Abstract
Neuronally coexpressed ELAV/Hu proteins comprise a family of highly related RNA binding proteins which bind to very similar cognate sequences. How this redundancy is linked to in vivo function and how gene-specific regulation is achieved have not been clear. Analysis of mutants in Drosophila ELAV/Hu family proteins ELAV, FNE, and RBP9 and of genetic interactions among them indicates that they have mostly independent roles in neuronal development and function but have converging roles in the regulation of synaptic plasticity. Conversely, ELAV, FNE, RBP9, and human HuR bind ELAV target RNA in vitro with similar affinities. Likewise, all can regulate alternative splicing of ELAV target genes in nonneuronal wing disc cells and substitute for ELAV in eye development upon artificially increased expression; they can also substantially restore ELAV's biological functions when expressed under the control of the elav gene. Furthermore, ELAV-related Sex-lethal can regulate ELAV targets, and ELAV/Hu proteins can interfere with sexual differentiation. An ancient relationship to Sex-lethal is revealed by gonadal expression of RBP9, providing a maternal fail-safe for dosage compensation. Our results indicate that highly related ELAV/Hu RNA binding proteins select targets for mRNA processing through alteration of their expression levels and subcellular localization but only minimally by altered RNA binding specificity.
INTRODUCTION
RNA binding proteins (RBPs) are key regulators of gene expression. Through regulation of alternative splicing and polyadenylation, they expand the proteome and control spatiotemporal expression by affecting mRNA transport, turnover, localization, and translatability (1, 2). In the brain, alternative mRNA processing is particularly abundant and substantially contributes to the complexity of this organ (3, 4). Many RBPs comprise highly related gene families, but they seem to discriminate only marginally between short cognate binding sequences (5). Although redundancy can be evolutionarily stable over extended periods of time (6), it is not clear if highly related RBPs act redundantly in vivo, regulating mostly the same genes in the same biological processes, or if they have diverged such that they regulate genes involved in different biological processes. Detailed analysis of the functions of highly related RBPs in animal models is required to decipher the underlying mechanisms of how highly related RBPs achieve target specificity.
ELAV (embryonic-lethal abnormal visual system)/Hu proteins comprise a family of RBPs broadly coexpressed in the nervous system and widely used neuronal markers (7, 8). ELAV/Hu proteins are prototype RBPs which harbor three highly conserved RNA recognition motifs (RRMs), whereby the first two RRMs are arranged in tandem and the third RRM is separated by a less conserved hinge region. Humans have four ELAV/Hu protein-encoding genes (HuB, HuC, HuD, and HuR), while Drosophila has three (elav, fne, and Rbp9), which derive from a common ancestor but have duplicated independently in vertebrates and arthropods (9). In mice, all Hu proteins are expressed in largely overlapping patterns in mature neurons (10). In addition, HuB is also expressed in gonads, and HuR is ubiquitous. Expression of ELAV and FNE in Drosophila starts with the birth of neurons, while RBP9 is first detected in late larval neurons (11–13). RBP9 is also expressed in gonads. The closest relative of ELAV family proteins in flies is Sex-lethal (Sxl), the master regulator of sexual differentiation and dosage compensation (14).
Due to its nuclear localization, the founding member of the ELAV/Hu family of RBPs, Drosophila ELAV, has initially been associated with gene-specific regulation of alternative splicing and polyadenylation, but it can also regulate mRNA stability (15–21). In contrast, human Hu RBPs have mostly been associated with regulating the stability of mRNAs, their localization, and their translatability but were recently also shown to regulate alternative pre-mRNA processing (22–29). Although ELAV/Hu family RBPs bind to short, uridine-rich motifs, which are ubiquitously found in introns and untranslated regions (UTRs), they seem to have a complement of dedicated target genes (24–27), and their activities are not restricted to a specific process in the life of an mRNA (8). Since ELAV/Hu RBPs can shuttle between the nucleus and cytoplasm (30), they likely also exert gene-specific functions depending on their cellular localization.
Although ELAV family RBPs are broadly coexpressed in the brain of Drosophila, initial characterization of mutants of individual elav family genes revealed a number of distinct developmental and behavioral phenotypes. elav is required for axonal targeting in the embryonic central nervous system (CNS), for synaptic growth, for photoreceptor survival, and for neuronal migration in the optic lobe (19, 31, 32), and fne is required for mushroom body development and male courtship performance (33) while Rbp9 supports blood-brain barrier integrity and the extended life span of flies (34, 35). Since these phenotypes have not been comprehensively analyzed in mutants of all elav family genes or in combinations thereof, it has not been clear if and to what extent they have overlapping functions. Our results indicate that ELAV family RBPs in Drosophila exert specific functions in the development, maintenance, and functioning of the nervous system but that they converge in the regulation of synaptic growth in ELAV- and FNE/RBP9-independent pathways. Intriguingly, however, FNE, RBP9, human Hu RBPs and closely related Sxl can regulate alternative splicing of ELAV target genes in nonneuronal wing disc cells, and all ELAVs can direct eye development by GAL4/upstream activation sequence (UAS)-mediated artificially increased expression. When placed under the control of the elav promoter and UTRs, ELAV family RBPs can substitute for ELAV function at an organismal level. ELAV/Hu RBPs can also interfere with sexual differentiation, and an ancient relationship to Sxl is revealed by gonadal expression of RBP9, providing a maternal fail-safe for dosage compensation by the male-specific lethal (MSL) complex. Since ELAV/Hu RBPs bind RNA rather indiscriminately and can substantially rescue elav mutants under the regulatory control of the elav gene, these results indicate that selection of target genes is mainly achieved through alteration of expression levels and subcellular localization and only marginally by altered RNA binding specificity.
MATERIALS AND METHODS
Fly genetics and recombinant DNA technology.
