Abstract
We investigated genetic overlap between Alzheimer’s disease (AD) and Parkinson’s disease (PD). Using summary statistics (p-values) from large recent genomewide association studies (GWAS) (total n = 89,904 individuals), we sought to identify single nucleotide polymorphisms (SNPs) associating with both AD and PD. We found and replicated association of both AD and PD with the A allele of rs393152 within the extended MAPT region on chromosome 17 (meta analysis p-value across 5 independent AD cohorts = 1.65 × 10−7). In independent datasets, we found a dose-dependent effect of the A allele of rs393152 on intra-cerebral MAPT transcript levels and volume loss within the entorhinal cortex and hippocampus. Our findings identify the tau-associated MAPT locus as a site of genetic overlap between AD and PD and extending prior work, we show that the MAPT region increases risk of Alzheimer’s neurodegeneration.
INTRODUCTION
Alzheimer’s disease (AD) and Parkinson’s disease (PD) are the two most common neurodegenerative disorders. Neuropathologically, AD is characterized by the presence of extracellular amyloid-β (Aβ) plaques and intracellular tau-associated neurofibrillary tangles whereas PD involves deposition of α-synuclein containing Lewy bodies.1 Though AD and PD are considered distinct neurodegenerative entities, there is evidence for Lewy body pathology in AD 2 and Alzheimer’s-type pathology in PD 3 suggesting overlap between these two disorders. Importantly, although tau-associated pathology is considered a hallmark of AD, genome-wide association studies (GWAS) in PD have identified several polymorphisms in and around the tau encoding microtubule-associated protein gene (MAPT) 4,5 indicating that similar biochemical perturbations may contribute to both AD and PD. 6 Furthermore, prior reports investigating the genetic relationship between MAPT and AD risk have been conflicting, with some studies finding a positive association 7–8 and other studies showing no association 8–9, indicating that the role of the MAPT gene in influencing Alzheimer’s neurodegeneration is still largely unknown.
Combining GWAS from two disorders provides insights into genetic pleiotropy (defined as a single gene or variant being associated with more than one distinct phenotype) and could elucidate shared pathobiology. Here, using summary statistics (p-values and minor allele frequencies) from large genetic studies 11–15, we sought single nucleotide polymorphisms (SNPs) associating with both AD and PD.
METHODS
Participant Samples
We obtained complete GWAS results in the form of summary statistics from the PD International Parkinson’s Disease Genetics Consortium (IPDGC) and AD Alzheimer’s Disease Genetics Consortium (ADGC). The PD GWAS summary statistic results from IPDGC consisted of 5,333 cases and 12,019 controls obtained from 5 studies with genotyped and imputed data at 7,689,524 SNPs (Table 1a, for additional details see reference 11). The AD GWAS summary statistic data from ADGC consisted of 11,840 cases and 10,931 controls obtained from 15 studies with genotyped and imputed data at 2,324,889 SNPs (Table 1a, for additional details see reference 12). The ADGC GWAS summary statistic data were co-varied for age, sex and number of APOE alleles. There was no overlap between the ADGC and the IPDGC cases/controls.
Table 1.
| a: Characteristics of Parkinson’s disease (IPDGC) and primary Alzheimer’s disease (ADGC) genome-wide association studies evaluated in this manuscript. | ||||
|---|---|---|---|---|
| IPDGC | ADGC | |||
| Cases | Controls | Cases | Controls | |
| N | 5333 | 12019 | 11840 | 10931 |
| Age at assessment (mean) | 57.6 | 67.8 | 80.6 | 76.7 |
| % Women | 41 | 48.7 | 61 | 58.5 |
| % APOE ε4 carriers | N/A | N/A | 51.6 | 26.7 |
| b: Demographic, clinical, and imaging data for all ADNI participants evaluated in this study. AD = Alzheimer’s disease, MCI = mild cognitive impairment, HC = cognitively normal older adults, MMSE = Mini-mental status exam, CDR-SB = Clinical Dementia Rating-Sum of Boxes score | |||
|---|---|---|---|
| HC (n = 174) |
MCI (n = 311) |
AD (n = 135 ) |
|
| Age, Mean (SD) | 76.3 (5.1) | 75.0 (7.3) | 75.4 (7.7) |
| Female, % | 48 | 36 | 48 |
| Education Years, Mean (SD) | 16.1 (2.7) | 15.7 (2.9) | 14.9 (2.9) |
| CDR-SB, Mean (SE) | 0.03 (0.11) | 1.6 (0.9) | 4.2 (1.5) |
| APOE ε4 carriers (%) | 25 | 57 | 69 |
| Entorhinal cortex APC, Mean (SD) | −0.57 (2.5) | −2.10 (1.6) | −2.92 (1.7) |
| Hippocampus APC, Mean (SD) | −0.90 (1.1) | −2.19 (1.7) | −3.45 (1.9) |
To test for replication, we also assessed the p-values of the PD genome-wide significant SNPs in four separate AD cohorts, namely the Genetic and Environmental Risk in Alzheimer's Disease (GERAD) sample, a cohort of AD cases and controls drawn from the population of Iceland (deCODE cohort), a small cohort of mild cognitive impairment or AD cases and controls drawn from the population of Norway (Oslo), and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. The AD GWAS summary statistic results from the GERAD consortium were obtained from 13 studies and consisted of 3,941 cases (62.7% female) and 7,848 controls (55.6 % female) with genotyped data at 529,205 SNPs (for additional details see reference 13). A total of 5571 controls from the PD IPDGC GWA were also present in the AD GERAD GWA. The AD GWAS summary statistic data drawn from the Icelandic population (deCODE) included 3,759 AD cases (65.8 % female) and 8,888 older controls (57.8% females) greater than 85 years of age (for additional details see references 14 and 15). The AD GWAS summary statistic data from the CHARGE consortium were obtained from 4 studies and included 1,315 AD cases (62.1% female) and 21,766 controls (56.9 % female) (for additional details see reference 27). The AD GWAS summary statistic data drawn from the Norwegian population (Oslo) included 434 individuals classified as AD or mild cognitive impairment (57% female) and 1,830 controls (49% female) (for additional details please see Supplemental Information).
These studies addressed potential concerns of population stratification by limiting analysis to individuals of European descent, including principal components of genetic variation in the regression tests and controlling post-hoc for genomic inflation with genomic control (for additional details see references 11–15,27).
For the gene expression analyses, we used publicly available, genotyping (performed on the Affymetrix GeneChip Human Mapping 500K Array Set platform) and RNA expression data (performed on the Illumina HumanRefseq-8 Expression BeadChip system) from neuropathologically confirmed 176 late-onset AD cases (mean age = 83.4 years, standard deviation = 6.6) and 188 controls (mean age = 81.2 years, standard deviation = 9.1) from the Gene Expression Omnibus (GEO) data set GSE15222. 16 We additionally evaluated genotype and imaging data obtained from 620 older participants (174 healthy older controls, 311 individuals with mild cognitive impairment (MCI) and 135 individuals with probable AD) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI – see Table 1b and Supplemental Methods). We restricted our analyses to those participants with available genotype and quality-assured baseline and follow-up MRI scans (6 months to 3.5 years, mean of 2.02 years, standard deviation 0.80 years) available as of April 2011. We assessed longitudinal sub-regional change in gray matter volume (atrophy) on serial 2471 T1-weighted MRI scans using a modified version of the FreeSurfer software package (for additional details see Supplemental Methods).
