Highlights
-
•
Alginate inhibits proteolytic activity of pepsin but not trypsin.
-
•
Level of pepsin inhibition correlates with alginate F[M].
-
•
An in vitro model gut system was used to model alginate inhibition of proteolysis.
-
•
Proteolysis inhibited in gastric phase of digestion, but not small intestinal phase.
-
•
pH dependent ionic interactions reduce substrate availability at pH 2.5.
Keywords: Alginate, Pepsin, Trypsin, Proteolysis, Model gut system
Abstract
Alginates are widely used in the food and medical industries, including as a Gastro-Oesophagul Reflux treatment. This work investigates the inhibitory effects of alginate on the reflux aggressors trypsin and pepsin and the role of alginate-substrate binding, pH and alginate structure on inhibition. Alginates were shown to reduce pepsin activity by up to 53.9% (±9.5SD) in vitro. Strong positive correlation between alginate mannuronate residue frequency and levels of pepsin inhibition was observed. Limited inhibition of trypsin was shown. Viscometric observations of pH dependent interactions between alginate and protein suggest a mechanism whereby pH dependent ionic interactions reduce substrate availability to enzyme at acidic pH. To understand how dietary protein digestion is affected by alginate, proteolytic digestion was investigated in an in vitro model of the upper digestive tract. Significant inhibition of proteolysis was shown in the gastric phase of digestion, but not the small intestinal phase.
1. Introduction
Alginates have previously been shown to inhibit pepsin activity in vitro, Sunderland, Dettmar, and Pearson (2000) showed alginates could inhibit pepsin activity by 52% in vitro. Strugala, Kennington, Campbell, Skjak-Braek, and Dettmar (2005), showed significant correlations between alginate structure and levels of inhibition, with high frequency of mannuronic acid residues (F[M]) alginates tending to inhibit better than those high in guluronic acid residues (F[G]). However the mechanism of pepsin inhibition is poorly understood, as is how alginate effects dietary proteolysis and the activity of small-intestinal proteases such as trypsin.
Pepsin has been shown to be a major aggressor in GORD (Gastro-Oesophagul Reflux disease) (Strugala, Avis, Jolliffe, Johnstone, & Dettmar, 2009) with in vivo addition of pepsin to the oesophagus resulting in reflux-like oesophagitis in animal models (Goldberg, Dodds, Gee, Montgomery, & Zboralske, 1969). Alginates are used in the treatment of reflux (Sunderland et al., 2000). The primary mechanism of alginate treatment of reflux is the formation of an alginate raft upon coming into contact with stomach acid; sodium or potassium bicarbonate in the formulation releases carbon dioxide which becomes trapped in the gel as a foam, allowing the alginate acid-gel raft to float on the top of the stomach contents and create a physical barrier to refluxate (Dettmar et al., 2006, Sengupta et al., 2015). The inhibitory action of alginate on pepsin is thought to be a secondary mechanism for the anti-reflux activity of alginate based agents (Strugala et al., 2009).
The damaging potential of the pancreatic protease trypsin has also been demonstrated in gastroduodenal refluxate. With the use of proton pump inhibitors in the treatment of reflux disease, gastric pH becomes elevated. During a gastro-duodenal reflux event, if trypsin passes through the stomach at pH 4.0 or above (or rapidly through at low pH of 2 or less) it can retain proteolytic activity and cause damage (Pearson et al., 2011). We therefore seek to investigate the effects of alginate on trypsin activity and examine a potential role of alginate in the management of trypsin mediated damage in gastro-duodenal reflux.
Furthermore, alginates are widely used in the food and medical industries and have a range of bioactive properties, as reviewed elsewhere (Brownlee et al., 2005). Alginates have been shown to be potent inhibitors of pancreatic lipase activity and are being investigated as a potential tool for the management of obesity (Balasubramaniam et al., 2013). It is therefore important to understand the secondary effects alginate may have on protein digestion.
In this work we therefore seek to investigate the inhibitory action of alginate on trypsin and pepsin in vitro and to characterise the effects of varying alginate structure on any inhibition observed. We also investigate the pH dependency of viscometric interactions between alginate and protein substrates. Finally the inhibitory effects of alginate on protein digestion are investigated in a model gut system.
2. Methods
2.1. Materials
All alginate samples tested were kindly supplied by FMC Biopolymer and Technostics Ltd (Hull, UK). Bovine serum albumin (BSA) was purchased from VWR Jencons. Unless otherwise stated, all other chemicals and reagents were purchased from Sigma–Aldrich (Poole, UK). The structures of the alginate samples were characterised by 13C NMR neighbour diad analysis and the full characteristics of all samples are shown in Table 1.
Table 1.
Codes and molecular characteristics for alginates used in this study presented alongside levels of pepsin and trypsin inhibition. F(G) is the fraction of the alginate polymer composed of guluronate and F(M) the fraction of mannuronate. n(G > 1) is the number of consecutive guluronate residues above 1.
| F(G) | F(M) | F(GG) | F(MM) | F(MGM) | F(GGG) | n(G > 1) | Pepsin inhibition |
Trypsin inhibition |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 mg/ml | 2.5 mg/ml | 1.25 mg/ml | 5 mg/ml | 2.5 mg/ml | 1.25 mg/ml | ||||||||
| FMC 2 | 0.66 | 0.34 | 0.54 | 0.23 | 0.076 | 0.51 | 15.47 | 26.00 ± 8.78 | 19.99 ± 7.46 | −3.07 ± 13.75 | 11.46 ± 6.54 | 2.74 ± 2.86 | 1.53 ± 3.02 |