Fly breeding, genetics, and recombinant DNA technology were done according to standard procedures as described previously (36). The fne null allele, fneΔ, was generated by FLP/FRT-mediated recombination between the following two transposon insertion lines: PBac{WH}fnef06439 and PBac{WH}hecf06077 (see Fig. S2 in the supplemental material) (37, 38). Whole-eye clones of the elave5 null allele were generated as described previously (39) using an elave5 w FRT19B chromosome. Larvae and adult animals were obtained by using the elavts1 temperature-sensitive allele transheterozygous with elave5 and reared at the permissive temperature (18°C) for 3 days and then shifted to the restrictive temperature (25°C). To avoid unrelated effects from the genetic background of homozygously viable alleles, they were out-crossed to a lethal allele in the case of elav or to small chromosomal deficiencies Df(1)ED7165 for fne or Df(2L)ED206 for Rbp9. Further details about mutant alleles and gene expression patterns can be found in FlyBase (www.flybase.org). Transgenic flies were obtained by ϕC31-mediated transformation as described previously (40) using landing sites at 57F (RBP9 genomic construct, PBac{y+-attP-3B}VK22), 76A (UAS construct, PBac{y+-attP-3B}VK00002), and 55C (genomic rescue construct, P{y[lsqb]+t7.7[rsqb]=CaryP}attP1). The additional UAS-HA-elav construct was inserted in 86F (M{3xP3-RFP.attP′}ZH-86Fa). Three copies of a myc epitope tag (3×myc) was cloned into Pacman CH322-140N12, which contains a genomic fragment encompassing the entire Rbp9 gene, by using the two BamHI sites harbored in the beginning of the N-terminal auxiliary domain of Rbp9, generating the following sequence: AGCACCACCGGATCAGGAGAACAAAAATTAATTTCAGAAGAAGACTTAAGTACTGAGCAGAAGCTAATAAGCGAGGAGGATCTATCCGGAGAACAAAAATTAATTTCAGAAGAAGACTTACCGGCTACGGCC. The genomic region of the fne gene was obtained by PCR amplification of a modified pUC containing an attB site for integration and a green fluorescent protein (GFP) marker for identification of transgenic flies using primers pUC P fne F (CCCGAAAGTGAAAGTGAAGCGATTTTGCGACGCCTCCCGAAGCTACACCCGAAAATCAACTTCCAACGACAGAATTCGAGTTCAAGAAGAAGGCG) and pUC P3×P3 fne R (CCTTGGAATACCAAATGTCAACTTTGGTTCAAGGCCACCAGCAGTCGGTGGAAGACTTCCACCAAGACGTCCCGGCGGCCGCGTCTAGATAACTTCG) and then retrieved from the bacterial artificial chromosome (BAC) clone RP98-39D14 by recombineering according to the manufacturer's protocol (Genebridges). Two copies of a hemagglutinin (2×HA) tag were then inserted, using NcoI and BamHI sites, into the N terminus of FNE, generating the following sequence: ACCAACGCCATGGCAAGTACTTACCCCTACGACGTGCCCGACTACGCCCAGGGAAGTTACCCCTACGACGTGCCCGACTACGCCGATATTGTGAAGA. UAS constructs were generated by cloning the open reading frames (ORFs) into pUASTMSattB, which contains a modified polylinker and an attB site inserted between the BamHI and SphI sites before the UAS promoter and a simian virus 40 (SV40) trailer. In addition, a 29-nucleotide (nt) translation initiation site from the adh gene (GAATTCGAGATCTAAAGAGCCTGCTAAAGCAAAAAAGAAGTCACC), followed by the start sequence (ATGTCGACCGGCTCGAGC), and a 2×HA tag were introduced before the ORF; 60 nt of the elav UTR following the stop codon were also added to UAS-HA-elav constructs, but these 60 nt were omitted from other UAS constructs. To generate the UAS-HA-fne, UAS-HA-RBP9, and UAS-HA-Hu constructs, the elav ORF was swapped using flanking HindIII and XbaI sites and an additional SphI site in the vector to set up a three-way ligation. To express Sxl and Halfpipe (Hfp), UAS transgenes on the second and third chromosomes were used; UAS transgenes on the second chromosome were used to express B52 (41–43). Genomic rescue constructs were cloned into a modified pCaSpR containing the elav promoter (32) and modified to start with the ATG after exon 2 of elav. An HA tag was inserted in frame after codon 9 of elav, flanked by EcoRI and SgrAI sites (ATTCCATACCCCTACGACGTGCCCGACTACGCCGCC), followed by the ORF starting with the first codon after the ATG, an AscI site generated by transformation of the sequence after the stop codon, 1,176 nt of the elav 3′ UTR, including the NheI site, and an attB site. For the elav-NLS-HA-RBP9 construct, a nuclear localization signal (NLS) sequence (GGCGTGAGCCGCAAGCGCCCCCGCCCCGGCCCA) was inserted after codon 8 of elav before the HA tag using EcoRI and SgrAI sites (44). The eFVGU construct was generated by swapping the erect wing (ewg) ORF and UTR as in eFeG (32) with the elav ORF and UTR as described above using the NheI and EcoRI sites in eFeG.
EMSAs, RT-PCR, protein and Western analysis, antibody stainings, and histology.
Production of recombinant proteins, 32P-labeled in vitro transcripts, and electrophoretic mobility shift assays (EMSAs) were done as described previously (36). RNA extraction and reverse transcription-PCR (RT-PCR) were done as described previously (36). Polyacrylamide gels were dried, exposed to phosphorimager screens (Bio-Rad), and quantified with Quantity One software (Bio-Rad). cDNAs were amplified from ewg transcripts with the primer pair ewg4F and ewg5R and the pair ewg6F and ewg6R, from neuroglian (nrg) using primers nrg2F, nrg2S, and nrg3L, and from arm using primers armF and armR (20, 45). Protein gels and Western analysis were done according to standard protocols as described by Soller et al. (36) using rat anti-ELAV antibody (monoclonal antibody [MAb] 7E8A10 at 1:250) (Developmental Studies Hybridoma Bank [DHSB]), rat anti-HA antibody (3F10, 1:50; Roche), and mouse anti-alpha-tubulin (1;100,000; Sigma). In situ antibody stainings were done as described previously (32) using rat anti-HA (MAb 3F10, 1:20; Roche), mouse anti-ELAV (MAb 7D, 1:20, which recognizes 7 amino acids [aa] unique to ELAV; DHSB) (46), mouse anti-FasII (1D4, 1:100; DHSB), MAb BP102 (1:20; DHSB), and anti-GFP (1:500; Molecular Probes) and visualized with Alexa Fluor 488- and/or Alexa Fluor 647-coupled secondary antibodies (1:250; Molecular Probes) or by diaminobenzidine staining (1 mg/ml) in the presence of 0.01% H2O2 using horseradish peroxidase (HRP)-coupled secondary antibodies (1:10,000; Sigma). DAPI (4′,6′-diamidino-2-phenylindole) was used at 1 μg/ml. Paraffin sections were done as described previously (47).
For quantification of antibody stainings in wing imaginal discs, the full width of at least four discs per genotype was scanned, and fluorescence intensity quantification was done on the average z-series projection of the stack in ImageJ, as previously described by Toba and White (48).
For imaging of larval and adult brains, confocal z-stacks were taken using a 40× objective lens on a Leica SP5/SP2 instrument at multiple positions to ensure the complete capture of the imaged brain with sufficient spatial overlap between each position. The number of z-stacks and acquisition settings were kept constant for each brain that was being imaged. The average intensity overlay from the stacks was stitched together using the FIJI 2D stitching plug-in using ImageJ (49).
Behavioral analysis, longevity, and statistics.
A negative geotaxis assay was performed as described previously (50). Briefly, 20 adult flies per genotype were anesthetized with CO2, placed in the bottom of a closed 25-ml plastic pipette, and left to recover for 30 min. Flies were tapped to the bottom of the column and left to climb up for 45 s. The number of flies that climbed above the 25-ml mark (ntop) and the number remaining below the 2-ml mark (nbottom) were recorded. Recovery time between repeats was 1 min. A performance index (PI) was calculated as follows: PI = 0.5 × (ntotal + ntop − nbottom)/ntotal. Statistical analysis was done by analysis of variance (ANOVA), followed by planned pairwise comparisons using Fisher's protected least significant difference, performed using StatView.
For the analysis of longevity, 60 flies per genotype were aged for 60 days in groups of 20 per vial without live baker's yeast (Saccharomyces cerevisiae). Viable flies were transferred to fresh food medium every 3 to 5 days, and the number of dead flies was recorded.
RESULTS
Highly related and coexpressed Drosophila ELAV family RNA binding proteins exert distinct biological functions, but their roles converge in the regulation of synaptic growth.