Statistical analyses
We used stepwise gatekeeper hypothesis testing 17 to identify SNPs associating with both PD and AD. We restricted our analyses to only those SNPs assayed in both GWASs from the IPDGC and the ADGC Consortia. First, we identified ‘pruned’ SNPs (removing all SNPs with r2 > 0.2, within 1 Mb of a given SNP) that were significant at a genome-wide level (p < 5 × 10 −8) within PD. Next, we evaluated the p-values of these PD genome-wide significant SNPs within the AD ADGC GWAS (Apolipoprotein E (APOE), age and sex co-varied summary statistic p-values) and applied a Bonferroni correction to control for multiple comparisons. Note that since the SNPs were a priori selected independently of the p-values from AD ADGC the proper Bonferroni correction is in terms of the number of PD genome-wide significant SNPs. Therefore, the p-value threshold for detecting significant ADGC loci controls for the number of PD genome-wide significant SNPs rather than p < 5 × 10 −8. It is important to note that this stepwise gatekeeper hypothesis testing approach implies a strict control for family-wise error rate in a multiple testing framework. 17
RESULTS
Genetic overlap between AD and PD at the A allele of rs393152
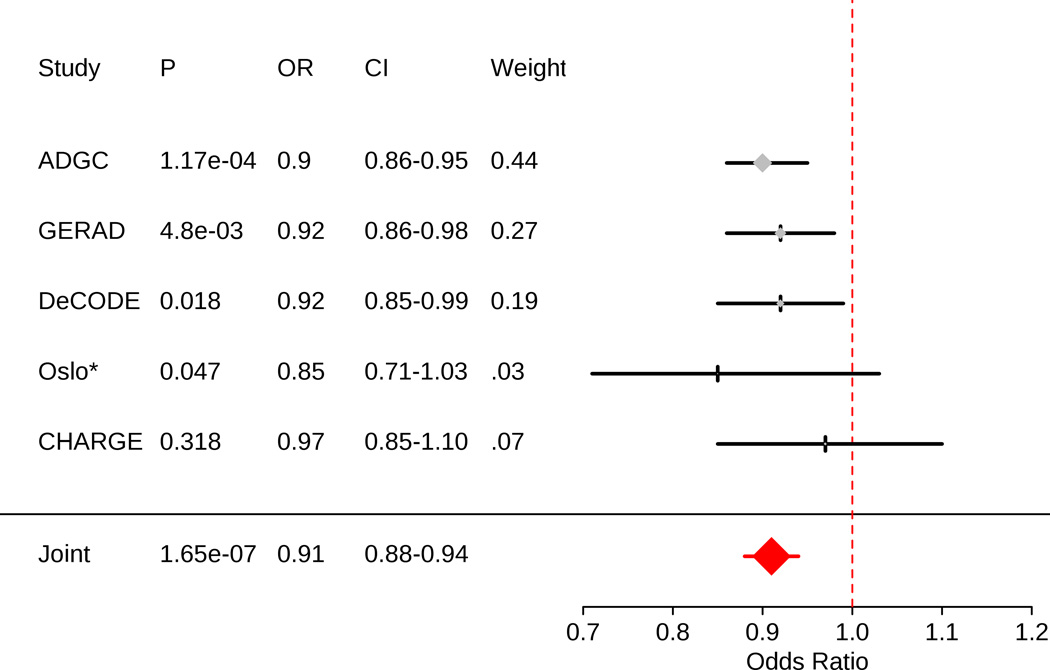
We found 8 SNPs on 4 chromosomes that were genome-wide significant in PD, thus requiring a Bonferroni corrected p-value significance threshold of 0.00625 (Table 2). Across all 8 SNPs, we found that the A allele of rs393152, within the CRHR1 region on chromosome 17 (within the extended MAPT locus) and with a minor allele frequency of 23.1%, significantly increased AD risk in the ADGC cohort (p-value = 1.17 × 10−4, odds ratio (OR) for the minor allele = 0.90, 95% confidence interval (CI) = 0.86–0.95) (Table 2) (Figure 1). In a replication analysis, we found that the A allele of rs393152 also significantly increased AD risk within the GERAD (one-tailed p-value = 0.0048, OR for the minor allele = 0.92, 95% CI = 0.86–0.98), deCODE (one-tailed p-value = 0.017, OR for the minor allele = 0.92, 95% CI = 0.85–0.99) and Oslo cohorts (one-tailed p-value = 0.047, OR for the minor allele = 0.85, 95% CI = 0.71–1.02). We replicated directionality of effect for the A allele of rs393152 within the CHARGE cohort (one-tailed p-value = 0.318, OR for the minor allele = 0.97, 95% CI = 0.85–1.10). We conducted an inverse variance weighted meta-analysis 18 and found a two-tailed meta-analysis p-value of 1.65 × 10−7 (meta analysis OR = 0.91, 95% CI = 0.88–0.94) (Figure 1).
Table 2.
Summary of evaluated loci.
| SNP | Chr | Nearest Gene |
Minor Allele Frequency |
Risk Allele for PD |
PD p-value | Risk Allele for AD |
ADGC p- value |
Other genes in genomic region defined by LD |
|---|---|---|---|---|---|---|---|---|
| rs9917256 | 2 | STK39 | 0.1365 | A | 1.62 × 10 −9 | A | 0.79 | |
| rs11248051 | 4 | GAK | 0.1299 | T | 3.50 × 10 −8 | T | 0.19 | DGKQ, TMEM175 |
| rs4698412 | 4 | BST1 | 0.4344 | A | 2.03 × 10−8 | G | 0.031 | |
| rs356220 | 4 | SNCA | 0.4869 | T | 1.47 × 10−25 | C | 0.014 | CR605611 |
| rs3857059 | 4 | SNCA | 0.0684 | G | 1.66 × 10−14 | A | 0.78 |
AK123890, MMRN1 |
| rs2197120 | 4 | SNCA | 0.1995 | G | 6.29 × 10−10 | G | 0.99 | AK123890 |
| rs12603319 | 17 | FBXW10 | 0.2192 | T | 1.144 × 10−8 | T | 0.39 | |
| rs393152 | 17 | CRHR1 | 0.231 | A | 2.22 × 10−18 | A | 1.17 × 10−4 |
ARHGAP27, KANSL1, LOC100128977, LOC5132, LOC644172, MAPT, MGC57346, PLEKHM1 |
Figure 1.
Forest plot for rs393152. Since rs393152 was not available within the Oslo cohort (*), we used a proxy SNP (rs17690703; r2 = .765, D'=1 in Hapmap2).
We evaluated the statistical power for detecting an association of rs393152 with AD across the discovery (ADGC) and the combined, meta-analysis AD cohorts (ADGC + GERAD + deCODE + Oslo + CHARGE). Using a GWAS threshold of p < 5 × 10 −8 the power within ADGC was 0.028 and within the meta-analysis cohort was 0.36, demonstrating that even the combined cohort consisting of 21,289 AD cases and 51,263 controls was underpowered to detect an association between AD and rs393152 using a standard GWAS approach. However, leveraging PD such that power is computed conditional on discovery in the PD sample (stepwise gatekeeper hypothesis testing), by using p < 0.00625 (where Bonferroni corrected p = 0.05/number of genome-wide significant SNPs in PD), the power within ADGC was 0.854 and within the meta-analysis cohort was 0.998 indicating that restricting evaluation to only PD-significant SNPs results in considerable increase in statistical power for AD gene discovery. We also calculated the sample size needed to detect rs393152 ((C−1 Θ−1(5 × 10−8)2/ Θ−1(0.00625)2), where Θ−1 is the inverse standard normal cumulative distribution function) and found that in comparison to our discovery cohort, 4.5 times as many subjects would be needed to detect rs393152 using a standard GWAS approach at the same alpha /Type I error.