| FMC 3 | 0.68 | 0.32 | 0.57 | 0.21 | 0.069 | 0.53 | 14.66 | 11.38 ± 10.63 | 3.95 ± 13.84 | −2.43 ± 8.87 | 4.39 ± 3.12 | 7.83 ± 1.48 | 5.88 ± 6.64 |
| FMC 4 | 0.51 | 0.49 | 0.34 | 0.32 | 0.124 | 0.3 | 8.97 | 25.46 ± 8.69 | 17.80 ± 6.19 | 10.96 ± 5.67 | 4.92 ± 6.55 | 7.68 ± 2.55 | 3.52 ± 6.13 |
| FMC 5 | 0.53 | 0.47 | 0.36 | 0.31 | 0.123 | 0.32 | 9.67 | 33.95 ± 5.25 | 25.20 ± 6.92 | 12.78 ± 8.99 | 10.44 ± 1.99 | 7.45 ± 2.59 | 7.87 ± 2.68 |
| FMC 6 | 0.52 | 0.48 | 0.35 | 0.31 | 0.122 | 0.3 | 8.15 | 19.23 ± 7.81 | 13.06 ± 9.93 | 1.91 ± 9.78 | 3.85 ± 3.41 | 8.25 ± 5.77 | 3.54 ± 3.87 |
| FMC 7 | 0.42 | 0.58 | 0.24 | 0.4 | 0.133 | 0.2 | 6.47 | 29.43 ± 5.44 | 16.89 ± 6.22 | 11.78 ± 6.48 | 7.29 ± 2.91 | 5.60 ± 0.79 | −1.67 ± 2.49 |
| FMC 9 | 0.45 | 0.55 | 0.26 | 0.36 | 0.136 | 0.2 | 5.78 | 42.57 ± 5.25 | 24.73 ± 8.84 | 14.54 ± 10.16 | −10.31 ± 10.99 | −10.02 ± 25.23 | −4.31 ± 12.13 |
| FMC 10 | 0.42 | 0.58 | 0.21 | 0.37 | 0.14 | 0.14 | 3.96 | 39.41 ± 6.71 | 13.38 ± 14.73 | 2.52 ± 11.24 | −1.70 ± 12.13 | 5.37 ± 5.24 | −1.13 ± 9.47 |
| FMC 12 | 0.35 | 0.65 | 0.19 | 0.49 | 0.111 | 0.13 | 4.54 | 38.13 ± 4.52 | 20.59 ± 10.28 | 11.79 ± 6.57 | −2.83 ± 11.61 | 0.35 ± 9.64 | 4.97 ± 17.73 |
| FMC 13 | 0.34 | 0.66 | 0.17 | 0.49 | 0.124 | 0.12 | 4.63 | 45.99 ± 5.97 | 31.42 ± 10.03 | 13.63 ± 4.92 | 4.23 ± 8.16 | 2.59 ± 13.33 | 2.72 ± 11.59 |
| LF120 | 0.424 | 0.576 | 0.24 | 0.391 | 0.156 | 0.183 | 4.7 | 44.73 ± 10.98 | 28.46 ± 10.68 | 1.21 ± 7.74 | 6.22 ± 7.71 | 3.79 ± 10.77 | 3.50 ± 6.66 |
| LFR560 | 0.633 | 0.367 | 0.505 | 0.239 | 0.096 | 0.45 | 9.9 | 32.42 ± 5.31 | 18.34 ± 6.49 | 9.94 ± 7.85 | −6.76 ± 11.41 | 0.77 ± 10.58 | 0.25 ± 8.08 |
| LF10L | 0.45 | 0.553 | 0.257 | 0.362 | 0.153 | 0.19 | 4.4 | 42.66 ± 6.23 | 22.84 ± 5.84 | 8.18 ± 7.93 | −5.86 ± 8.66 | −4.38 ± 7.84 | −9.05 ± 6.65 |
| H120L | 0.45 | 0.551 | 0.276 | 0.379 | 0.15 | 0.22 | 5.9 | 46.09 ± 9.53 | 24.37 ± 11.90 | 9.81 ± 6.54 | 3.07 ± 5.70 | 4.66 ± 2.60 | 2.53 ± 5.56 |
| SF120 | 0.664 | 0.336 | 0.545 | 0.218 | 0.083 | 0.484 | 9.6 | 21.93 ± 5.42 | 8.83 ± 5.56 | 0.45 ± 5.31 | −7.20 ± 7.96 | 2.23 ± 4.00 | 10.05 ± 5.19 |
| SF200 | 0.68 | 0.322 | 0.573 | 0.218 | 0.079 | 0.537 | 16.7 | 13.10 ± 6.03 | 15.93 ± 6.37 | 14.11 ± 13.58 | −0.89 ± 2.46 | −1.35 ± 3.10 | −5.52 ± 2.53 |
| SF/LF | 0.66 | 0.336 | 0.548 | 0.22 | 0.081 | 0.506 | 13.8 | 15.56 ± 5.40 | 4.08 ± 6.86 | −5.26 ± 6.79 | 0.90 ± 7.99 | −2.88 ± 10.97 | −1.73 ± 2.90 |
| SF60L | 0.411 | 0.589 | 0.219 | 0.393 | 0.155 | 0.133 | 3.3 | 43.87 ± 7.36 | 15.50 ± 8.88 | 4.12 ± 11.67 | −2.11 ± 14.83 | −2.05 ± 17.15 | −9.14 ± 23.66 |
2.2. N-Terminal proteolytic assay
Proteolytic activity was determined using an adapted version of the N-terminal assay developed by Lin, Means, and Feeney (1969) and modified by Hutton, Allen, Pearson, Ward, and Venables (1986). The activity assay was scaled down to a 96-well microplate as reported in Ali, Parikh, Chater and Pearson (2013). Alginate was prepared in 0.05 M phosphate (pH 2.5) as alginate was shown to alter the pH of the reaction mixture when made up in distilled water as per Strugala et al. (2005).
For pepsin activity, 10 μg/ml pepsin (prepared 10 min prior to assay) and 10 mg/ml succinyl albumin were each made up in 0.05 M phosphate buffer at pH 2.5. In order to prevent precipitation, alginate samples were prepared at 10 mg/ml in the basic side of the buffer and then diluted at a 1:1 ratio with the acidic side of the buffer to give a concentration of 5 mg/ml in 0.05 M phosphate buffer. Trinitrobenzosulfonic acid (TNBS) was prepared at 2 μl/ml in deionised water. Sodium bicarbonate was prepared 10% w/v. Pentosan polysulphate (SP54) at 5 mg/ml was used as a positive inhibition control (Bianchi and Cook, 1964, Cook and Drill, 1967).
For trypsin activity assay, 5 μg/ml trypsin (prepared 10 min prior to assay) and 10 mg/ml succinyl albumin were each made up in 0.066 mM Sorensen's phosphate buffer at pH 7. Alginate samples were prepared at 5 mg/ml in 0.066 M Sorensen's phosphate buffer. Trinitrobenzosulfonic acid (TNBS) and sodium bicarbonate were prepared as above. Soybean trypsin inhibitor was used as a positive inhibition control.
Eighteen alginates were tested for their ability to inhibit pepsin and trypsin activity. All alginate samples were tested at three concentrations; 5, 2.5 and 1.25 mg/ml. This gave concentrations in the reaction mixture of 1.36, 0.68 and 0.34 mg/ml, respectively.
Thirty microlitres of alginate sample was pre-incubated with 50 μl succinyl albumin substrate for 60 min on a shaker. At T0 30 μl enzyme solution or buffer blank was added as appropriate and the plate was incubated for 30 min at 37 °C. After 30 min, 50 μl sodium bicarbonate and 50 μl TNBS was added, mixed and the plate was incubated for 15 min at 55 °C. At T45, 50 μl SDS (10% w/v) and 50 μl 1 M hydrochloric acid were added and the plate was left to stand until all wells had stopped effervescing, and absorbance was measured at 340 nm using a Biotek 96 well microplate reader (Elx808 Biotek, Bedfordshire, UK).
To calculate percentage pepsin inhibition the following formula was used:
2.3. Alginate-protein viscosity interactions
Alginates were made up at 2.5 mg/ml as control or with casein or BSA at 10 mg/ml in deionised water and titrated across the pH 1.5–9 using HCl and NaOH. Samples were incubated at room temperature for 30 min and then left to stand for a further 30 min to allow precipitate to settle. Two millilitres of the sample supernatent was decanted, and viscosity was measured.
Sample viscosity was measured using a cup and bob viscometer (Contraves, Switzerland). All samples were tested in triplicate and relative viscosity (ηrel) was calculated from percentage deflection. Specific viscosity (ηsp) was then calculated from relative viscosity.