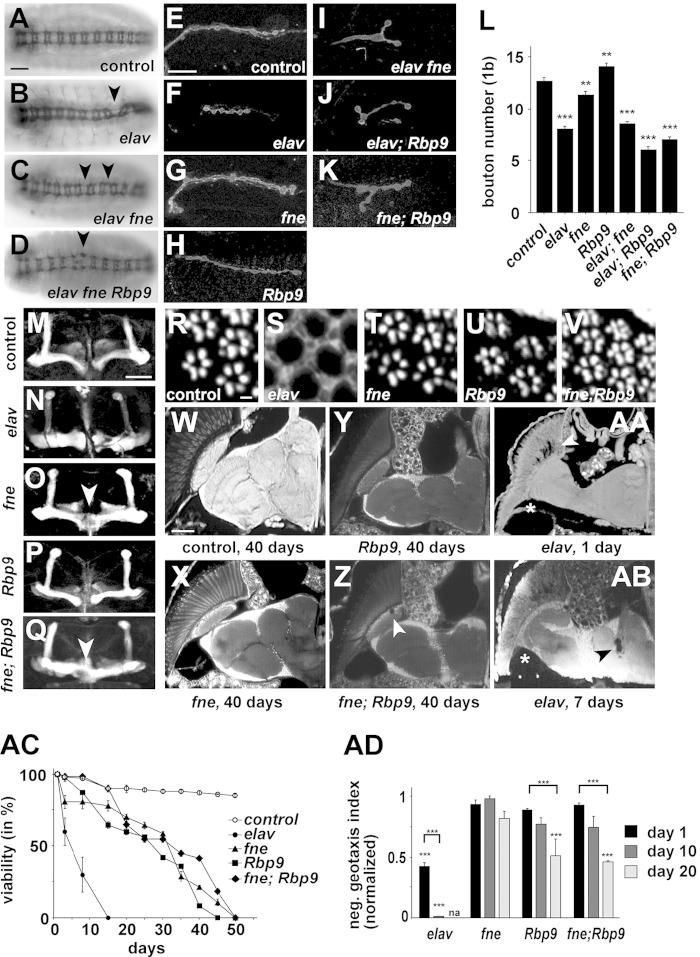
Drosophila ELAV family RBPs are neuronally coexpressed and are highly homologous in their RNA binding domains, ranging in similarity from 90 to 93% in RRM1, 76 to 90% in RRM2, and 90 to 98% in RRM3 (see Table S1 in the supplemental material). Compared to human Hu RBPs, they share 84 to 91%, 76 to 82%, and 84 to 90% similarity in RRMs 1 to 3, respectively. Although expression patterns of ELAV, FNE, and RBP9 had been determined individually previously, there is only limited information about their overlapping expression (11, 12). Since we were unable to obtain highly specific antibodies for FNE and RBP9, we generated epitope-tagged genomic constructs to assess their coexpression. Analysis of the expression from these constructs in transgenic flies and comparison with the expression pattern of ELAV revealed that FNE and ELAV, as well as RBP9 and ELAV, are coexpressed in all neurons in the adult brain (see Fig. S1 in the supplemental material). ELAV is mostly nuclear, FNE is about equally distributed between the nucleus and cytoplasm, and RBP9 is mostly cytoplasmic (see Fig. S1). To determine the extent of highly homologous ELAV family RBPs acting redundantly in Drosophila, we analyzed mutants of elav (elave5 null and elavts1 temperature-sensitive alleles [13]), fne (null allele) (see Fig. S2 in the supplemental material), and Rbp9 (Rbp9P2690 null allele [51]) and combinations thereof for developmental and behavioral phenotypes assigned to one of the ELAV family genes in mutants of the other two (Fig. 1) (19, 31–35).
FIG 1.
Mutants of Drosophila ELAV family RBPs display distinct phenotypes but converge in the regulation of synaptic growth. (A to D) Axonal projections in control, elav (elave5/Y), elav fne (elave5 fneΔ/Y), and elav fne Rbp9 (elave5 fneΔ/Y; Rbp9P2690) embryos were stained with MAb BP102. Arrowheads indicated projection defects and/or irregular positioning of neuromeres. Scale bar, 25 μm. (E to L) Neuromuscular junctions at muscle 13 of control, elav (elave5/elavts1), fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), elav fne (elave5 fneΔ//elavts1 fneΔ), elav; RBP9 (elave5/elavts1; Rbp9P2690/Df(2L)ED206) and fne; Rbp9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) third-instar larvae were stained with anti-HRP, and type 1b boutons were quantified (n = 15 to 30) (L). Scale bar, 25 μm. elave5/elavts1 mutants were raised at the permissive temperature during embryogenesis. Statistically significant differences compared to the control values are indicated by asterisks (**, P < 0.01; ***, P < 0.001). (M to Q) Mushroom bodies of control, elav (elave5/elavts1) (flies raised at the permissive temperature during embryogenesis), fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), and fne; Rbp9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) adult flies were stained with anti-Fas2. Arrowheads indicate fused beta lobes. Scale bar, 25 μm. (R to V) Photoreceptors of control, elav (elave5; whole-eye clone), fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), and fne RBP9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) adult flies from paraffin sections were visualized by autofluorescence. Scale bar, 5 μm. (W to AB) Horizontal paraffin sections of adult heads from control, fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), and fne; Rbp9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) 40-day-old adult flies and from elav (elave5/elavts1) (flies raised at the permissive temperature during embryogenesis) 1-day-old (AA) and 7-day-old (AB) adult flies were visualized by autofluorescence. Arrowheads indicate vacuolization, and the asterisks indicate the nonrotated medulla. Scale bar, W 50 μm. (AC) Longevity of control, elav (elave5/elavts1) (flies raised at the permissive temperature during embryogenesis), fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), and fne; Rbp9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) flies is shown as the mean from three replicates (20 flies each) with the standard error. (AD) Negative geotaxis of 1-day-, 10-day-, and 20-day-old elav (elave5/elavts1) (flies raised at the permissive temperature during embryogenesis), fne (fneΔ/Df (1)ED7165), Rbp9 (Rbp9P2690/Df(2L)ED206), and fne; Rbp9 (fneΔ/Df (1)ED7165; Rbp9P2690/Df(2L)ED206) adult flies is shown as the mean from three experiments with the standard error normalized to the performance of control flies (set to 100%). Statistically significant differences of comparisons to control fly values are indicated (***, P < 0.001).
The elave5 null mutant is embryonic lethal, while fneΔ and Rbp9P2690 null mutants or fneΔ; Rbp9P2690 double mutants are viable. Raising elav temperature-sensitive mutants (elave5/elavts1) during embryogenesis at the permissive temperature renders them weakly adult viable (23%). Double mutants of elave5/elavts1 with either fneΔ or Rbp9P2690 result in larval/pupal lethality, and triple mutants are embryonic lethal. Mutants of elave5 exert defects in axonal wiring during embryonic nervous system development, resulting in irregular positioning of neuromeres and thinning of commissures and connectives, but this phenotype does not worsen in elave5 fneΔ double null or elave5 fneΔ; Rbp9P2690 triple null mutant combinations (Fig. 1A to D), arguing for a unique function of elav in this process.
Next, we analyzed elav family mutants and combinations for defects in synaptic growth at third-instar neuromuscular junctions (NMJs) (Fig. 1E to L). Here, a strong reduction in the number of synaptic connections is observed in elave5/elavts1 mutants, which does not significantly decrease in the absence of fne and Rbp9 (Fig. 1I, J, and L). Mutants of fneΔ and Rbp9P2690 have slightly reduced and slightly increased numbers of synaptic boutons, respectively, but, strikingly, fneΔ; Rbp9P2690 double mutants show a dramatic reduction of synaptic connections, suggesting that they act in the same pathway (Fig. 1K and L). The lack of genetic interactions of elav with fne and Rbp9 further suggests that they regulate synaptic growth independently.
For the development of adult mushroom bodies, fne is required for restricting axonal extension of the beta lobe (33), but this phenotype was not observed in elave5 or Rbp9P2690 mutants and did not get worse in fneΔ; Rbp9P2690 double mutants (Fig. 1M to Q), indicating a unique function of fne in this process.
During pupal development, elav is required for rotation of the medulla, and in adult flies it is required for maintenance of photoreceptor and central brain neurons (Fig. 1R to AB) (31). These phenotypes were not observed in fneΔ, Rbp9P2690 and fneΔ; Rbp9P2690 mutants except for occasional vacuolizations observed in the lamina of fneΔ; Rbp9P2690 double mutants (10 out of 12). Furthermore, elave5/elavts1 mutants had a much more dramatically reduced life span than fneΔ, Rbp9P2690 and fneΔ; Rbp9P2690 mutants (Fig. 1AC), but the lack of a genetic interaction between fne and Rbp9 suggests nonoverlapping functions. Similarly, adult locomotion as assayed with a negative geotaxis assay was impaired in an age-dependent manner in Rbp9P2690 but not in fneΔ flies (Fig. 1AD), excluding overlapping functions. In contrast, elave5/elavts1 mutants showed much more reduced locomotion and were also ataxic (Fig. 1AD).