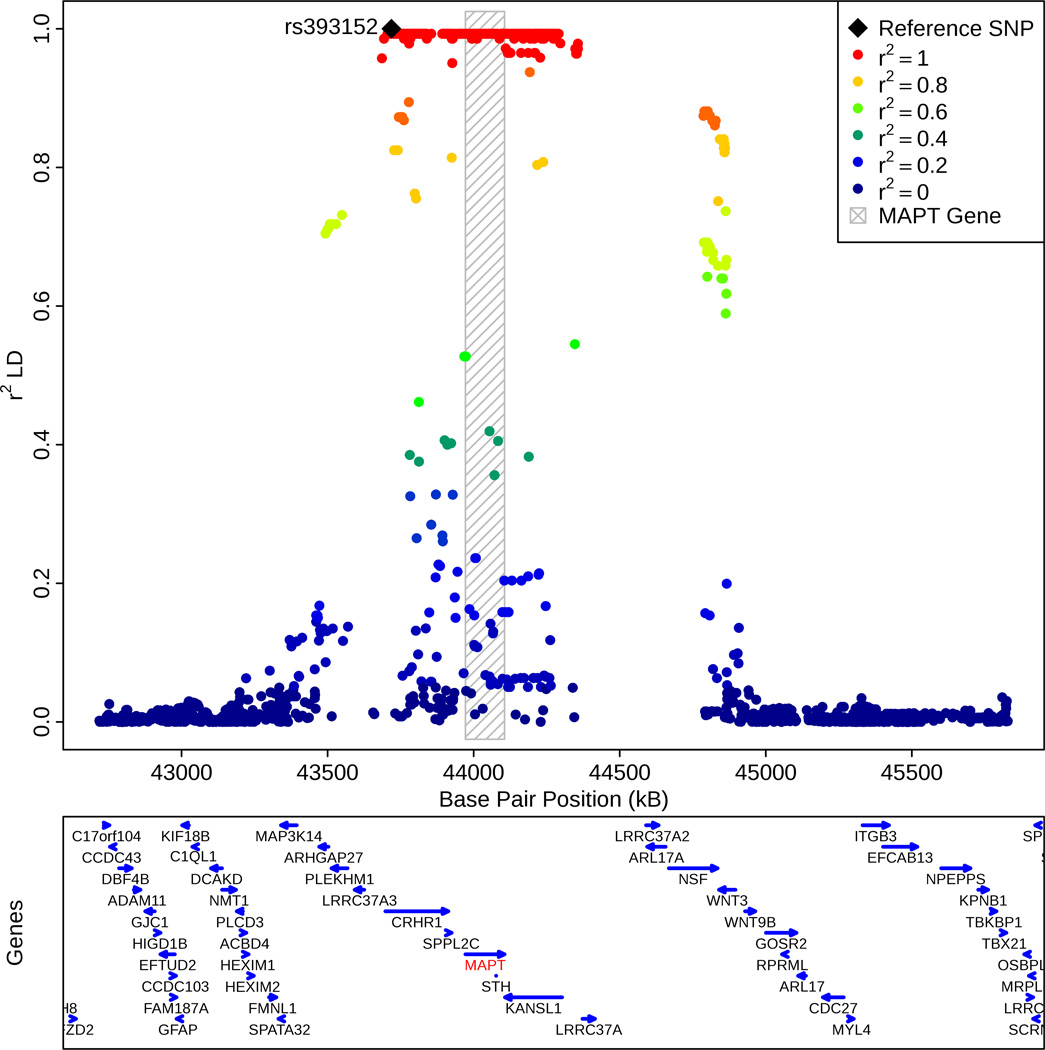
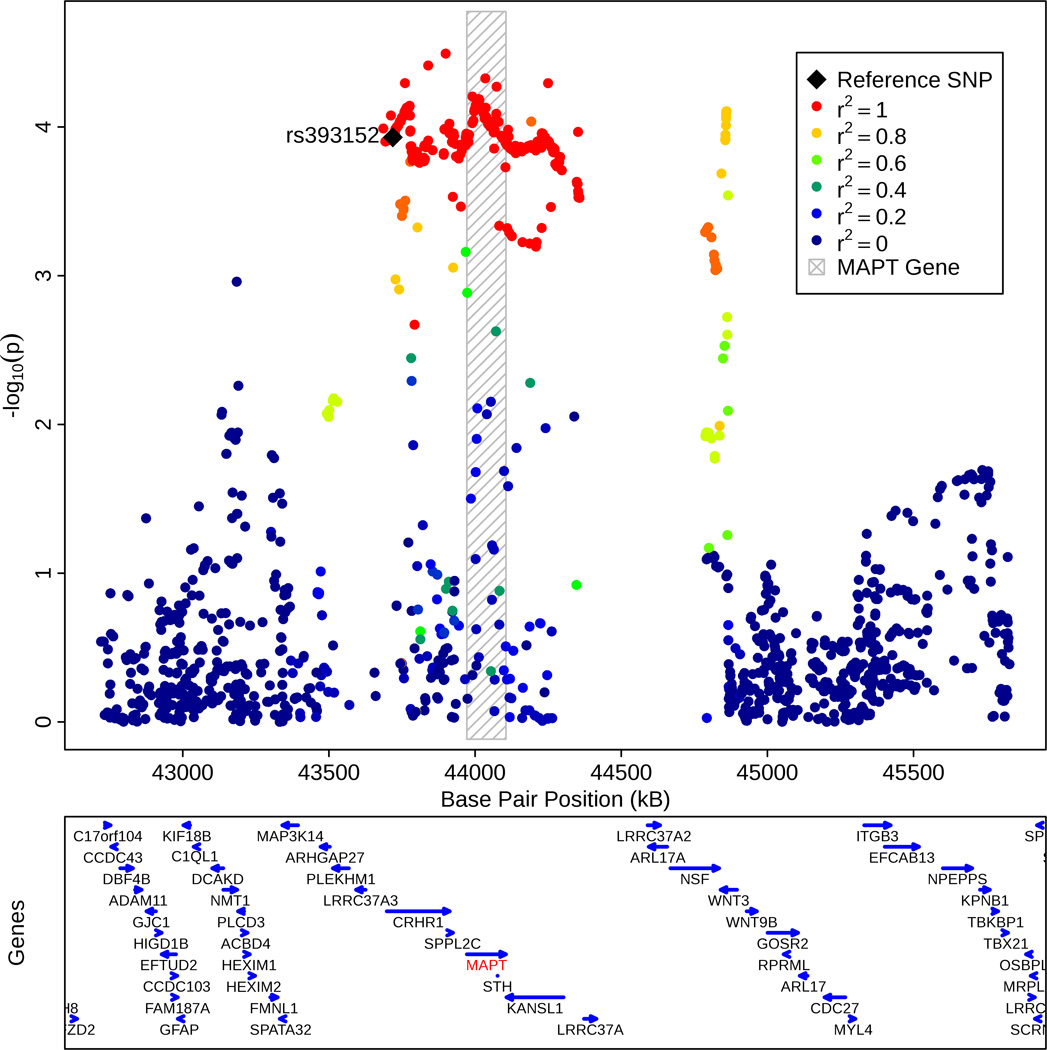
Based on the 1000 Genomes Project LD structure, we found that rs393152 was in r2 LD > 0.8 with a number of variants within the MAPT gene on chromosome 17 (Figure 2a). Fine mapping showed that rs1981997 constituted the peak of the AD association signal within MAPT (r2 = 1.0 with rs393152 in HapMap 2; Figure 2b). Across the ADGC (risk allele = A, two tailed p-value = = 9.54 × 10−5, OR = 0.90, 95% CI = 0.85–0.95), GERAD (one tailed p-value = 0.006, OR = 0.92, 95% CI = 0.86–0.98, deCODE (one tailed p-value = 0.018, OR = 0.92, 95% CI = 0.84–0.99), Oslo (one tailed p-value = 0.047, OR = 0.85, 95% CI = 0.71–1.03) and CHARGE (one-tailed p-value = 0.0327, OR = 0.96, 95% CI = 0.84–1.08) cohorts, the leading SNP in the MAPT region, rs1981997, demonstrated a similar meta-analysis p-value to rs393152 (two-tailed meta-analysis p-value of 1.29 × 10−7, see Supplemental Figure 4) providing further evidence that our AD/PD pleiotropic variant was tagging the MAPT gene and not a false positive result. We also note that rs393152 has been previously shown to tag the H1 haplotype at the MAPT locus (r2 = 0.761). 5 Because of the extensive LD structure in this region, we cannot exclude the possibility that other genes, besides MAPT, are the pathologically relevant genes. However, MAPT is biologically the most plausible candidate.
Figure 2.