2.4. Model gut
Model gut analysis was conducted in an artificial simulation of the upper GI tract. The system was set up and solutions prepared as described in Houghton et al. (2014). Analysis of protein digestion was conducted as described below.
2.4.1. Protein digestion
In order to distinguish effects on protein digestion from the gastric and pancreatic phases of digestion, gastric and pancreatic proteolysis assays are described separately.
2.4.2. Gastric protein digestion
In the gastric phase 0.5 g BSA is added to the salivary diluents at T[−10] and the assay was run until the end of the gastric phase at T[60].
2.4.3. Small intestinal protein digestion
One gram of BSA was added to synthetic saliva at T[−10] and gastric pepsin was omitted from the gastric diluent to prevent any protein digestion in the gastric phase.
2.4.4. Background control
For background controls 10 ml synthetic saliva was prepared without substrate BSA.
2.4.5. Proteolysis analysis
Undigested polypeptides were removed from samples by trichloroacetic acid (TCA) precipitation and centrifugation. Protein digestion was measured by assaying amino acids and short oligopeptides remaining in the supernatant with the Pierce BCA Total Protein assay kit.
The Pierce BCA assay is known to under-report amino acid and oligo-peptide metabolites of protein digestion (Wiechelman, Braun, & Fitzpatrick, 1988). The authors observed that 37.76% of BSA is reported in the BCA assay after exhaustive overnight digestion with pepsin at pH2 (1 mg/ml) and trypsin at pH7 (1 mg/ml) at 37 °C (this can be corrected for by a factor of 2.65). It was also shown that due to bile binding of protein metabolites, only 60.33% of digested protein is detected (this can be corrected for by a factor of 1.66 in the small intestinal phase, resulting in a correction factor of 4.4 in the small intestinal phase to correct for both the under-reporting and bile binding effects) (data not included).
2.5. Statistical analysis
All data were analysed using GraphPad Prism 4 statistical software. Statistical analyses of levels of inhibition compared to control was done using a Student's t-test with a significance level of p = 0.05. All samples were tested in triplicate and are reported as mean and standard deviation.
3. Results
3.1. In vitro protease activity assay
At pH 2.5, all alginates tested showed the ability to inhibit pepsin activity at the highest concentration of 5 mg/ml (Table 1). Average pepsin activity was reduced by 6.8% ± 6.1 SD at 1.25 mg/ml, by 18.3% ± 7.5 at 2.5 mg/ml and by 31.9% ± 6.1 at 5 mg/ml. The strongest inhibitor at 5 mg/ml was H120L which has an F[G] of 0.45 and reduced pepsin activity to 53.9% ± 9.5 of control activity. The weakest inhibitor at 5 mg/ml was FMC3 which has an F[G] of 0.68 and reduced pepsin activity to 88.6% ± 10.6 SD of control activity. Full characteristics for alginates are shown in Table 1.
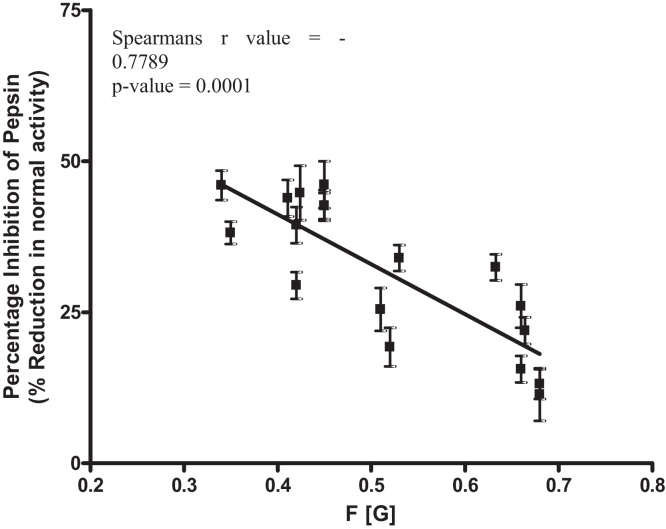
Alginate structure was correlated with percentage pepsin inhibition. A significant negative correlation was shown between pepsin inhibition and alginate F[G] at 5 mg/ml (Fig. 1). This indicates that at 5 mg/ml, an increasing proportion of mannuronic acid residues, and decreasing proportion of guluronic acid residues yielded higher levels of pepsin inhibition.
Fig. 1.
Correlation of alginate G-residue frequency (F[G]) and level of pepsin inhibition with 5 mg/ml alginate. Pepsin activity is shown as a percentage of control pepsin activity. The error bars show the standard deviation of six replicates (n = 6). The line of best fit indicates a negative correlation which is significant with a Spearman r value of −0.7789 and a p value of 0.0001.
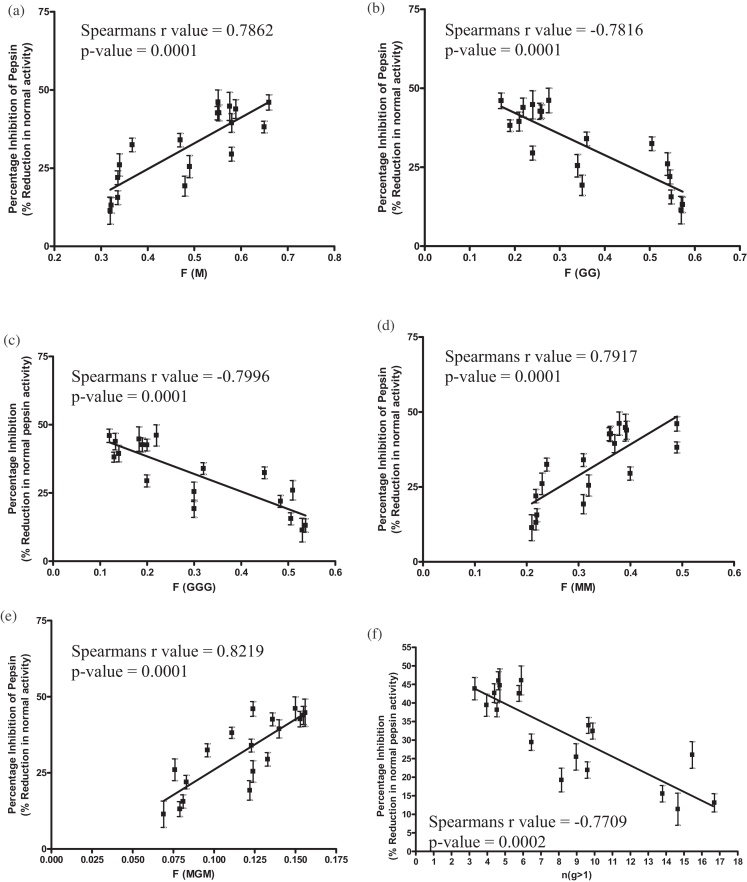
The structure and biophysical properties of alginates are not just dictated by F[G] frequency, but also by the arrangement of contiguous blocks of M and G residues. Levels of pepsin inhibition were compared against the frequency of the structural patterns; F[M], F[GG], F[MM], F[GGG], F[MGM] and F[GM/MG] and also against n(G > 1), the G-block length (Fig. 2). Similar significant relationships between structure and inhibition were observed, where higher levels of mannuronic acid brought about significantly higher levels of pepsin inhibition. As the G-block length increases, a significant reduction in the inhibition of pepsin was observed, showing a negative correlation between inhibition and n(G > 1).
