool for the rapid screening of the eight OP pesticides in agricultural products. Compared with synthesizing competitors, isolating competitors from phage display libraries is an efficient method for accelerating the development of highly sensitive competitive immunoassays. To the best of our knowledge, this is the first time that a phage-borne peptide has been applied in the multi-analyte detection of OP pesticides.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31301690), the Doctoral Program of Higher Education Research Fund (20130097120006), the Youth Innovation Fund in Science & Technology of Nanjing Agricultural University (KJ2013008) and the National High Technology Research and Development Program of China (2012AA101401-1). Partial funding was also received from the National Institute of Environmental Health Sciences Superfund Research Program (P42 ES04699) and the National Institute of Occupational Health Sciences Western Center for Agricultural Health Research (U50 OH07550).
References
- 1.Roex EW, Keijzers R, van Gestel CA. Aquat Toxicol. 2003;64:451–460. doi: 10.1016/s0166-445x(03)00100-0. [DOI] [PubMed] [Google Scholar]
- 2.Piao YZ, Kim YJ, Kim YA, Lee HS, Hammock BD, Lee YT. J Agric Food Chem. 2009;57:10004–10013. doi: 10.1021/jf901998y. [DOI] [PubMed] [Google Scholar]
- 3.Wang CM, Li XB, Liu YH, Guo YR, Xie R, Gui WJ, Zhu GN. J Agric Food Chem. 2010;58:5658–5663. doi: 10.1021/jf904575k. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, González-Techera A, González-Sapienza GG, Ahn KC, Gee SJ, Hammock BD. Environ Sci Technol. 2008;42:2047–2053. doi: 10.1021/es702219a. [DOI] [PubMed] [Google Scholar]
- 5.Shan G, Huang H, Stoutamire DW, Gee SJ, Leng G, Hammock BD. Chem Res Toxicol. 2004;17:218–225. doi: 10.1021/tx034220c. [DOI] [PubMed] [Google Scholar]
- 6.Ahn KC, Watanabe T, Gee SJ, Hammock BD. J Agric Food Chem. 2004;52:4583–4594. doi: 10.1021/jf049646r. [DOI] [PubMed] [Google Scholar]
- 7.Lee WY, Lee EK, Kim YJ, Park WC, Chung T, Lee YT. Anal Chim Acta. 2006;557:169–178. [Google Scholar]
- 8.Wang CM, Liu YH, Guo YR, Liang CZ, Li XB, Zhu GN. Food Chem. 2009;115:365–370. [Google Scholar]
- 9.Hua XD, Yang JF, Wang LM, Fang QK, Zhang GP, Liu FQ. PLoS one. 2012;7:e53099. doi: 10.1371/journal.pone.0053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith GP. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Li PW, Lei JW, Zhang Q, Zhang W, Li CM. RSC Adv. 2013;3:22367–22372. [Google Scholar]
- 12.Dufner P, Jermutus L, Minter RR. Trends Biotechnol. 2006;24:523–529. doi: 10.1016/j.tibtech.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Sompuram SR, Kodela V, Ramanathan H, Wescott C, Radcliffe G, Bogen SA. Clin Chem. 2002;48:410–420. [PubMed] [Google Scholar]
- 14.Menendez A, Scott JK. Anal Biochem. 2005;336:145–157. doi: 10.1016/j.ab.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 15.Du B, Qian M, Zhou ZL, Wang P, Wang L, Zhang XP, Wu M, Zhang P, Mei B. Biochem Biophys Res Commun. 2006;342:956–962. doi: 10.1016/j.bbrc.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Goldman ER, Pazirandeh MP, Charles PT, Balighian ED, Anderson GP. Anal Chim Acta. 2002;457:13–19. [Google Scholar]
- 17.Kim YG, Lee CS, Chung WJ, Kim EM, Shin DS, Rhim JH, Lee YS, Kim BG, Chung J. Biochem Biophys Res Commun. 2005;329:312–317. doi: 10.1016/j.bbrc.2005.01.137. [DOI] [PubMed] [Google Scholar]
- 18.Bratkovic T. Cell Mol Life Sci. 2010;67:749–67. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens CL, Cwirla SE, Lee RYW, Whitehorn E, Chen EYF, Bakker A, Martin EL, Wagstrom C, Gopalan P, Smith CW, Tate E, Koller KJ, Schatzm PJ, Dower WJ, Barrett RW. J Biol Chem. 1995;270:21129–21136. doi: 10.1074/jbc.270.36.21129. [DOI] [PubMed] [Google Scholar]
- 20.Ball WJ, Jr, Wang Z, Malik B, Kasturi R, Dey P, Short MK, Margolies MN. J Mol Biol. 2000;301:101–115. doi: 10.1006/jmbi.2000.3934. [DOI] [PubMed] [Google Scholar]
- 21.Wang YR, Wang H, Li PW, Zhang Q, Kim HJ, Gee SJ, Hammock BD. J Agric Food Chem. 2013;61:2426–2433. doi: 10.1021/jf4004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thirumala-Devi K, Miller JS, Reddy G, Reddy DVR, Mayo MA. J Appl Microbiol. 2001;90:330–336. doi: 10.1046/j.1365-2672.2001.01249.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu RR, Yu Z, He QH, Xu Y. Food Control. 2007;18:872–877. [Google Scholar]
- 24.Lai WH, Fung DYC, Xu Y, Liu RR, Xiong YH. Food Control. 2009;20:791–795. [Google Scholar]
- 25.He QH, Xu Y, Zhang CZ, Li YP, Huang ZB. Food Control. 2014;39:56–61. [Google Scholar]
- 26.Cardozo S, González-Techera A, Last JA, Hammock BD, González-Sapienza GG. Environ Sci Technol. 2005;39:4234–4241. doi: 10.1021/es047931l. [DOI] [PubMed] [Google Scholar]
- 27.Shan GM, Wengatz I, Stoutamire DW, Gee SJ, Hammock BD. Chem Res Toxicol. 1999;12:1033–1041. doi: 10.1021/tx990091h. [DOI] [PubMed] [Google Scholar]
- 28.Jin RY, Guo YR, Wang CM, Wu JX, Zhu GN. Food Sci. 2009;74:T1–T6. doi: 10.1111/j.1750-3841.2008.01002.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao EC, Zhao WT, Han LJ, Jiang SR, Zhou ZQ. J Chromatogr A. 2007;1175:137–140. doi: 10.1016/j.chroma.2007.10.069. [DOI] [PubMed] [Google Scholar]
- 30.Reverberi R, Reverberi L. Blood Transfus. 2007;5:227–240. doi: 10.2450/2007.0047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]