(a) Regional linkage disequilibrium (LD) plot demonstrating the relationship between rs393152 on chromosome 17 and loci greater than and less than 1 MB. The bottom panel indicates the location of genes in the region. Linkage Disequilibrium measured in the 1000 genomes European Populations using plink v1.07.
(b) Regional association plot illustrating the association signal within the MAPT region on chromosome 17. The bottom panel indicates the location of genes in the region. Linkage Disequilibrium measured in the 1000 genomes European Populations using plink v1.07.
Non-polygenic pleiotropy between AD and PD
We further investigated whether the observed genetic overlap between AD and PD was polygenic and generalizable across a number of loci or non-polygenic and driven by the MAPT locus alone. Using recently developed statistical methods to evaluate pleiotropic effects 19–22, we investigated relative ‘enrichment’ of pleiotropic SNPs in AD (APOE, age and sex co-varied summary statistic p-values from ADGC) as a function of significance in PD (summary statistic p-values from IPDGC) (for additional details see Supplemental Methods). Removing the MAPT-associated genetic signal, consisting of all SNPs in r2 > 0.2 (based on 1000 Genomes Project LD structure) within 1 Mb of MAPT variants, resulted in considerable attenuation of genetic enrichment (Supplemental Figured 1a–d) indicating that the observed pleiotropy between AD and PD was non-polygenic and likely confined to the MAPT region. Similarly, after ‘pruning’ (removing SNPs in r2 > 0.2) all available ADGC SNPs, we found a single pleiotropic locus on chromosome 17 between AD and PD that was in r2 = 1.0 with MAPT. Though some genetic enrichment was still present after removing the MAPT-associated SNPs, we found a similar pattern in PD SNP enrichment conditioned on AD (Supplementary Figure 2).
AD-PD pleiotropic locus correlates with MAPT transcript levels
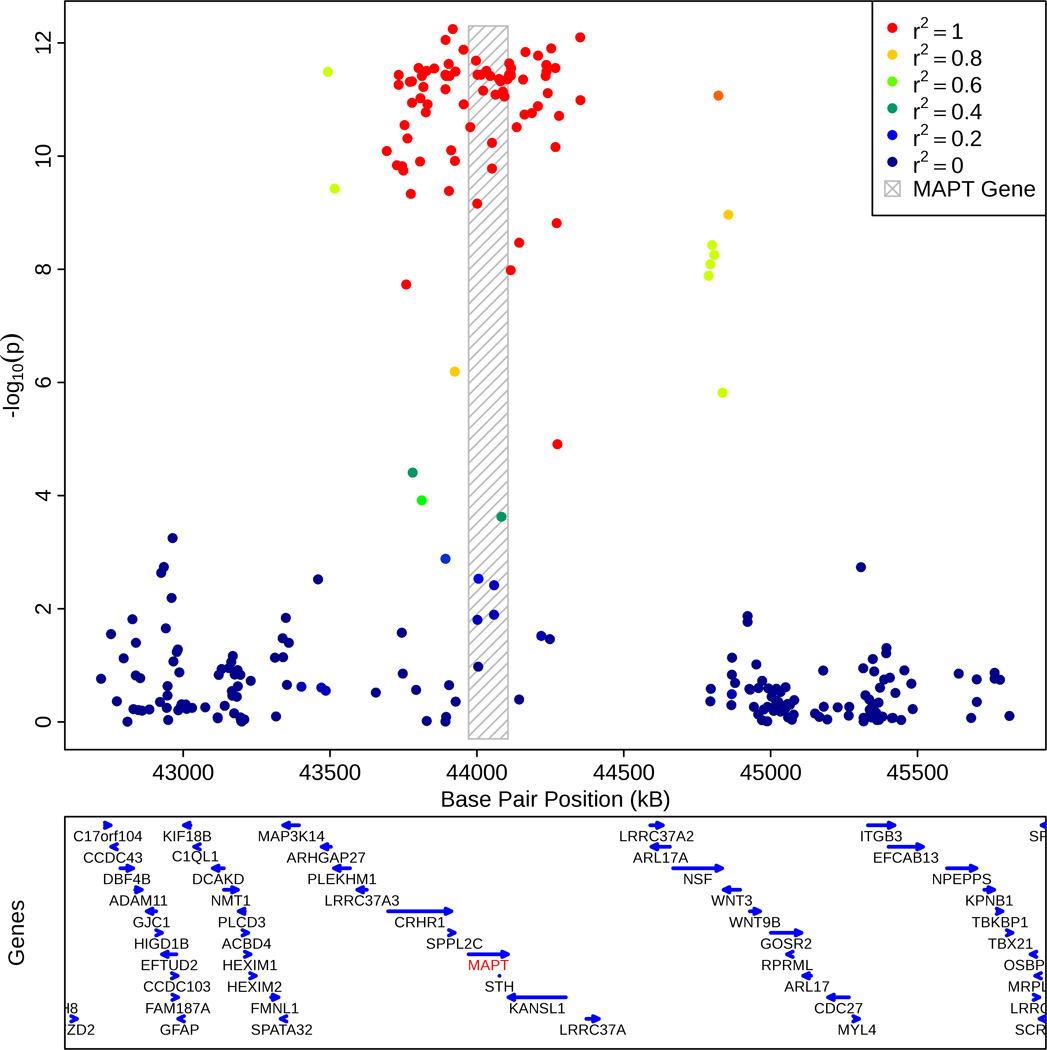
We assessed the relationship between the AD-PD pleiotropic locus on chromosome 17 and MAPT transcript levels within the brain (target id = GI_8400714-A and reference sequence = NM_016841. 1 in GSE15222, for additional details see references 16 and 32). Since rs393152 was not available in the GEO dataset, we focused on rs422112 within the CRHR1 locus on chromosome 17, the best available proxy (closest distance and r2 > 0.98) for rs393152. We used an additive model with minor allele (T) counts coded as 0, 1, and 2. Given the allele frequencies and near complete LD between rs393152 and rs422112, the ‘A’ allele of rs393152 tags the ‘C’ allele of rs422112 and the ‘G’ allele of rs393152 tags the ‘T’ allele of rs422112. Using linear regression, co-varying for the effects of age at death, APOE ε4 carrier status, diagnosis (AD cases vs. controls), brain tissue region (frontal, parietal, temporal, or cerebellar), postmortem interval, institute source of sample, and hybridization date, we evaluated the relationship between rs422112 and MAPT transcript expression levels. Across all cases and controls, we found a strong association between the T allele of rs422112 and decreased MAPT transcript expression levels (standardized β-coefficient = −0.27, t-statistic = −6.61, p-value = 1.45 × 10−10) which corresponds to presence of the A allele of rs393152 and increased MAPT transcript expression (Figure 3). Subgroup analyses demonstrated similar results within the AD cases and controls (see Supplemental Results). We further assessed the specificity of our findings by evaluating the relationship between the AD-PD pleiotropic locus and transcript levels of synaptophysin (SYP), a neuronal protein, and synuclein (SNCA), a neural protein associated with tau and PD. In contrast to MAPT transcript levels, we found no relationship between rs422112 and transcript levels of either SYP or SNCA (see Supplemental Results and Figure 3). We additionally performed a ‘locus wide association study’ testing all SNPs in the MAPT region for association with MAPT transcript expression levels. SNPs in r2 = 1.0 with rs393152 constituted the peak of the association signal (p < 1.0 ×10−8) with MAPT transcript expression levels (Figure 4). We also evaluated the relationship between SNPs in LD with rs393152 and transcript levels of other chromosome 17 genes within the larger MAPT region that were available within GSE15222. 16 As illustrated in Supplemental Figures 3a–f, SNPs in LD with rs393152 did not demonstrate significant association with transcript levels of other genes within the MAPT region further illustrating the specificity of our MAPT findings.
Figure 3.