Fig. 2.
Correlation of alginate structural patterns; (a) F[M], (b) F[GG], (c) F[GGG], (d) F[MM], (e) F[MGM], and (f) n(G > 1) the G-block length against level of pepsin inhibition with 5 mg/ml alginate. Pepsin activity is shown as a percentage of control pepsin activity. The error bars show the standard deviation of six replicates (n = 6).
Molecular weight data was only available for eight of the alginates tested. Alginates can vary greatly in molecular weight and the Technostics alginates used in this study ranged from 34,700 to 387,000 Da. However no significant correlation between molecular weight and pepsin inhibition could be demonstrated as shown in (data not shown).
Four of the 19 samples had small but significant effects on trypsin activity. FMC 5 was the only alginate sample to show a significant inhibition of trypsin activity at all three concentrations, 10.4% ± 2 at 5 mg/ml, 7.5% ± 2.5 at 2.5 mg/ml and 7.9% ± 2.7 at 1.25 mg/ml (Table 1). FMC3 and FMC4 showed a decrease of 7.8% ± 1.5 and 7.7% ± 2.6, respectively, at 2.5 mg/ml. However neither alginate showed a significant affect at the other concentrations tested. FMC 7 showed significant inhibition at 5 and 2.5 mg/ml of 7.3% ± 2.9 and 5.6% ± 0.8, respectively, however showed no significant affect at 1.25 mg/ml.
3.2. pH dependent alginate–protein interactions
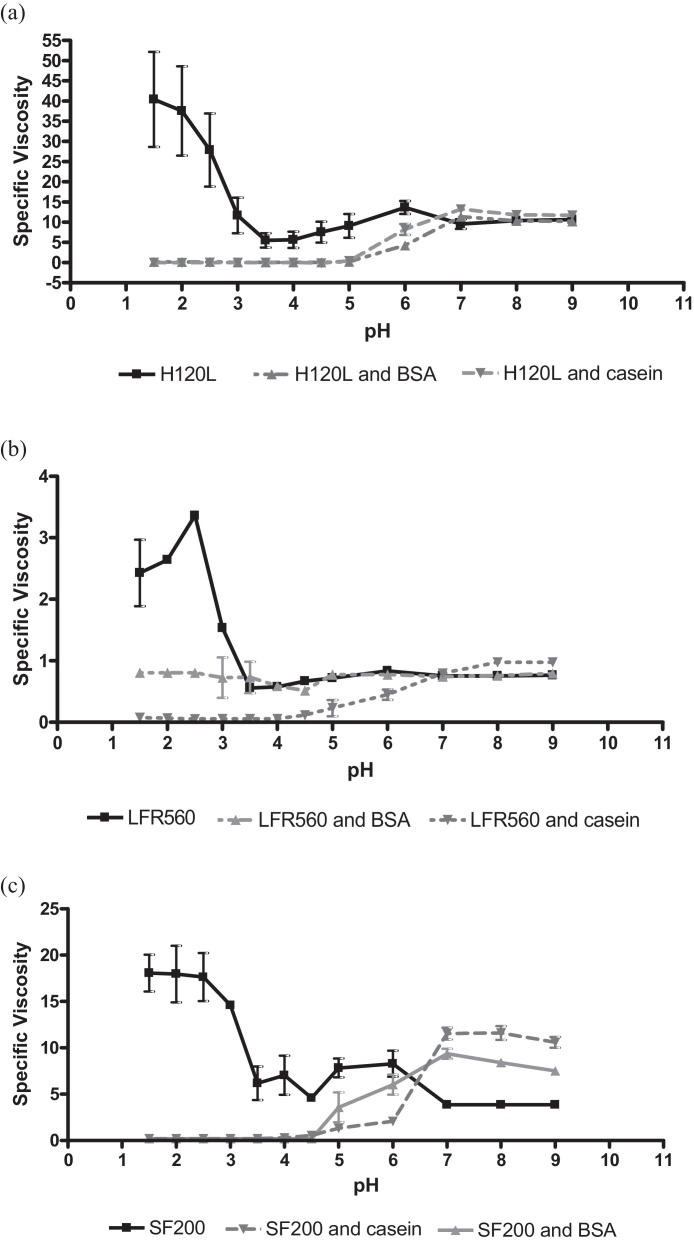
In the control alginate solution the samples showed behaviour typical of a pH dependent gel with specific viscosity increasing at lower pH's as an acid gel was formed. Addition of BSA or casein to the mixture caused a visible precipitate to form in samples titrated to acidic pH, but not in samples at neutral pH. From the pH dependent specific viscosity plot of H120L and BSA/Casein in Fig. 3a, at a pH around neutral there was little or no difference in viscosity of supernatant with the addition of BSA or casein. However in the samples with BSA and casein present, at lower pHs the viscosity of the supernatant approached zero as a precipitate had formed between alginate and the protein which has settled to the bottom of the tube, bringing the viscous alginate component out of solution.
Fig. 3.
(a–c) pH dependent viscosity interaction of alginates with BSA (10 mg/ml) and casein (10 mg/ml) across the pH range (n = 3). (a) H120L (2.5 mg/ml) molecular weight = 397,000, (b) LFR560 (2.5 mg/ml) molecular weight = 34,700, (c) SF200 (2.5 mg/ml) molecular weight = 387,000.
Four alginates were tested in this way. SF120L behaved in a similar manner as H120L (data not included). As can be seen from Fig. 3b, LFR560 had a maximum specific viscosity of between 3 and 4 as compared to the other alginates with maximum specific viscosities in the range 15–45. This is due to LFR560 being a low molecular weight alginate (34,700 Da) as compared to alginates H120L, SF120 and SF200 with molecular weights up to an order of magnitude larger (195,000–397,000).
In all samples the same formation of precipitate and lower supernatant viscosity was observed. In the case of SF200, at neutral pH the addition of BSA and casein to the mixture caused an approximate doubling of supernatant viscosity suggesting a synergistic interaction between alginate SF200 and protein at neutral pH (Fig. 3c). The interaction between SF200 and BSA/casein at lower pH was consistent with the other tested samples.
LFR560 was the only sample in which the sample retained some of its viscosity at low pH after the addition of BSA, this suggests that there is an interaction by which some alginate remains. This result may indicate a size-dependent interaction between BSA and alginate, although LFR560 was the only low molecular weight alginate available for testing.