Taking these results together, individual ELAV family genes have mostly distinct roles during neuronal development, maintenance, and function, shown by the absence of genetic interactions, but their roles converge in the regulation of synaptic plasticity.
Alternative splicing of known ELAV targets is unaffected in fneΔ; Rbp9P2690 null mutants.
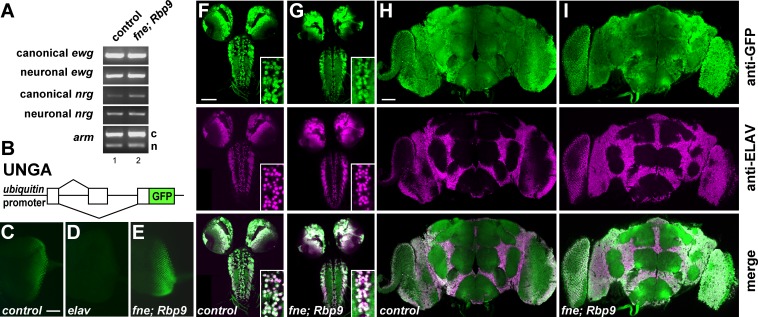
The major target of ELAV, ewg, has a prominent function in regulating synaptic growth at third-instar NMJs (32, 52). Since FNE and RBP9 also affect this process, we wondered if FNE and RBP9 regulate alternative splicing of the ELAV target genes ewg, nrg, and arm in a subset of Drosophila neurons. Although in the absence of ELAV the neuronal isoforms of these genes are completely absent in photoreceptor neurons (16, 53), this analysis has not been comprehensively extended to all parts of the brain, leaving the possibility that FNE and/or RBP9 assist or substitute for ELAV in the regulation of these genes. No obvious reduction in the neuronal isoform of these three genes was detected in fne; Rbp9 double mutants by RT-PCR from adult brains (Fig. 2A). Potentially, loss of FNE and RBP9 could affect alternative splicing only in a few cells in the brain, which would not be detected by RT-PCR. To visualize alternative splicing at a cellular resolution, we used an nrg GFP reporter, UNGA (21) (Fig. 2B), which is ELAV dependent (Fig. 2C and D). In the absence of FNE and RBP9, all neurons expressing ELAV also alternatively splice the nrg GFP reporter UNGA in larval photoreceptor neurons and in larval and adult brains (Fig. 2E to I), which was also observed in photoreceptor neurons and larval brains of individual mutants of fne or Rbp9 (see Fig. S3 in the supplemental material).
FIG 2.
Loss of FNE and RBP9 does not affect alternative splicing of ELAV target genes erect wing, neuroglian, and armadillo. (A) Analysis of neuronal alternative splicing in the ewg, nrg, and arm genes in fne; Rbp9 double mutants by RT-PCR. n, neuronal isoform; c, canonical isoform. (B) Schematic of the ELAV-responsive nrg GFP reporter UNGA. (C to E) Alternative splicing of nrg from the UNGA reporter, visualized by anti-GFP staining, is not affected in photoreceptor neurons of fne; Rbp9 mutants but is dramatically reduced in elavedr mutants. Scale bar, 50 μm. (F to I) Alternative splicing of nrg from the UNGA reporter is not affected in neurons of the third-instar larval or adult brain in fne; Rbp9 mutants, which were visualized with anti-GFP staining (top row) and in comparison to anti-ELAV staining (middle and bottom rows). Note the complete overlap between ELAV expression and GFP from the spliced UNGA reporter in fne; Rbp9 mutants (bottom rows of panels F to I). Scale bar, 100 μm.
Recombinant FNE, RBP9, and HuR bind to ELAV target RNA with similar affinities.
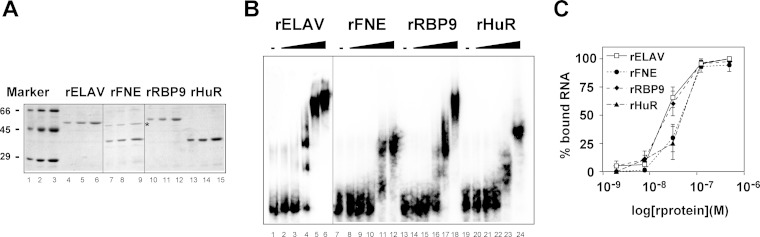
Next, we determined the RNA binding specificities of ELAV family members in vitro using the well-characterized ELAV binding sequence in the ewg gene (pA-I), which comprises 135 bp (20, 36, 40). For these binding experiments, we generated recombinant proteins in Escherichia coli for ELAV, FNE, and RBP9 and also for human HuR because it is functionally closest to ELAV family proteins in Drosophila (Fig. 3A; see also Fig. S4 and Table S1 in the supplemental material). Surprisingly, all proteins bound ewg pA-I RNA in a narrow affinity range and, similar to ELAV, also cooperatively formed multimeric complexes in electrophoretic mobility shift assays (EMSAs) (Fig. 3B and C). Multimeric complexes of recombinant FNE (rFNE) and rHuR assembled on pA-I RNA run faster, in accordance with their size (Fig. 3A and B), which has previously been observed with the N-terminally truncated form of ELAV, RBD60 (13). Binding constants for rELAV, rFNE, rRBP9, and rHuR were 22 nM, 47 nM, 23 nM, and 49 nM, respectively (Fig. 3C).
FIG 3.
Binding of recombinant ELAV/Hu family RBPs to RNA of the ELAV target ewg. (A) SDS-polyacrylamide gel showing Coomassie blue-stained recombinant ELAV family RBPs used for binding assays. For each of the recombinant proteins, 0.5 μg, 1 μg, and 2 μg were loaded. Marker proteins were bovine serum albumin (66 kDa), ovalbumin (45 kDa), and carbonic anhydrase (30 kDa). A bacterial protein copurifying with rFNE (lanes 7 to 9) is indicated by the star next to lane 9. (B) EMSA gel with RNA from the ELAV binding site in ewg (pA2-I). Uniformly 32P-labeled RNAs (100 pM) were incubated with recombinant proteins (2 nM, 8 nM, 32 nM, 125 nM, and 500 nM) and separated on 4% native polyacrylamide gels. (C) Graphic representation of EMSA data. The percentage of bound RNA [(input RNA − unbound RNA)/input RNA × 100] is plotted against the concentration of recombinant proteins (in molar units) presented as log values.
FNE, RBP9, and Hu proteins can regulate alternative splicing of ELAV target genes.