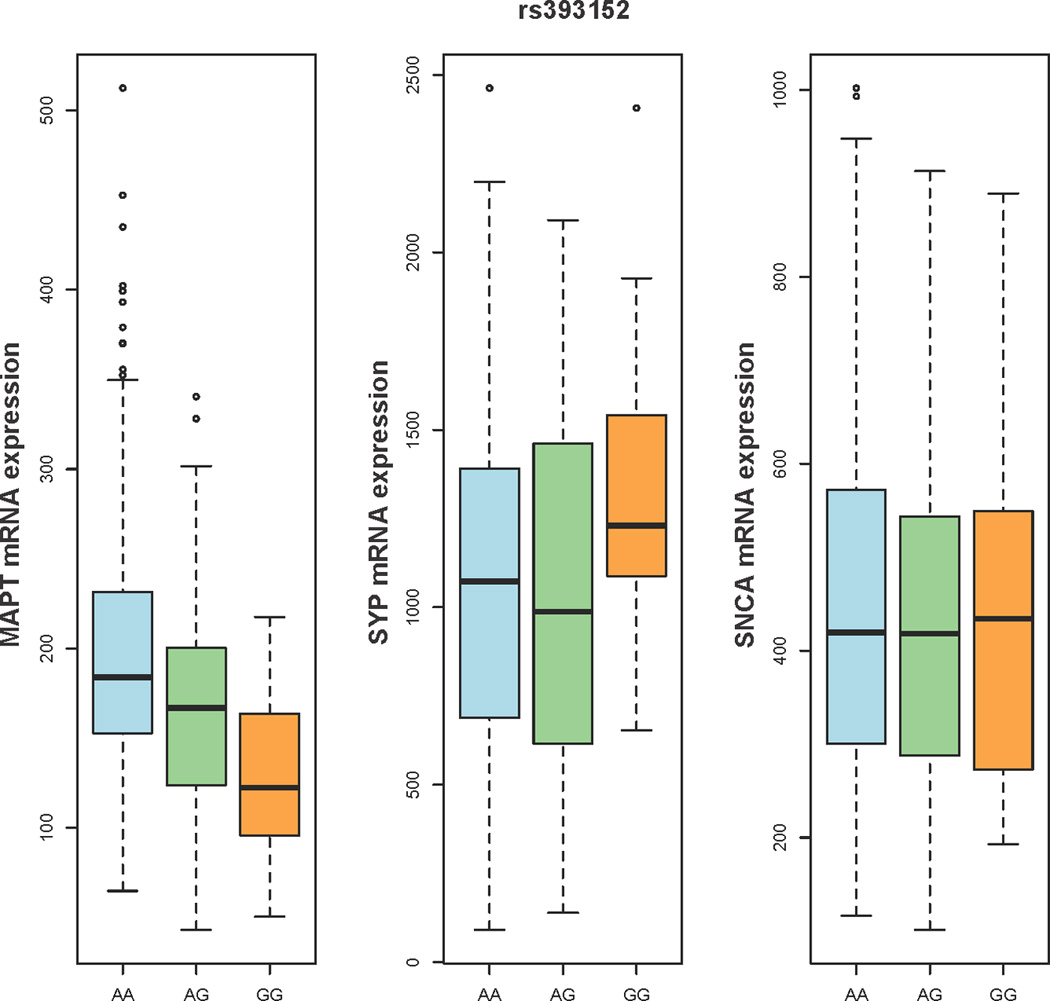
Box plots illustrating the relationship between rs393152 alleles (x-axis) and gene expression levels of MAPT, SYP, and SNCA (y-axis). For each plot, thick black lines show the median value. Regions above and below the black line show the upper and lower quartiles, respectively. The dashed lines extend to the minimum and maximum values with outliers shown as open circles. For MAPT, a proxy SNP was used (please see Results for additional details). As illustrated, the A allele of rs393152 demonstrated a selective dose-dependent effect on the level of intracranial MAPT transcript.
Figure 4.
Regional association plot demonstrating the relationship between MAPT transcript expression levels (y-axis) and SNPs in LD with rs393152 on chromosome 17. The bottom panel indicates the location of genes in the region. Linkage Disequilibrium measured in the 1000 genomes European Populations using plink v1.07. As illustrated, SNPs in r2 LD =1 with rs393152 constituted the peak of the association signal with MAPT transcript expression levels.
AD-PD pleiotropic locus correlates with longitudinal brain atrophy
Using linear mixed effects models, we assessed the relationship of rs393152 with longitudinal brain atrophy specifically within the entorhinal cortex and hippocampus, two medial temporal lobe regions selectively affected in the earliest stages of AD. 23 These models co-varied for the effects of baseline age, sex, education, group status (healthy older control vs. MCI vs. AD), disease severity (Clinical Dementia Rating-Sum of Box score), and APOE ε4 carrier status. We used an additive model with major allele (A) counts coded as 0,1 2. Across all available ADNI participants, we found that the A allele of rs393152 was significantly associated with increased atrophy rates (volume loss) of the entorhinal cortex (standardized β-coefficient = −0.003, SE = 0.001, p-value = 0.0071) and hippocampus (standardized β-coefficient = −0.003, SE = 0.001, p-value = 0.0031).
AD-PD pleiotropic locus demonstrates larger effect among APOE ε4 non-carriers
We further assessed the relationship between rs393152, MAPT transcript expression levels, and medial temporal lobe atrophy separately among APOE ε4 carriers (presence of at least one ε4 allele) and non-carriers (absence of at least one ε4 allele). Using the linear mixed effects model framework described above, we found a stronger effect between rs393152 and MAPT transcript expression levels among APOE ε4 non-carriers (standardized β-coefficient = −0.22, SE = 0.04, p-value = 1.1 × 10−6) than the APOE ε4 carriers (standardized β-coefficient = −0.14, SE = 0.04, p-value = 0.001). Similarly, we found a stronger effect between rs393152 and medial temporal lobe atrophy among APOE ε4 non-carriers (entorhinal cortex: standardized β-coefficient = −0.002, SE = 0.001, p-value = 0.04; hippocampus: standardized β-coefficient = −0.003, SE = 0.001, p-value = 0.01) than among APOE ε4 carriers (entorhinal cortex: standardized β-coefficient = −0.003, SE = 0.002, p-value = 0.07; hippocampus: standardized β-coefficient = −0.003, SE = 0.002, p-value = 0.07) (Figure 5).
Figure 5.
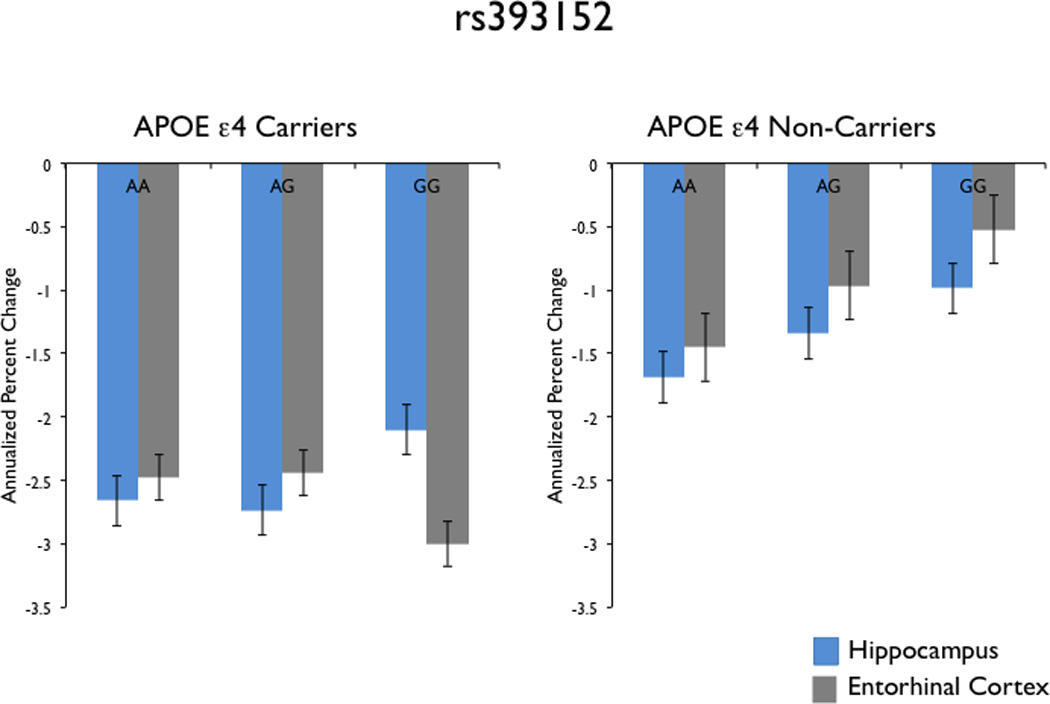
Bar plots demonstrating the relationship between rs393152 alleles (x-axis) and volume loss (annualized percent change – y-axis) of the hippocampus (blue) and entorhinal cortex (gray) among APOE ε4 carriers (left panel) and APOE ε4 non-carriers (right panel). As illustrated, the A allele of rs393152 demonstrated a selective dose-dependent relationship with medial temporal lobe atrophy only among APOE ε4 non-carriers.