3.3. Protein digestion in the model gut
3.3.1. Gastric protein digestion
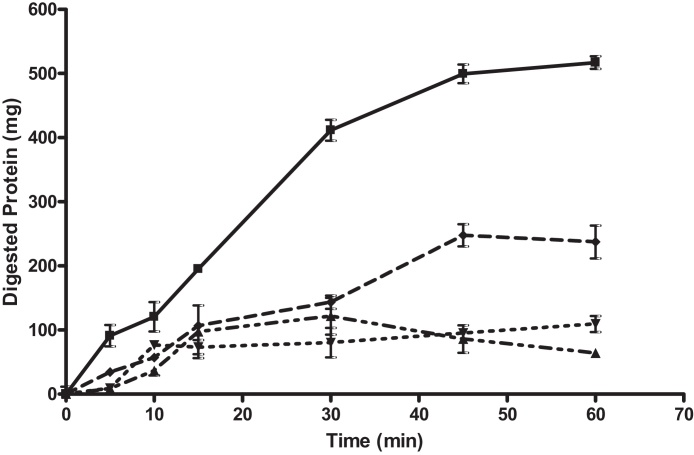
Pentosan polysulphate (SP54) was used as the positive inhibition control. At all tested concentrations of 50, 100 and 200 mg pentosan polysulphate there was significant inhibition of gastric proteolysis at all time points from T[5] onwards (Fig. 4) At T[5], 50, 100 and 200 mg of pentosan polysulphate significantly inhibited the gastric digestion of protein by 62.5% (P = 0.005), 90.1% (P = 0.003) and 90.5% (P = 0.002), respectively. At T[60] by the end of the gastric phase, 50, 100 and 200 mg of pentosan polysulphate significantly inhibited the gastric digestion of protein by 54.1% (P = 0.0001), 78.9% (P = 0.001) and 87.6% (P = 0.0004), respectively.
Fig. 4.
Bovine serum albumin digestion in gastric phase of a model gut system with and without SP54. The graph shows total protein recovered from model gut system after TCA (trichloroacetic acid) precipitation to stop enzyme activity and remove undigested polypeptides. 0.5 g BSA was digested alone (control digestion) and in the presence of varying concentrations of SP54. Control digestion is represented as (■) and digestion with SP54 at 125 mg as (♦), 250 mg (▾) and 500 mg (▴). All samples were tested in triplicate, errors are shown as standard deviation. At T[60] by the end of the gastric phase, 125, 250 and 500 mg of pentosan polysulphate significantly inhibited the gastric digestion of protein by 54.1% (P = 0.0001), 78.9% (P = 0.001) and 87.6% (P = 0.0004), respectively (t-test).
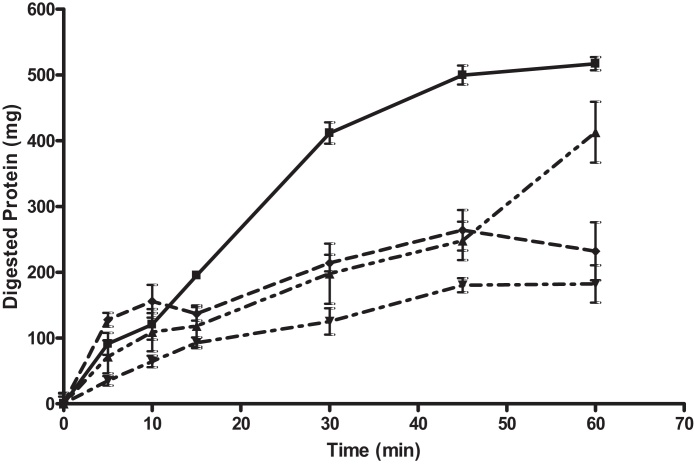
Four alginates from across the F[G] range were tested, and all FMC3, FMC13, SF120 and H120L produced significant inhibition of protein digestion in the gastric phase. By the end of the simulated gastric phase, FMC13 was the weakest of the four alginates tested (Fig. 5). By T[60] after an hour of simulated digestion with 125, 250 and 500 mg of FMC13, proteolytic digestion was reduced by 23.4% (P = 0.021), 52.2% (P = 0.04) and 43.5% (P = 0.013), respectively, as compared to a control.
Fig. 5.
Bovine serum albumin digestion in gastric phase of a model gut system with and without FMC13 alginate. The graph shows total protein recovered from model gut system after TCA (trichloroacetic acid) precipitation to stop enzyme activity and remove undigested polypeptides. 0.5 g BSA was digested alone (control digestion) and in the presence of varying concentrations of FMC13. Control digestion is represented as as (■) and digestion with FMC13 at 125 mg as (♦), 250 mg (▾) and 500 mg (▴). All samples were tested in triplicate, errors are shown as standard deviation. At T[60] by the end of the gastric phase, 125, 250 and 500 mg of FMC13 alginate significantly inhibited the gastric digestion of protein by 23.4% (P = 0.021), 52.2% (P = 0.040) and 43.5% (P = 0.013), respectively (t-test).
At timepoints T[30] and T[45] there was a larger percentage inhibition as compared to control. At T[30] with 125, 250 and 500 mg of FMC13 proteolytic digestion was reduced by 52.8% (P = 0.004), 75.7% (P = 0.004) and 62.7 (P = 0.001), respectively. At T[45] after 45 min of simulated digestion with 125, 250 and 500 mg of FMC13 proteolytic digestion by 52.8% (P = 0.004), 70.9% (P = 0.002) and 73.06 (P = 0.018).
FMC3 showed a similar inhibition profile to FMC13 (Fig. 6). At T[30], protein digestion was reduced by 51.9% (P = 0.0002), 69.6% (P = 0.013) and 48.0% (P = 0.016) with 125, 250 and 500 mg of FMC3 alginate, respectively. At T[45], protein digestion was reduced by 50.4% (P = 0.005), 64.0% (P = 0.015) and 47.2% (P = 0.0004) as compared to control with 125, 250 and 500 of FMC3 alginate, respectively. By the final timepoint at T[60], protein digestion was reduced by 20.2% (P = 0.029), 64.8% (P = 0.024) and 55.1% (P = 0.035) as compared to control with 125, 250 and 500 mg of FMC3 alginate, respectively.
Fig. 6.
Bovine serum albumin digestion in gastric phase of a model gut system with and without FMC3 Alginate. The graph shows total protein recovered from model gut system after TCA (trichloroacetic acid) precipitation to stop enzyme activity and remove undigested polypeptides. 0.5 g BSA was digested alone (control digestion) and in the presence of varying concentrations of FMC3. Control digestion is represented as as (■) and digestion with FMC3 at 125 mg as (▴), 250 mg (▾) and 500 mg (♦). All samples were tested in triplicate, errors are shown as standard deviation. At T[60] by the end of the gastric phase, 125, 250 and 500 mg of FMC3 alginate significantly inhibited the gastric digestion of protein by 52.8% (P = 0.004), 70.9% (P = 0.001499) and 73.06 (P = 0.01846) respectively (t-test).
With SF120 at T[15], the highest levels of inhibition were achieved with the highest concentration of 500 mg SF120 (Fig. 7). At T[30], protein digestion was reduced by 35.4% (P = 0.010), 47.3% (P = 0.033) and 62.1% (P = 0.002) as compared to control with 125, 250 and 500 mg of SF120 alginate, respectively. At T[45], protein digestion was reduced by 60.8% (P = 0.003), 37.5% (P = 0.003) and 70.2% (P = 0.019) with 125, 250 and 500 mg of SF120 alginate respectively. By the final timepoint at T[60], protein digestion was reduced by 32.9% (P = 0.0025), 30.8% (P = 0.007) and 50.5% (P = 0.001) with 125, 250 and 500 mg of SF120 alginate, respectively.