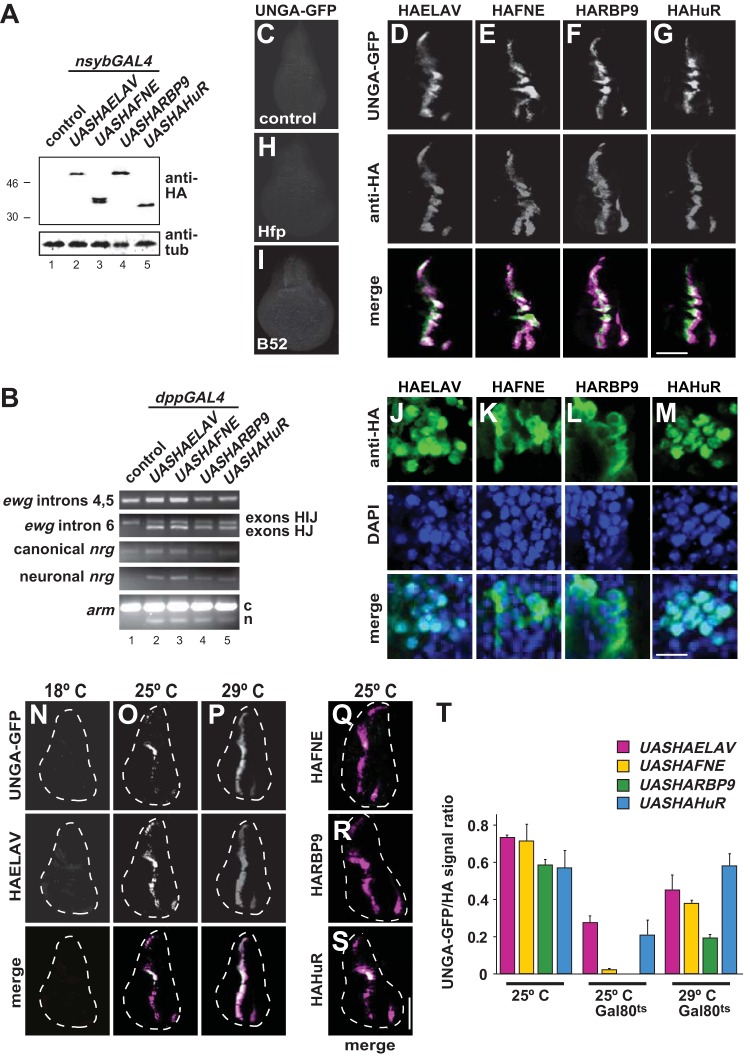
Expression of ELAV in nonneuronal wing discs results in neuronal splicing of ELAV target genes (16). Since recombinant FNE, RBP9, and HuR bound ELAV target RNA with similar affinities, we wanted to know if they could also regulate elav target genes when expressed in wing discs. ELAV family RBPs were expressed from hemagglutinin-tagged (HA) UAS transgenes (Fig. 4A). Expression in nonneuronal wing disc tissue using dpp-GAL4 results in neuronal splicing of ewg, nrg, and arm, based on RT-PCR or the UNGA reporter (Fig. 4B and C to G). The more distantly related poly(U) RBP Halfpipe (Hfp) or the SR protein B52 did not induce alternative splicing of the nrg reporter UNGA (Fig. 4H and I). Consistent with a role in alternative splicing regulation, ELAV and HuR predominantly localized to the nucleus. FNE showed no distinct localization, while RBP9 was predominantly present in the cytoplasm (Fig. 4J to M). Expression of HuB and HuC from UAS transgenes with dpp-GAL4 promoted neuron-specific splicing of UNGA (see Fig. S4 in the supplemental material), but HuD was not detectable and did not induce GFP expression although expression with elav-GAL4c155 resulted in lethality. Since expression of HuB and HuC was also undetectable or resulted in lethality with some neuronal GAL4 drivers, we focused on HuR.
FIG 4.
Elevated levels of FNE, RBP9, and HuR can regulate alternative splicing of ELAV targets. (A) Expression of HA-tagged ELAV (e.g., HAELAV), FNE, RBP9, and HuR from UAS-containing transgenes in adults with nsyb-GAL4 by Western blotting detection with anti-ELAV antibodies. (B) Neuronal alternative splicing of ELAV targets ewg intron 6 from exon H to J, nrg and arm induced by expression of HA-tagged ELAV, FNE, RBP9, and HuR from UAS-containing transgenes in wing discs with dpp-GAL4 as assessed by RT-PCR. c, canonical; n, neuronal. (C to I) Neuronal alternative splicing of the nrg GFP reporter UNGA upon expression of HA-tagged ELAV, FNE, RBP9, and HuR from UAS-containing transgenes in wing discs with dpp-GAL4. Staining with anti-GFP and anti-HA is as indicated on the left. Due to temporally regulated expression of dpp-GAL4 and because expression of ELAV proteins precedes GFP expression, signals of ELAV proteins and GFP do not entirely overlap. Note that the distantly related poly(U) binding protein Hfp (H) and the SR protein B52 (I) do not induce UNGA splicing. Scale bar, 150 μm. (J to M) Cellular localization of HA-tagged ELAV, FNE, RBP9, and HuR from UAS-containing transgenes in wing discs with dpp-GAL4. Staining with anti-HA and DAPI is as indicated on the left. Scale bar, 10 μm. (N to S) Neuronal alternative splicing of the nrg GFP reporter UNGA upon expression of HA-tagged ELAV, FNE, RBP9, and HuR from UAS-containing transgenes in wing discs with dpp-GAL4 in the presence of temperature-sensitive inhibitor of GAL4, GAL80ts, expressed from a UAS transgene at 18°C, 25°C, and 29°C. Staining in panels N to P with anti-GFP and anti-HA is as indicated on the left. Merged images are shown in the bottom row of panels N to P and in panels Q to S. Scale bar, 150 μm. (T) Quantification of UNGA-splicing shown as means with the standard error from five wing discs.
Ectopic expression of all ELAV/Hu family RBPs in nonneuronal wing discs induced neuron-specific alternative splicing of the known ELAV targets, and they behaved indiscriminately, likely due to expression levels saturating for UNGA regulation (Fig. 4T, 25°C). To reduce concentrations of ELAV/Hu RBPs, a temperature-sensitive inhibitor of GAL4 expressed under a UAS promoter, UAS-GAL80ts, was used. This genetic configuration resulted in reduced expression of ELAV and concomitant neuronal alternative splicing of the UNGA reporter at 25°C (Fig. 4N to T). At this temperature, FNE and RBP were not able to induce the UNGA reporter, while induction by HuR was comparable to that by ELAV (Fig. 4O, S, and T), indeed revealing different thresholds in vivo.
FNE, RBP9, and Hu proteins can partially substitute for ELAV in eye development.
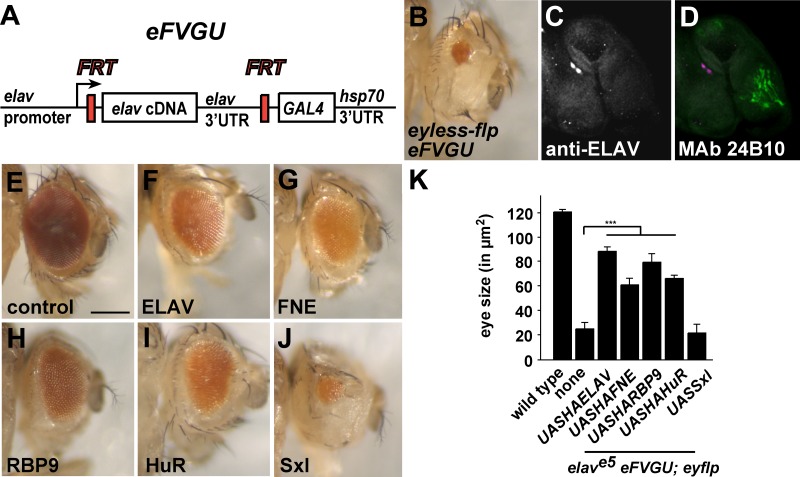
Since FNE, RBP9, and HuR can regulate alternative splicing of ELAV target genes, we wanted to know whether they could substitute for ELAV in eye development as broader expression interferes with organismal viability. For this experiment, we used an elav flip-out rescue construct, eFVGU, where the ELAV gene ORF is flanked with FRT sites followed by GAL4, leading to artificially increased expression. When elav is removed with eyflp in the eye primordium, GAL4 will be expressed, which can then drive UAS transgenes (Fig. 5A). In the absence of ELAV only a tiny eye developed (Fig. 5B to D). In contrast, the presence of ELAV, FNE, RBP9, or HuR but not ELAV's closest relative, Sex-lethal (Sxl), rescued eye development substantially (Fig. 5E to P). Pan-neural expression of ELAV with the GAL4 UAS system, however, was unable to rescue viability of elav null mutants.