DISCUSSION
In this study, we leveraged gene variants associating with PD to search for variants that associate with AD. We found a gene variant that was in strong LD with markers in the MAPT gene on chromosome 17 and that was previously associated with PD. This SNP was significantly associated with longitudinal atrophy of the entorhinal cortex and hippocampus and demonstrated a strong association with MAPT transcript levels within the brain. Considered together, our findings point to the tau-associated MAPT locus as a site genetic overlap between AD and PD.
These results indicate that leveraging the genetic signal in one phenotype may improve statistical power for gene discovery in a second, related phenotype. Rather than evaluating all possible AD susceptibility loci, we restricted our analyses to only those 8 SNPs that were below genome-wide threshold in PD. As such, detection of AD susceptibility loci only among genome-wide significant PD susceptibility loci obviates the need for applying a p < 5 × 10 −8 threshold and constitutes stepwise gatekeeper hypothesis testing. 17 This two-stage stepwise gatekeeper framework is conceptually similar to the ‘proxy-phenotype’ method, which has recently been utilized to identify common variants associated with cognitive performance. 24 It is important to note that this approach does not lower the statistical ‘bar’ for gene discovery and maintains a constant Type I error rate. By exploiting statistical power from PD, we were able to identify one SNP within the CRHR1 locus on chromosome 17 (meta-analysis p-value = 1.65 × 10−7, OR = 0.91, 95% CI = 0.88–0.94) that was significantly associated with increased AD risk. Importantly, use of this stepwise, pleiotropic approach, where power is computed conditional on discovery in the PD sample, resulted in considerable improvement in statistical power for AD gene detection. In contrast, using a standard GWAS approach, neither the discovery ADGC cohort nor the combined meta-analysis cohort were sufficiently powered to detect rs393152. Given the comparable sample sizes with our current study, it is likely that the original AD GWASs 12–13, 25–26 and even the recent meta-analysis (stage 1) 27 were underpowered to detect MAPT-associated signal in AD.
There are several indications that the detected pleiotropy within chromosome 17 represents biological signal and not analysis artifacts or type 1 error. First, the use of APOE co-varied SNPs from the ADGC minimizes concerns that the detected SNPs represent spurious association resulting from the known large effect of APOE on AD risk (for an example of this, see reference 28). Importantly, our findings indicate the presence of genetic signal independent of the chromosome 19 APOE cluster. Second, rs393152 was significantly associated with AD risk in three independent AD replication cohorts and demonstrated equivalent effect sizes in all five AD cohorts. Third, the identified pleiotropic locus was in r2 LD > 0.8 with a number of variants within the tau-encoding MAPT gene on 17q21 indicating that the detected signal was specific to the MAPT region. Fourth, the leading AD-associated SNP in the MAPT region (rs1981997, r2 LD = 1.0 with rs393152 in the HapMap 2) demonstrated a similar meta-analysis p-value to rs393152 providing further evidence that our AD/PD pleotropic SNP was not a false positive result. Finally, the A allele of rs393152 showed a dose-dependent effect specifically with MAPT transcript levels within the brain and was significantly associated with longitudinal medial temporal lobe atrophy, an established endophenotype of Alzheimer’s neurodegeneration.
These single locus results point to shared pathobiology between AD and PD. Although we cannot exclude the possibility that other genes at this chromosome 17 locus are the pathologically relevant genes, our data are biologically plausible and consistent with prior experimental evidence establishing the role of MAPT in neurodegenerative diseases. 29 The pleiotropic variant we found, rs393152, tags the H1 haplotype at the MAPT locus5, which has been associated with a number of tauopathies including corticobasal degeneration (CBD), progressive supranuclear palsy (PSP), and PD. 5,30 Furthermore, broadly consistent with a prior study 31, our results suggest non-extensive, non-polygenic pleiotropy between AD and PD localized to the MAPT cluster on chromosome 17.
Despite a number of prior studies 7–10, the role of MAPT in AD is still unclear. Extending prior work suggesting a significant relationship between the MAPT H1 7 (within the GERAD cohort) and H2 8 (within the ADGC cohort) haplotypes and AD risk, our findings indicate that the A allele of rs393152, which tags the H1 haplotype at the MAPT locus 5, increases risk for AD. Building on prior research demonstrating a robust association between a variant in the H2 haplotype and reduced MAPT brain expression levels 8, we found a dose-dependent effect of the A allele of rs393152 (Figure 3) on intracranial MAPT gene expression. In contrast, we found no association between rs393152 and transcript levels of either synaptophysin or synuclein indicating the specificity of the relationship between the identified AD-PD pleiotropic locus and MAPT transcript expression. Our gene expression findings are consistent with prior work demonstrating a significant relationship between the H1 haplotype and MAPT levels. 32–33 However, a previous study 34 of exon levels from multiple human brain regions found no association between the H1c subhaplotype and MAPT expression indicating that additional work using large samples is needed to systematically investigate the H1/H2 sub-haplotypes and MAPT brain expression levels. Additionally, building on prior work detecting smaller gray matter volumes within cognitively normal 35 and cognitively impaired 36 MAPT carriers, we found a significant relationship between the A allele of rs393152 and longitudinal atrophy of the entorhinal cortex and hippocampus, two medial temporal lobe regions selectively affected with tau-associated neurofibrillary pathology in the earliest stages of AD. Considered together, this suggests that the PD-associated MAPT cluster influences Alzheimer’s neurodegeneration likely via tau-related mechanisms.
From an AD perspective, these results highlight the importance of considering tau. Recent evidence indicates that dominantly inherited mutations in MAPT cause forms of frontotemporal dementia with parkinsonism 29, a rare MAPT variant (p.A152T) increases risk for AD and frontotemporal dementia syndromes 37 and tau modulates Aβ-associated Alzheimer’s neurodegeneration. 38 Consistent with this work, our present results indicate that tau-associated polymorphisms impact MAPT transcript levels and affect medial temporal lobe volume loss. When considered together with prior CSF 39–41, and imaging research 42–43, our findings suggest that data from GWAS, expression quantitative trait loci, and structural imaging measures may better elucidate underlying pathobiology than any of these markers by themselves. These results also demonstrate the utility of using entorhinal cortex and hippocampal atrophy rates as endophenotypes to identify and confirm AD risk variants.
In this study the diagnosis of AD and PD was based on clinical evaluations, without histopathological confirmation. Post-mortem evidence indicates the co-occurrence of α-synuclein, tangle and amyloid pathology. 44 Therefore, one concern is that concomitant Parkinson’s pathology may have contributed to our MAPT associated effect in AD. In a small cohort of autopsy confirmed AD cases and controls, we replicated the directionality and magnitude of the A allele of rs393152 (Supplemental Figure 5) indicating that our AD-associated findings are not due to concomitant PD pathology. Furthermore, building on prior genetic work 45, among APOE ε4 non-carriers, we found a stronger relationship between rs393152 and both gene expression levels and medial temporal lobe atrophy (Figure 5) suggesting that MAPT may predominantly influence Alzheimer’s neurodegeneration in a smaller subset of individuals who do not possess APOE ε4 alleles. As a caveat, we note that since we primarily evaluated summary statistics from the discovery and replication cohorts, additional work with raw genotype data is needed to determine whether the AD-associated MAPT effect varies based on APOE ε4 carrier status. Another concern is the potential ‘contamination’ of PD samples with other tauopathies (such as PSP and CBD) strongly associated with MAPT. Using neuropathologically confirmed PD cases, a recent study 46 found a significant association between rs393152 and idiopathic PD indicating that our current findings are unlikely due to contamination with unrecognized cases of PSP or CBD.