Fig. 7.
Bovine serum albumin digestion in gastric phase of a model gut system with and without SF120 alginate. The graph shows total protein recovered from model gut system after TCA (trichloroacetic acid) precipitation to stop enzyme activity and remove undigested polypeptides. 0.5 g BSA was digested alone (control digestion) and in the presence of varying concentrations of SF120. Control digestion is represented as as (■) and digestion with FMC3 at 125 mg as (▴), 250 mg (▾) and 500 mg (♦). All samples were tested in triplicate, errors are shown as standard deviation. At T[60] by the end of the gastric phase, 125, 250 and 500 mg of SF120 alginate significantly inhibited the gastric digestion of protein by 32.9% (P = 0.0025), 30.8% (P = 0.007) and 50.5% (P = 0.001), respectively (t-test).
At higher concentrations of alginate H120L (250 and 500 mg) there was a significant increase in the rate of protein digestion at T[5], and with 500 mg an increase also at T[10] (data not shown). At T[5] there was an increase in protein digestion of 71.8% with 250 mg H120L and of 154% with 500 mg, although neither of these increases were statistically significant. At T[10] there was an increase in the digested protein yield of 145% with 500 mg H120L, which was statistically significant (P = 0.038). From T[30] onwards the data for H120L follows a similar trend to what was seen with the other alginate samples, whereby there is a reduced level of protein digestion at all timepoints for all alginates.
3.3.2. Pancreatic phase protein digestion
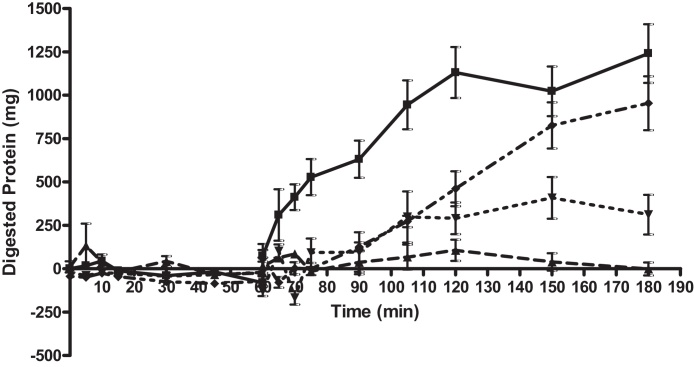
Fig. 8 shows a control digestion of 1 g of BSA in the model gut system with soybean trypsin inhibitor (SBTI) was used as the positive inhibition control. Three concentrations of SBTI were tested in the model gut in order to validate the detection of inhibition of proteolysis. The addition of SBTI to the digestive mixture reduced proteolytic activity and reduced the amount of digested protein recovered from the assay in the small intestinal phase. From T[70] onwards the addition of 250 and 500 mg yielded statistically significant inhibition at all timepoints until the end of the assay. Inhibition levels of 23%, 75% and 100% were observed with 125, 250 and 500 mg of SBTI.
Fig. 8.
Bovine serum albumin (BSA) digestion in the small-intestinal phase of a model gut system with and without SBTI. The graph shows total protein recovered from model gut system after TCA (trichloroacetic acid) precipitation to stop enzyme activity and remove undigested polypeptides. 1 g BSA was digested alone (control digestion) and in the presence of varying concentrations of SBTI. Control digestion is represented as as (■) and digestion with SBTI at 125 mg as (♦), 250 mg (▾) and 500 mg (▴). All samples were tested in triplicate, errors are shown as standard deviation. With 500 mg SBTI, from T[75] until T[180] inhibition of proteolytic activity was between 90.6 and 100% and statistically significant at all time-points (p < 0.05). With 250 mg SBTI, statistically significant inhibition of over 60.1% was achieved at all timepoints after T[70] (p < 0.05). With 125 mg of SBTI, statistically significant inhibition of proteolytic digestion was achieved between T[70] and T[120] ranging from 59.25 to 100% (p < 0.05), however at T[150] and T[180] the reduction in protein digestion relative to control could not be shown to be statistically significant.
Four alginates were tested for their effects on protein digestion in the small-intestinal phase of the model gut system. While there were variations in levels of protein digestion with the addition of alginate, none of these deviations from the relative control time-point could be shown to be statistically significant (data not shown).
4. Discussion
Alginates have been shown to have the ability to modify the activity of multiple digestive enzymes in vitro and affect the digestion profile of major macronutrients. Some of these functional effects have been shown to be linked to structural characteristics of alginates (Sunderland et al., 2000, Wilcox, 2010, Wilcox, et al. 2014).
Through the use of an N-terminal proteolysis assay it was possible to determine that alginate was a potent inhibitor of pepsin activity, but had no significant inhibitory effect on trypsin.
The high F[M] alginate H120L reduced pepsin activity to the highest extent and it was shown that the potency of inhibition correlated with alginate structure. A strong positive correlation between alginate F[M] and levels of pepsin inhibition, supported the findings of Strugala et al. (2005). And we have shown that an increasing proportion of contiguous G-blocks was shown to be negatively associated with inhibition of pepsin; [n(G > 1)], F[GG] and F[GGG] all negatively correlating with pepsin inhibition.
Only a small number of the tested alginate samples were observed to have had a statistically significant inhibition of trypsin. The catalytic mechanisms of pepsin and trypsin are distinct, it is therefore possible that alginate is able to interact with and disrupt the catalytic mechanism of pepsin, but not of trypsin. Pepsins are aspartate proteases, and broad specificity endopeptidases with a preferance for cleavage between hydrophobic amino acids (Powers, Harley, & Myers, 1977). Trypsin on the other hand is a serine protease. Serine proteases are usually endopeptidases and preferentially cleave within the poplypeptide chain, prefererentially cleaving on the carboxyl side of lysine and arginine (Hedstrom, 2002a, Hedstrom, 2002b).
In pepsin mediated proteolysis, the two aspartate residues (Asp32 and 215 in pig pepsin) form an acid base pair in the active site cleft, holding a water molecule which facilitates nucleophillic attack on the peptide bond. The extensive hydrogen bonding network is required to maintain the basic Asp32 in the COO− state. Nucleophillic attack by the water molecule on the peptide bond NH–CO generates –NH2 and –COOH (Szecsi, 1992).
Looking at the mechanism of other pepsin inhibitors can be instructive of how alginate may inhibit pepsin inhibition. Pepstatin is a linear peptide inhibitor of aspartic proteases including pepsin, it is a competitive pepsin inhibitor which blocks the active site by forming a network of hydrogen bonds and charge–charge interactions with active-site residues (El Aouad et al., 2011). The inhibitor complexes with the enzyme and prevents substrate binding (Fujinaga, Chernaia, Tarasova, Mosimann, & James, 1995).
Mannuronic and guluronic acid residues are rich in hydroxyl groups which would be capable of forming hydrogen bond interactions with these same active site residues. The formation of these hydrogen bonds is likely to rely on the flexibility of the alginate chains in solution and the report (Smidsrod & Skjak-Braek, 1990) that GG rich alginates are the least flexible and MG rich the most could explain why GG rich alginates are the worst inhibitors. Furthermore the C O group of the carboxyl group is able to participate in hydrogen bonding, and to a lesser extent form charge–charge interactions.