FIG 5.
Rescue of eye development by ELAV/Hu family RBPs in elav mutant eyes. (A) Schematic of the eFVGU elav rescue construct. FRT-mediated recombination results in loss of elav and GAL4 expression under the elav promoter. (B to D) Eye and eye discs of elave5 eFVGU; eyflp males. Neurons shown in panels C and D were stained with anti-ELAV and MAb 24B10, respectively. (E to J) Eyes of wild-type and elave5 eFVGU; eyflp males expressing ELAV/Hu family RBPs or Sxl from UAS transgenes. (K) Quantification of the eye size shown in panels B and E to J. Statistically significant rescue compared to results in the absence of a UAS transgene is indicated (***, P < 0.001).
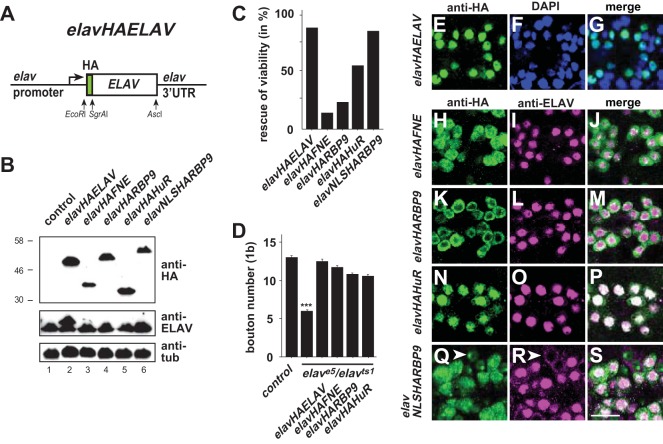
FNE, RBP9, and HuR rescue ELAV function under endogenous control of the elav gene.
Mutants of ELAV family RBPs in Drosophila show distinct phenotypes, but the proteins showed little discrimination at the level of RNA binding or ELAV target gene regulation when overexpressed. We therefore reasoned that distinct functions of these proteins are tightly linked to their expression levels. Indeed, during embryogenesis and larval life, ELAV is the dominantly expressed ELAV family protein. In contrast, FNE and RBP9 expression is high during pupal development, and RBP9 shows predominant expression in adults (see Fig. S5 in the supplemental material). Since the GAL4/UAS system leads to increased and also delayed expression, we wanted to more accurately test if distinct functions depend on expression levels. Therefore, we exchanged the ELAV gene ORF in an HA-tagged elav rescue construct harboring its UTRs with the ORFs coding for FNE, RBP9, and HuR (Fig. 6A) and inserted these constructs into the same genomic locus, resulting in the same expression levels. Expression levels of RBP9 and HuR in these transgenic flies were comparable to those with expression of ELAV, while FNE seemed to be less stable (Fig. 6B). The elav-HA-ELAV transgene rescued viability of the strong hypomorphic allele elavts1 (Fig. 6C). Strong rescue was also obtained with elav-HA-HuR, while elav-HA-RBP9 and elav-HA-FNE rescued viability less well. Although only the elav-HA-HuR transgene showed a marginal rescue of the elave5 null allele (5% with two copies; n = 100), all four transgenes rescued synaptic growth defects when elave5/elavts1 animals were raised at the permissive temperature during embryonic development (Fig. 6D). Since RBP9 rescued viability less well than HuR, cellular localization could be the reason for this. RBP9 localized predominantly to the cytoplasm, while HuR was predominantly nuclear, and FNE was equally present in the nucleus and cytoplasm (Fig. 6E to P). When RBP9 was targeted to the nucleus by including an NLS in the transgene (Fig. 6Q), the elav-NLS-HA-RBP9 transgene was comparable to elav-HA-ELAV in its ability to rescue the elavts1 allele (Fig. 6C) but did not rescue the elave5 null allele (n = 498). Accordingly, nuclear localization is increased in all neurons. In a subset of neurons, however, NLSRBP9 was predominantly nuclear, while ELAV became cytoplasmic (Fig. 6Q to S).
FIG 6.
FNE, RBP9, and HuR can replace neuronal ELAV function under the control of the elav gene. (A) Schematic of the elav rescue construct elav-HA-ELAV. (B) Expression of HA-tagged ELAV (e.g., HAELAV), FNE, RBP9, and HuR under the control of the elav gene in adult flies was determined by Western blotting detection with anti-HA antibodies. In lane 2, HA-tagged ELAV has a larger size due to the presence of the HA tag. (C) Rescue of adult viability of strong hypomorph elavts1 by expression of HA-tagged ELAV, FNE, RBP9, HuR, and NLSRBP9 under the control of the elav gene (n = 200 to 400). (D) Rescue of synaptic growth in elave5/elavts1 flies (raised at the permissive temperature during embryonic development) by expression of HA-tagged ELAV, FNE, RBP9, and HuR under the control of the elav gene is shown as the mean plus standard error of the mean of the number of type 1b boutons at muscle 13 (n = 15 to 28). (E to S) Cellular localization of HA-tagged ELAV, FNE, RBP9, HuR, and NLSRBP9 under the control of the elav gene in larval ventral nerve cord midline neurons. Staining with anti-HA, DAPI, and anti-ELAV is as indicated at the top of the panels. Arrowheads point toward neurons, where NLSHARBP9 is predominantly nuclear, while ELAV becomes cytoplasmic. Scale bar, 10 μm.
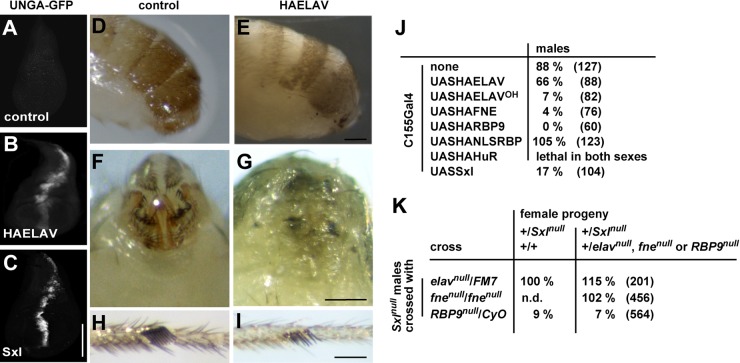
ELAV-related Sxl can regulate alternative splicing of ELAV target genes, and ELAV can interfere with sexual differentiation.
The closest relative of ELAV family RBPs in flies is Sxl, which is the master regulator of sex determination and dosage compensation in Drosophila but a neuronal protein in other Diptera (54, 55). Sxl has 63% and 64% similarity in RRM1 and RRM2, respectively (see Table S1 in the supplemental material), but does not have the third RRM implicated in multimerization although multiple Sxl proteins bind cooperatively to target RNA (48, 56). We therefore asked if Sxl can induce neuron-specific alternative splicing of the UNGA reporter in wing discs. Indeed, Sxl also induced neuron-specific alternative splicing of the UNGA reporter in wing discs (Fig. 7A to C). Since Sxl is able to substitute for ELAV, we next tested if ELAV/Hu family RBPs can interfere with Sxl's role in sexual differentiation. For this experiment, ELAV/Hu family RBPs (ELAV, FNE, RBP9, and HuR) were expressed in the pattern of doublesex (dsx), the main effector for sexual differentiation (57). Expression of ELAV/Hu family RBPs with dsx-GAL4 yielded pharate adults, which showed no genital differentiation in both sexes and impaired development of male sex combs while male pigmentation was not affected, and flies looked normal otherwise when dissected from the pupal case (Fig. 7D to I). Males expressing Hfp or B52 in the dsx pattern did not show impaired development of sex-specific features.
FIG 7.