From a translational perspective, this work illustrates that data from large GWAS and a pleiotropic framework can provide important insights into the relationships between various diseases. Complementary to recently developed polygenic pleiotropic methods 19–22, the analytic framework used in this manuscript is useful for detecting non-polygenic pleiotropy and can be integrated with other biomarkers to test biologically driven hypotheses. The combination of genetic, molecular, and neuroimaging measures may be additionally helpful for detecting and quantifying the biochemical effects of therapeutic interventions.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs. Irene Litvan, Subhojit Roy and Marilyn Albert for helpful comments on an earlier version of this manuscript. This research was supported by grants from the National Institutes of Health (R01AG031224, K01AG029218, K02NS067427, T32 EB005970, UO1AG032984, U24-AG041689, and R01 MH100351), the Research Council of Norway (#213837, #225989, #223273, #237250/EU JPND), the South East Norway Health Authority (2013-123), the Norwegian Health Association and the KG Jebsen Foundation. AJS was supported by NIH grants RC2DA029475 and R01HD061414 and the Robert J. Glushko and Pamela Samuelson Graduate Fellowship. Please see Supplemental Acknowledgements for ADGC and ADNI funding sources.
Footnotes
DISCLOSURES
Dr. Anders M. Dale is a Founder of and holds equity in CorTechs Labs, Inc, and serves on its Scientific Advisory Board. He is also a member of the Scientific Advisory Board of Human Longevity, Inc. (HLI), and receives funding through research agreements with General Electric Healthcare (GEHC) and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Dr. Linda K. McEvoy has stock options in CorTechs Labs, Inc.
Dr. James B. Brewer holds stock options in CorTechs Labs, Inc and serves on the advisory board and receives financial support from the Eli Lilly Biomarker Unit (Amyvid). Dr. Brewer also receives research support from General Electric and Janssen Alzheimer Immunotherapy.
REFERENCES
- 1.Nussbaum RL, Ellis CE. Alzheimer's disease and Parkinson's disease. N Engl J Med. 2003;348:1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 2.Uchikado H, DelleDonne A, Ahmed Z, Dickson DW. Lewy bodies in progressive supranuclear palsy represent an independent disease process. J Neuropathol Exp Neurol. 2006;65:387–395. doi: 10.1097/01.jnen.0000218449.17073.43. [DOI] [PubMed] [Google Scholar]
- 3.Mann DM, Jones D. Deposition of amyloid (A4) protein within the brains of persons with dementing disorders other than Alzheimer's disease and Down's syndrome. Neurosci Lett. 1990;109:68–75. doi: 10.1016/0304-3940(90)90539-l. [DOI] [PubMed] [Google Scholar]
- 4.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 5.Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Krüger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shulman JM, De Jager PL. Evidence for a common pathway linking neurodegenerative diseases. Nat Genet. 2009;41:1261–1262. doi: 10.1038/ng1209-1261. [DOI] [PubMed] [Google Scholar]
- 7.Gerrish A, Russo G, Richards A, Moskvina V, Ivanov D, Harold D, Sims R, Abraham R, Hollingworth P, Chapman J, Hamshere M, Pahwa JS, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Johnston JA, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Kölsch H, Heun R, Schürmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Davies G, Harris SE, Starr JM, Deary IJ, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Jones L, Holmans PA, O'Donovan MC, Owen MJ, Williams J. The role of variation at AβPP, PSEN1, PSEN2, and MAPT in late onset Alzheimer's disease. J Alzheimers Dis. 2012;28:377–387. doi: 10.3233/JAD-2011-110824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen M, Kachadoorian M, Quicksall Z, Zou F, Chai HS, Younkin C, Crook JE, Pankratz VS, Carrasquillo MM, Krishnan S, Nguyen T, Ma L, Malphrus K, Lincoln S, Bisceglio G, Kolbert CP, Jen J, Mukherjee S, Kauwe JK, Crane PK, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Parisi JE, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N. Association of MAPT haplotypes with Alzheimer's disease risk and MAPT brain gene expression levels. Alzheimers Res Ther. 2014;6:39. doi: 10.1186/alzrt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee O, Kauwe JS, Mayo K, Morris JC, Goate AM. Haplotype-based association analysis of the MAPT locus in late onset Alzheimer's disease. BMC Genet. 2007;8:3. doi: 10.1186/1471-2156-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham R, Sims R, Carroll L, Hollingworth P, O'Donovan MC, Williams J, Owen MJ. An association study of common variation at the MAPT locus with late-onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 150B:1152–1155. doi: 10.1002/ajmg.b.30951. [DOI] [PubMed] [Google Scholar]
- 11.International Parkinson Disease Genomics Consortium. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simón-Sánchez J, Schulte C, Lesage S, Sveinbjörnsdóttir S, Stefánsson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jönsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, McCorquodale DS3rd, Cuello C, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson V, NACC-Neuropathology Group. Heward CB, Reiman EM, Stephan D, Hardy J, Myers AJ. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dmitrienko A, Wiens BL, Tamhane AC, Wang X. Tree-structured gatekeeping tests in clinical trials with hierarchically ordered multiple objectives. Statistics in Medicine. 2007;26:2465–2478. doi: 10.1002/sim.2716. [DOI] [PubMed] [Google Scholar]
- 18.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, Zuber V, Bettela F, Ripke S, Kelsoe JR, Kendler KS, O'Donovan MC, Sklar P, McEvoy L, Desikan RS, Lie BA, Djurovic S, Dale AM. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: implications for immune related disease mechanisms, Molecular Psychiatry. 2014 Jan 28; [Google Scholar]
- 20.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, Kendler KS, O'Donovan MC, Rujescu D, Werge T, Sklar P, Roddey JC, Chen CH, McEvoy L, Desikan RS, Djurovic S Dale AMPsychiatric Genomics Consortium (PGC); Bipolar Disorder and Schizophrenia Working Groups. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, Rujescu D, Werge T, van de Bunt M, Morris AP, McCarthy MI, Roddey JC, McEvoy LK, Desikan RS Dale AM International Consortium for Blood Pressure GWAS; Diabetes Genetics Replication and Meta-analysis Consortium; Psychiatric Genomics Consortium Schizophrenia Working Group. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JZ, Hov JR, Folseraas T, Ellinghaus E, Rushbrook SM, Doncheva NT, Andreassen OA, Weersma RK, Weismüller TJ, Eksteen B, Invernizzi P, Hirschfield GM, Gotthardt DN, Pares A, Ellinghaus D, Shah T, Juran BD, Milkiewicz P, Rust C, Schramm C, Müller T, Srivastava B, Dalekos G, Nöthen MM, Herms S, Winkelmann J, Mitrovic M, Braun F, Ponsioen CY, Croucher PJ, Sterneck M, Teufel A, Mason AL, Saarela J, Leppa V, Dorfman R, Alvaro D, Floreani A, Onengut-Gumuscu S, Rich SS, Thompson WK, Schork AJ, Næss S, Thomsen I, Mayr G, König IR, Hveem K, Cleynen I, Gutierrez-Achury J, Ricaño-Ponce I, van Heel D, Björnsson E, Sandford RN, Durie PR, Melum E, Vatn MH, Silverberg MS, Duerr RH, Padyukov L, Brand S, Sans M, Annese V, Achkar JP, Boberg KM, Marschall HU, Chazouillères O, Bowlus CL, Wijmenga C, Schrumpf E, Vermeire S, Albrecht M, UK-PSCSC Consortium; International IBD Genetics Consortium. Rioux JD, Alexander G, Bergquist A, Cho J, Schreiber S, Manns MP, Färkkilä M, Dale AM, Chapman RW, Lazaridis KN, Franke A, Anderson CA, Karlsen TH International PSC Study Group. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 24.Rietveld CA, Esko T, Davies G, Pers TH, Turley P, Benyamin B, Chabris CF, Emilsson V, Johnson AD, Lee JJ, de Leeuw C, Marioni RE, Medland SE, Miller MB, Rostapshova O, van der Lee SJ, Vinkhuyzen AA, Amin N, Conley D, Derringer J, van Duijn CM, Fehrmann R, Franke L, Glaeser EL, Hansell NK, Hayward C, Iacono WG, Ibrahim-Verbaas C, Jaddoe V, Karjalainen J, Laibson D, Lichtenstein P, Liewald DC, Magnusson PK, Martin NG, McGue M, McMahon G, Pedersen NL, Pinker S, Porteous DJ, Posthuma D, Rivadeneira F, Smith BH, Starr JM, Tiemeier H, Timpson NJ, Trzaskowski M, Uitterlinden AG, Verhulst FC, Ward ME, Wright MJ, Davey Smith G, Deary IJ, Johannesson M, Plomin R, Visscher PM, Benjamin DJ, Cesarini D, Koellinger PD. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P European Alzheimer's Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 26.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM CHARGE Consortium; GERAD1 Consortium; EADI1 Consortium. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P European Alzheimer's Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer's Disease (GERAD); Alzheimer's Disease Genetic Consortium (ADGC); Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013 Dec;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, Lee JH, Bird TD, Bennett DA, Diaz-Arrastia R, Goate AM, Farlow M, Ghetti B, Sweet RA, Foroud TM, Mayeux R NIA-LOAD/NCRAD Family Study Group. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 30.Pittman AM, Fung HC, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Hum Mol Genet. 2006;15:R188–R195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- 31.Moskvina V, Harold D, Russo G, Vedernikov A, Sharma M, Saad M, Holmans P, Bras JM, Bettella F, Keller MF, Nicolaou N, Simón-Sánchez J, Gibbs JR, Schulte C, Durr A, Guerreiro R, Hernandez D, Brice A, Stefánsson H, Majamaa K, Gasser T, Heutink P, Wood N, Martinez M, Singleton AB, Nalls MA, Hardy J, Owen MJ, O’Donovan MC, Williams J, Morris HR, Williams NM. Analysis of Genome-Wide Association Studies of Alzheimer Disease and of Parkinson Disease to Determine If These 2 Diseases Share a Common Genetic Risk. JAMA Neurol. 2013 Aug 5; doi: 10.1001/jamaneurol.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, Lees A, Leung D, McKeith IG, Perry RH, Morris CM, Trojanowski JQ, Clark C, Karlawish J, Arnold S, Forman MS, Van Deerlin V, de Silva R, Hardy J. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, Malphrus KG, Palusak R, Lincoln S, Bisceglio G, Georgescu C, Kouri N, Kolbert CP, Jen J, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N Alzheimer's Disease Genetics Consortium. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, Luk C, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, Singleton AB, Cookson MR, Pittman AM, de Silva R, Weale ME, Hardy J, Ryten M. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canu E, Boccardi M, Ghidoni R, Benussi L, Testa C, Pievani M, Bonetti M, Binetti G, Frisoni GB. H1 haplotype of the MAPT gene is associated with lower regional gray matter volume in healthy carriers. Eur J Hum Genet. 2009;17:287–294. doi: 10.1038/ejhg.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goñi J, Cervantes S, Arrondo G, Lamet I, Pastor P, Pastor MA. Selective brain gray matter atrophy associated with APOE ε4 and MAPT H1 in subjects with mild cognitive impairment. J Alzheimer's Dis. 2013;33:1009–1019. doi: 10.3233/JAD-2012-121174. [DOI] [PubMed] [Google Scholar]
- 37.Coppola G, Chinnathambi S, Lee JJ, Dombroski BA, Baker MC, Soto-Ortolaza AI, Lee SE, Klein E, Huang AY, Sears R, Lane JR, Karydas AM, Kenet RO, Biernat J, Wang LS, Cotman CW, Decarli CS, Levey AI, Ringman JM, Mendez MF, Chui HC, Le Ber I, Brice A, Lupton MK, Preza E, Lovestone S, Powell J, Graff-Radford N, Petersen RC, Boeve BF, Lippa CF, Bigio EH, Mackenzie I, Finger E, Kertesz A, Caselli RJ, Gearing M, Juncos JL, Ghetti B, Spina S, Bordelon YM, Tourtellotte WW, Frosch MP, Vonsattel JP, Zarow C, Beach TG, Albin RL, Lieberman AP, Lee VM, Trojanowski JQ, Van Deerlin VM, Bird TD, Galasko DR, Masliah E, White CL, Troncoso JC, Hannequin D, Boxer AL, Geschwind MD, Kumar S, Mandelkow EM, Wszolek ZK, Uitti RJ, Dickson DW, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Ross OA, Rademakers R, Schellenberg GD, Miller BL, Mandelkow E, Geschwind DH Alzheimer's Disease Genetics Consortium. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer's diseases. Hum Mol Genet. 2012;21:3500–3512. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 40.Li G, Sokal I, Quinn JF, Leverenz JB, Brodey M, Schellenberg GD, Kaye JA, Raskind MA, Zhang J, Peskind ER, Montine TJ. CSF tau/Abeta42 ratio for increased risk of mild cognitive impairment: a follow-up study. Neurology. 2007;69:631–639. doi: 10.1212/01.wnl.0000267428.62582.aa. [DOI] [PubMed] [Google Scholar]
- 41.Desikan RS, McEvoy LK, Thompson WK, Holland D, Brewer JB, Aisen PS, Sperling RA, Dale AM Alzheimer's Disease Neuroimaging Initiative. Amyloid-β--associated clinical decline occurs only in the presence of elevated P-tau. Arch Neurol. 2012;69:709–713. doi: 10.1001/archneurol.2011.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desikan RS, McEvoy LK, Thompson WK, Holland D, Roddey JC, Blennow K, Aisen PS, Brewer JB, Hyman BT, Dale AM Alzheimer's Disease Neuroimaging Initiative. Amyloid-β associated volume loss occurs only in the presence of p-tau. Ann Neurol. 2011;70:657–661. doi: 10.1002/ana.22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desikan RS, McEvoy LK, Holland D, Thompson WK, Brewer JB, Aisen PS, Andreassen OA, Hyman BT, Sperling RA, Dale AM for the Alzheimer's Disease Neuroimaging Initiative. Apolipoprotein E {varepsilon}4 Does Not Modulate Amyloid-β-Associated Neurodegeneration in Preclinical Alzheimer Disease. AJNR Am J Neuroradiol. 2013;34:505–510. doi: 10.3174/ajnr.A3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutherland GT, Siebert GA, Kril JJ, Mellick GD. 2011 Knowing me, knowing you: can a knowledge of risk factors for Alzheimer's disease prove useful in understanding the pathogenesis of Parkinson's disease? J Alzheimers Dis. 2011;25(3):395–415. doi: 10.3233/JAD-2011-110026. [DOI] [PubMed] [Google Scholar]
- 45.Myers AJ, Kaleem M, Marlowe L, Pittman AM, Lees AJ, Fung HC, Duckworth J, Leung D, Gibson A, Morris CM, de Silva R, Hardy J. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum Mol Genet. 2005;14:2399–2404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 46.Charlesworth G, Gandhi S, Bras JM, Barker RA, Burn DJ, Chinnery PF, Gentleman SM, Guerreiro R, Hardy J, Holton JL, Lees A, Morrison K, Sheerin UM, Williams N, Morris H, Revesz T, Wood NW. Tau acts as an independent genetic risk factor in pathologically proven PD. Neurobiol Aging. 2012;33:838. doi: 10.1016/j.neurobiolaging.2011.11.001. e7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.