The idea of a direct inhibitory interaction between alginate and pepsin was also argued by Sunderland et al. (2000) who showed in an alginate–pepsin centrifugation experiment that pepsin was pulled out of the solution by alginate upon centrifugation. This suggested direct binding of pepsin as a possible mechanism of inhibition.
Carboxyl groups have been shown to be important in the inhibition of lipase by pectin. This provides an example of how alginate may inhibit pepsin activity directly (Isaksson, Lundquist, & Ihse, 1982). The carboxyl groups of pectin are believed to be involved in the protanation of active site serine residue of the lipase enzyme. The protonation of the hydroxyl group of serine blocks the initiation of this charge relay system, thereby inactivating the enzyme (Wilcox, 2010). The importance of carboxyl groups to pectin inhibition of lipase has been shown as increasing levels of methyl esterification are correlated with reduced lipase inhibition. As it is the carboxyl group that becomes esterified, an increase in methyl esterification necessarily means a decrease in the number of carboxyl groups. Similar to pectins, alginates are rich in carboxylic acid groups.
Lipase and trypsin share similar active site mechanisms and similar pH optima, however alginate is able to inhibit the action of pancreatic lipase, but not trypsin. All trypsin enzymes have a negatively charged substrate binding pocket, and bind basic positively charged amino acids. As alginates are large negatively charged polymers, they would be repelled from the trypsin substrate binding site due to charge:charge repulsion and have poor accessibility to the active site binding pocket due to size (Katona, Berglund, Hajdu, & Graaf, 2002). While trypsin binding sites and alginate would both be negatively charged at pH 7.0, at pH 2.0 alginate would be mainly uncharged due to protonation of the carboxyl groups, allowing the potential for the hydrophobic faces of the sugar rings to interact with the hydrophobic binding pocket of pepsin. This may be a reason why alginate inhibits pepsin and lipase activity without affecting trypsin.
The residues of the catalytic triad are spread across the active site cleft. With Ser195 on one side and Asp102 and His57 on the other (Hedstrom, 2002a, Hedstrom, 2002b). With the substrate co-ordinated in place by forming an anti-parallel beta-sheet across the protein binding site, the electronegatively charged base His57 can act to accept the hydrogen from the hydroxyl group of Ser195. This allows Ser195 to act as a nucleophile, attacking the carbonyl carbon of the peptide bond, forming an acyl-enzyme intermediate with the substrate (Polgar, 2005). The carbonyl carbon is δ+ as a dipole is formed over the C O bond with the electrons pulled towards the electronegative oxygen, leaving the carbon susceptible to nucleophillic attack from serine.
SBTI inhibits trypsin activity by strongly binding across the active site and blocking substrate binding with Arg63′-Ile64′ of SBTI mimicking the scissile peptide bond with the positively charged Arg63′ occupying the primary specificity pocket of trypsin (Blow, Janin, & Sweet, 1974). Evidently, as a polysaccharide, an alginate molecule would not be able to mimic binding of a protein substrate.
Due to the distinctly different inhibition profiles for pepsin and trypsin, the manner in which alginates and protein substrates interact across the pH range was investigated viscometrically. Profound interactions between alginate and protein were observed at acidic pHs, but no pattern of interaction was observed at neutral pH; with all alginate samples tested, a protein–alginate co-precipitate was formed at acidic pH, but not at a neutral pH.
SP54, heparin sulphate, and other highly sulphated polysaccharides are known to inhibit pepsin activity (Bianchi and Cook, 1964, Cook and Drill, 1967) and protein–carbohydrate interactions are common in biology, and widely reported in vitro (Dickinson, 1998). Interactions between casein and carrageenans have been observed due to electrostatic interactions forming between the sulphate groups of carrageenan and positively charged regions of the casein polymer (Grindrod and Nickerson, 1968, Payens, 1972). As the pH is lowered, protein is taken below its iso-electric point, resulting in a loss of negative charges and formation of positive charges. The positively charged protein can then form interactions with negative charges on the carbohydrate and carbohydrate–protein complexes form, leading to precipitation (De Jong & Hubertus, 2008). This non-specific protein binding raises the possibility that in addition to interactions at the active site, non-specific inhibitor–substrate and inhibitor–enzyme interactions could be involved in pepsin inhibition.
Alginate is a negatively charged polymer, and as such would be capable of forming electrostatic interactions with proteins that have become positively charged after being taken below their pKa (Then et al., 2012). Alginate may associate with protein through hydrogen bonding at hydroxyl groups; charge–charge interactions with δ-carboxyl groups and the negatively charged COO– group of the alginate, although this group would become protonated at low pH. As with the carrageenan–casein interactions, these reactions would be sensitive to structure, pH, concentration and levels of counter-ions.
As alginates can form acid gels in the presence of gastric contents, it is important to consider if the inhibition of pepsin would be altered between alginate in solution or as a gel. If the levels of inhibition are compared with alginate in solution using the N-terminal proteolytic assay and the model gut containing all the pertinent gastric secretions, they are found to be similar. This indicates that if alginate comes out of solution, it can still inhibit, presumably by trapping substrate and enzyme in the gel, and by any alginate remaining solubilised inhibiting by binding to the enzyme or substrate.
5. Conclusions
The observation of a pH dependent interaction between alginate and protein suggests a possible mechanism by which alginate may prevent the action of pepsin without effecting trypsin in a similar way. The binding of alginate to protein at low pH and formation of a precipitate would remove protein from solution and make the substrate unavailable to pepsin. However, this precipitation interaction did not occur at neutral pH and therefore substrate remains available for trypsin digestion, this explains why little evidence of trypsin inhibition was observed.
Conflict of interest
There are no conflicts of interest to report.
Acknowledgements
This work was funded by the (BBG50092/1, BB/G00563X/1) BBSRC and Newcastle University.