Sxl can induce alternative splicing of ELAV target nrg, and ELAV family RBPs can interfere with sexual differentiation and dosage compensation. (A to C) Neuronal alternative splicing of the nrg GFP reporter UNGA in control wing discs and upon expression of UAS-HA-ELAV or UAS-Sxl with dpp-GAL4 stained with anti-GFP antibodies. Scale bar, 150 μm. (D to I) Expression of ELAV with dsx-GAL4 inhibits sexual differentiation of male genitals (side and back views in panels D and E panels F and G, respectively) and sex combs (H and I). Scale bars, 100 μm (E and G) and 50 μm (I). (J) Viability of males from neuronal overexpression of UAS transgenes with elav-GAL4C155, shown as percentage relative to females from the same cross. The total number of flies is shown in parentheses. (K) Viability of females from crosses of mutants in ELAV family proteins with Sxl7B0 null males, shown as a percentage relative to balancer-carrying females (elav) or to males (fne and Rbp9) from the same cross. The total number of flies is shown in parentheses.
Maternal RBP9 provides a fail-safe for Sxl-mediated dosage compensation.
When overexpressing ELAV, FNE, and RBP9 from UAS transgenes with neuronal elav-GAL4C155, we noticed pupal lethality of males (Fig. 7J). Similarly, overexpression of Sxl also resulted in male lethality. FNE and RBP9 overexpression was more effective in killing males, suggesting that cytoplasmic localization is required for this effect. Accordingly, routing RBP9 to the nucleus by including a nuclear localization signal (NLSRBP) relieved sex-specific toxicity, suggesting interference with Sxl's role in dosage compensation (Fig. 7J) (14). Similarly, males were effectively killed when a cytoplasmically localized ELAV derivative was expressed (ELAVOH) (44). We therefore reasoned that expression of RBP9 during oogenesis could cooperate with Sxl in translational suppression of male-specific lethal-2 (msl-2) to prevent dosage compensation in the early embryonic stages of female development. Indeed, removing one copy of RBP9 during oogenesis in combination with zygotic heterozygosity for Sxl results in female lethality (Fig. 7K). This effect is of maternal origin since there is no bias in female numbers in the RBP9 stock and since the reverse cross did not show female lethality. Also, female lethality was prevented in Sxl/+ daughters of Rbp9/CyO mothers, when msl-3, another protein of dosage compensation complex (located on the third chromosome) was zygotically removed (104% rescue; n = 102). Thus, maternal provision of RBP9 provides a fail-safe to prevent dosage compensation early in female development.
DISCUSSION
Neuronally coexpressed ELAV/Hu family RBPs are, like many other RBPs, highly conserved and show little discrimination in binding short U-rich motifs in vitro (5). Similar results were obtained for Drosophila ELAV proteins (ELAV, FNE, and RBP9) as well as for human HuR when the extended ELAV binding site in the ewg gene was used. In this ELAV target RNA, a number of short U-rich motifs are interspersed along the 135-nt binding site, but they do not have a fixed position, eluding RNA secondary structure to contribute to target selectivity (20, 36, 40). Likewise, when artificially expressed in nonneuronal larval wing disc cells or during eye development, ELAV/Hu proteins can regulate neuron-specific splicing of ELAV target genes and substitute for ELAV in eye development. The capacity to induce neuron-specific splicing events resides in a very narrow concentration range, and ELAV has only a slightly lower threshold. Accordingly, exchanging the ELAV gene ORF with other ELAV/Hu RBP gene ORFs in the elav gene can substantially substitute for ELAV function in transgenic Drosophila, but nuclear localization is required to provide functionality.
Concentration and localization of ELAV/Hu family proteins direct specificity of mRNA processing.
Our data show that the expression levels and cellular localization of ELAV/Hu proteins are important determinants for selection of target genes. Accordingly, broad overexpression of ELAV or other RNA binding proteins is lethal or results in developmental defects likely due to global misregulation of mRNA processing (41, 58). The functional significance of tissue-specific increased concentrations for alternative splicing has been shown for a number of RNA regulatory proteins (59–61). Also, quantitative variations of SR proteins and of antagonistic heterogeneous nuclear ribonucleoproteins (hnRNPs) have been shown to affect selection of alternative splice sites (62, 63).
The importance of the control of expression of ELAV family proteins is further indicated by the complexity of their genes in Drosophila. They are about 10 to 15 times bigger than the average Drosophila gene, have several promoters, and, most prominently, have unusually long 3′ UTRs (64, 65). Complex transcriptional control and extended UTRs are also found in Hu genes, suggesting that elaborate regulation of expression of ELAV/Hu genes is a key feature of their functioning (66).
In addition, ELAV/Hu proteins have also been found to cross-regulate each other. ELAV controls 3′ UTR extension by suppression of 3′ end processing at proximal poly(A) sites in its own gene and also in fne and Rbp9 (20, 67). Cross-regulation is also found in HuD, where Hu proteins regulate inclusion of an alternatively spliced exon (68). Furthermore, RBP9 and Sxl are required for translational repression of msl-2 to prevent dosage compensation of the X chromosome in female embryos, revealing an ancient relationship between the two proteins. Intriguingly, in flies more distantly related to Drosophila, such as the housefly Musca, Sxl is a neuronal protein but is not required for sex determination and dosage compensation (54, 55).
ELAV/Hu RBPs have both nuclear and cytoplasmic functions in mRNA processing. Hence, differential subcellular localization, e.g., through phosphorylation, provides an additional level to regulate target selection (69). ELAV localizes predominantly to the nucleus, and nuclear localization is required for viability (44), while RBP9 is predominantly cytoplasmic and FNE is about equally distributed between the nucleus and cytoplasm. Given the predominant localization of RBP9 to the cytoplasm, its capacity to regulate splicing in the nucleus is unexpected but could be explained by its shuttling between the nucleus and the cytoplasm, which has been described for human Hu proteins (30).
Distinct roles of Drosophila ELAV proteins in neuronal development and function but convergence in synaptic plasticity.
Mutants in the genes coding for ELAV family RBPs in Drosophila exert mostly distinct phenotypes in nervous system development, maintenance, and function, but all of them show synaptic growth defects. Except for fne and Rbp9 in synaptic growth regulation, no genetic interactions were detected leading to more severe developmental phenotypes. The fne; Rbp9 double mutants have fewer synaptic boutons than individual mutants, which is exactly what we would expect if they have overlapping functions. Since fne and Rbp9 did not genetically interact with elav, they seem to act independently of elav in synaptic growth regulation. Further, the overlapping roles of fne and Rbp9 seem specific to the regulation of synaptic growth since locomotion and life span phenotypes, which assess neuronal function more broadly, were similar in single and double mutants.
Although elav fne; Rbp9 triple mutants die as embryos and elav fne and elav; Rbp9 double mutants as late larvae, no genetic interactions were observed for any developmental phenotype. The lethality of these mutant combinations is likely due to the general weakness of elav mutants. Determination of the target genes of Drosophila ELAV family RBPs in the future will reveal whether they have overlapping roles in regulating neuronal function more broadly.
ELAV/Hu protein-regulated mRNA processing plays a major role in synaptic plasticity.
Although ELAV in Drosophila is expressed as soon as neurons are born, it is mostly not required for neuronal development, with the exception of a minor role in axonal wiring. The severe locomotion defects, including ataxia, of elav mutants suggest that ELAV is required in neurons for proper function or for refining neuronal connections. Indeed, the major target of ELAV, erect wing (ewg), regulates the number of synaptic connections made (32, 52). Essential roles in neuronal function have been found in HuC mutant mice. Here, synthesis of the neurotransmitter glutamate is affected, resulting in reduced neuronal excitability and impaired motor function (24). In contrast, HuD mutant mice have transient developmental defects in the cerebellum, reduced locomotion activity, and learning defects (70). Most intriguingly, however, HuC HuD double mutants have a much more severe neurological phenotype and die soon after birth, suggesting overlapping functions (24). This is further supported by shared sets of target genes of HuC and HuD affecting glutamate synthesis and genes coding for synaptic proteins. Similar observations have also been made for highly related NOVA1 and -2 in mice, which share an extended set of target genes involved in synaptic functions (71).