References
- Ali M.S., Parikh S., Chater P., Pearson J.P. Bile acids in laryngopharyngeal refluxate: Will they enhance or attenuate the action of pepsin? Laryngoscope. 2013;123(2):434–439. doi: 10.1002/lary.23619. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam V., Mustar S., Khalid N., Abd Rashed A., Noh M., Wilcox M., et al. Inhibitory activities of three Malaysian edible seaweeds on lipase and alpha-amylase. Journal of Phycology. 2013;25(5):1405–1412. [Google Scholar]
- Bianchi R., Cook D. Antipeptic and antiulcerogenic properties of a synthetic sulfated polysaccharide (Sn-263) Gastroenterology. 1964;47:409–414. [PubMed] [Google Scholar]
- Blow D., Janin J., Sweet R. Mode of action of soybean trypsin inhibitor (Kunitz) as a model for specific protein–protein interactions. Nature. 1974;249:54–57. doi: 10.1038/249054a0. [DOI] [PubMed] [Google Scholar]
- Brownlee I., Allen A., Pearson J., Dettmar P., Havler M., Atherton M., et al. Alginate as a source of dietary fiber. Critical Reviews in Food Science and Nutrition. 2005;45(6):497–510. doi: 10.1080/10408390500285673. [DOI] [PubMed] [Google Scholar]
- Cook D., Drill V. Pharmacological properties of pepsin inhibitors. Annals of the New York Academy of Sciences. 1967;140(2):724–733. [Google Scholar]
- De Jong, Hubertus G. 2008. New hunger suppressing food compositions. (Vol. WO/2009/082217) [Google Scholar]
- Dettmar P., Sykes J., Little S., Bryan J. Rapid onset of effect of sodium alginate on gastro-oesophageal reflux compared with ranitidine and omeprazole, and relationship between symptoms and reflux episodes. International Journal of Clinical Practice. 2006;60(3):275–283. doi: 10.1111/j.1368-5031.2006.00800.x. [DOI] [PubMed] [Google Scholar]
- Dickinson E. Stability and rheological implications of electrostatic milk protein–polysaccharide interactions. Trends in Food Science & Technology. 1998;9(10):347–354. [Google Scholar]
- El Aouad N., Monteiro M., Moreno C., Martin J., González I., Vicente F., et al. A new natural pepstatin from Kitasatospora (Actinomycetales) Planta Medica. 2011;77:59. [Google Scholar]
- Fujinaga M., Chernaia M., Tarasova N., Mosimann S., James M. Crystal structure of human pepsin and its complex with pepstatin. Protein Science. 1995;4(5):960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg H., Dodds W., Gee S., Montgomery C., Zboralske F. Role of acid and pepsin in acute experimental esophagitis. Gastroenterology. 1969;56(2):223–230. [PubMed] [Google Scholar]
- Grindrod J., Nickerson T. Effect of various gums on skimmilk and purified milk proteins. Journal of Dairy Science. 1968;51(6):834–841. [Google Scholar]
- Hedstrom L. An overview of serine proteases. Current Protocols in Protein Science. 2002 doi: 10.1002/0471140864.ps2110s26. (Chapter 21, Unit 21.10) [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chemical Reviews. 2002;102(12):4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Houghton D., Wilcox M., Brownlee I., Chater P., Seal C., Pearson J. Method for quantifying alginate and determining release from a food vehicle in gastrointestinal digesta. Food Chemistry. 2014;151:352–357. doi: 10.1016/j.foodchem.2013.11.070. [DOI] [PubMed] [Google Scholar]
- Hutton D., Allen A., Pearson J., Ward R., Venables C. Separation of pepsins in human gastric juice: Analysis of proteolytic and mucolytic activity. Biochemical Society Transactions. 1986;14 [Google Scholar]
- Isaksson G., Lundquist I., Ihse I. In vitro inhibition of pancreatic enzyme activities by dietary fiber. Digestion. 1982;24(1):54–59. doi: 10.1159/000198775. [DOI] [PubMed] [Google Scholar]
- Katona G., Berglund G., Hajdu J., Graaf L. Crystal structure reveals basis for the inhibitor resistance of human brain trypsin. Journal of Molecular Biology. 2002;315:1209–1218. doi: 10.1006/jmbi.2001.5305. [DOI] [PubMed] [Google Scholar]
- Lin Y., Means G., Feeney R. The action of proteolytic enzymes on N,N-dimethyl proteins. Basis for a microassay for proteolytic enzymes. The Journal of Biological Chemistry. 1969;244(3):789–793. [PubMed] [Google Scholar]
- Payens T. Light scattering of protein reactivity of polysaccharides especially of carrageenans. Journal of Dairy Science. 1972;55(2):141–150. doi: 10.3168/jds.S0022-0302(72)85452-3. [DOI] [PubMed] [Google Scholar]
- Pearson J., Parikh S., Orlando R., Johnston N., Allen J., Tinling S., et al. Review article: Reflux and its consequences – The laryngeal, pulmonary and oesophageal manifestations. Conference held in conjunction with the 9th International Symposium on Human Pepsin (ISHP) Kingston-upon-Hull, UK, 21–23 April 2010. Alimentary Pharmacology & Therapeutics. 2011;33(Suppl. 1):1–71. doi: 10.1111/j.1365-2036.2011.04581.x. [DOI] [PubMed] [Google Scholar]
- Polgar L. The catalytic triad of serine peptidases. Cellular and Molecular Life Sciences. 2005;62(19–20):2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J.C., Harley A.D., Myers D.V. Subsite specificity of porcine pepsin. Advances in Experimental Medicine and Biology. 1977;95:141–157. doi: 10.1007/978-1-4757-0719-9_9. [DOI] [PubMed] [Google Scholar]
- Sengupta I., Shah S., Shah N. A review on: Alginate forming in-situ gel for treating peptic ulcers and reflux disorders. Journal of Pharmaceutical Science and Bioscientific Research. 2015;5(2):172–179. [Google Scholar]
- Smidsrod O., Skjak-Braek G. Alginate as immobilization matrix for cells. Trends in Biotechnology. 1990;8(3):71–78. doi: 10.1016/0167-7799(90)90139-o. [DOI] [PubMed] [Google Scholar]
- Strugala V., Avis J., Jolliffe I., Johnstone L., Dettmar P. The role of an alginate suspension on pepsin and bile acids – Key aggressors in the gastric refluxate. Does this have implications for the treatment of gastro-oesophageal reflux disease? Journal of Pharmacology and Pharmacotherapeutics. 2009;61(8):1021–1028. doi: 10.1211/jpp/61.08.0005. [DOI] [PubMed] [Google Scholar]
- Strugala V., Kennington E.J., Campbell R.J., Skjak-Braek G., Dettmar P.W. Inhibition of pepsin activity by alginates in vitro and the effect of epimerization. International Journal of Pharmaceutics. 2005;304(1–2):40–50. doi: 10.1016/j.ijpharm.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Sunderland A., Dettmar P., Pearson J. Alginates inhibit pepsin activity in vitro; a justification for their use in gastro-oesophageal reflux disease (GORD) Gastroenterology. 2000;118(4):347. [Google Scholar]
- Szecsi P.B. The aspartic proteases. Scandinavian Journal of Clinical and Laboratory Investigation. 1992;210(Suppl.):5–22. [PubMed] [Google Scholar]
- Then C., Othman Z., Mustapha W., Sarmidi M., Aziz R., El Enshasy H. Production of alginate by Azotobacter vinelandii in semi-industrial scale using batch and fed-batch cultivation systems. Journal of Advanced Scientific Research. 2012;3(4):45–50. [Google Scholar]
- Wiechelman K., Braun R., Fitzpatrick J. Investigation of the bicinchoninic acid protein assay: Identification of the groups responsible for color formation. Analytical Biochemistry. 1988;175(1):231–237. doi: 10.1016/0003-2697(88)90383-1. [DOI] [PubMed] [Google Scholar]
- Wilcox M. Newcastle University; Newcastle Upon Tyne: 2010. Bioactive alginates. ICAMB. [Google Scholar]
- Wilcox M.D., Brownlee I.A., Richardson J.C., Dettmar P.W., Pearson J.P. The modulation of pancreatic lipase activity by alginates. Food chemistry. 2014;146:479–484. doi: 10.1016/j.foodchem.2013.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]