Our analysis of mutants in Drosophila ELAV family proteins revealed a major role of these RBPs in regulating synaptic growth. The role of ELAV family RBPs in structural synaptic plasticity is reminiscent of regulating higher-order brain functions, e.g., learning and memory. In accordance, HuD is upregulated upon learning in mice and also regulates GAP43 mRNA, which is required for learning and memory (72). It is thus conceivable that a major role of ELAV/Hu proteins is the altering of neuronal plasticity, whereby different ELAV proteins are used to integrate multiple signals to regulate an overlapping set of target genes.
A model for regulating gene expression by highly related RNA binding proteins.
The limited number of genes in higher eukaryotes requires elaborate regulatory networks to generate molecular, cellular, and functional complexity. A key feature of such regulatory networks is the integration of multiple signals to generate a gene expression output, as shown, e.g., for the regulation of synaptic growth in Drosophila (32, 52). It is conceivable, that highly related RBPs can regulate the same genes via overlapping or identical binding sites. Differential control in regulating the concentrations, activity to bind RNA, and cellular localization then serves to integrate cellular status via distinct signaling pathways (Fig. 8). An alternative route to bind target genes has been suggested through recruitment at the promoter and deposition by elongating RNA polymerase II (Pol II) (73). Our data from using heterologous promoters for expressing ewg and nrg reporters, however, argue against this possibility for these genes but might affect a minority of large genes (40, 48, 74).
FIG 8.
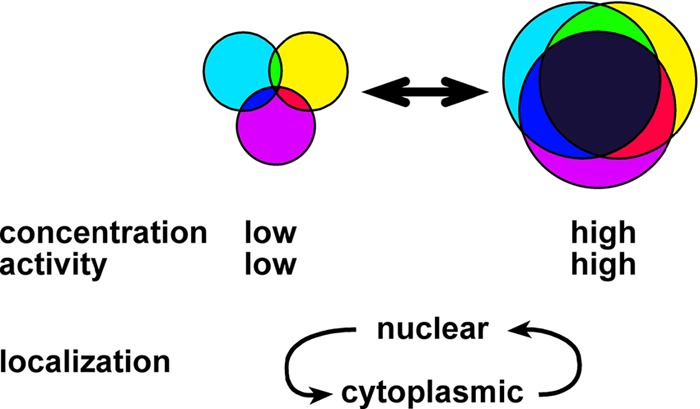
Model for target selectivity and functional diversification of ELAV/Hu family RBPs. Circles represent the complement of targets for ELAV, FNE, and RBP9, and overlapping areas indicate shared targets. Main determinants of target selectivity are concentration, binding activity, and subcellular localization.
In summary, our results demonstrate that ELAV/Hu proteins can exert overlapping functions due to their conserved recognition of highly similar RNA sequences. Their target specificity, however, is tuned by regulating cellular concentration and localization. Increased levels of RNA binding proteins, including ELAV/Hu proteins, have been found in many cancers, illustrating the importance for tight control (63, 75, 76). Thus, alterations of the expression levels, activity, or cellular localization of ELAV/Hu proteins have major implications for human health.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bloomington, Exilixis/Harvard, and Kyoto stock centers and S. Goodwin, J. Simpson, J. Kim, J. Horabin, H. Richardson, and G. Toba for fly lines, M.-L. Samson and Developmental Hybridoma Studies Bank for antibodies, FlyBase for RNA-Seq data, J. Colburn for discussions, and S. Vilain for comments on the manuscript.
For this work we acknowledge funding from the BBSRC and the Wellcome Trust.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00473-15.
REFERENCES
- 1.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 3.Darnell RB. 2013. RNA protein interaction in neurons. Annu Rev Neurosci 36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soller M. 2006. Pre-messenger RNA processing and its regulation: a genomic perspective. Cell Mol Life Sci 63:796–819. doi: 10.1007/s00018-005-5391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kafri R, Springer M, Pilpel Y. 2009. Genetic redundancy: new tricks for old genes. Cell 136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Hinman MN, Lou H. 2008. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soller M, White K. 2004. ELAV. Curr Biol 14:R53. doi: 10.1016/j.cub.2003.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Samson ML. 2008. Rapid functional diversification in the structurally conserved ELAV family of neuronal RNA binding proteins. BMC Genomics 9:392. doi: 10.1186/1471-2164-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okano HJ, Darnell RB. 1997. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci 17:3024–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YJ, Baker BS. 1993. The Drosophila gene rbp9 encodes a protein that is a member of a conserved group of putative RNA binding proteins that are nervous system-specific in both flies and humans. J Neurosci 13:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samson ML, Chalvet F. 2003. found in neurons, a third member of the Drosophila elav gene family, encodes a neuronal protein and interacts with elav. Mech Dev 120:373–383. doi: 10.1016/S0925-4773(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 13.Yao K-M, Samson M-L, Reeves R, White K. 1993. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol 24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- 14.Schutt C, Nothiger R. 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127:667–677. [DOI] [PubMed] [Google Scholar]
- 15.Koushika SP, Lisbin MJ, White K. 1996. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol 6:1634–1641. doi: 10.1016/S0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 16.Koushika SP, Soller M, White K. 2000. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol Cell Biol 20:1836–1845. doi: 10.1128/MCB.20.5.1836-1845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisbin MJ, Qiu J, White K. 2001. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev 15:2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogulja-Ortmann A, Picao-Osorio J, Villava C, Patraquim P, Lafuente E, Aspden J, Thomsen S, Technau GM, Alonso CR. 2014. The RNA-binding protein ELAV regulates Hox RNA processing, expression and function within the Drosophila nervous system. Development 141:2046–2056. doi: 10.1242/dev.101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simionato E, Barrios N, Duloquin L, Boissonneau E, Lecorre P, Agnes F. 2007. The Drosophila RNA-binding protein ELAV is required for commissural axon midline crossing via control of commissureless mRNA expression in neurons. Dev Biol 301:166–177. doi: 10.1016/j.ydbio.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Soller M, White K. 2003. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev 17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toba G, Qui J, Koushika SP, White K. 2002. Ectopic expression of Drosophila ELAV and human HuD in Drosophila wing disc cells reveals functional distinctions and similarities. J Cell Sci 115:2413–2421. [DOI] [PubMed] [Google Scholar]
- 22.Antic D, Lu N, Keene JD. 1999. ELAV tumor antigen, hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev 13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell Mol Life Sci 58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB. 2012. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. 2011. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uren PJ, Burns SC, Ruan J, Singh KK, Smith AD, Penalva LO. 2011. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J Biol Chem 286:37063–37066. doi: 10.1074/jbc.C111.266882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. 2008. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol Cell Biol 28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, Zhou HL, Hasman RA, Lou H. 2007. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem 282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 30.Fan XC, Steitz JA. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos A-R, Grossman D, White K. 1985. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. . J Neurogenet 2:197–218. [DOI] [PubMed] [Google Scholar]
- 32.Haussmann IU, White K, Soller M. 2008. Erect wing regulates synaptic growth in Drosophila by integration of multiple signaling pathways. Genome Biol 9:R73. doi: 10.1186/gb-2008-9-4-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanini D, Jallon JM, Rabinow L, Samson ML. 2012. Deletion of the Drosophila neuronal gene found in neurons disrupts brain anatomy and male courtship. Genes Brain Behav 11:819–827. doi: 10.1111/j.1601-183X.2012.00817.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Kim YJ, Kim-Ha J. 2010. Blood-brain barrier defects associated with Rbp9 mutation. Mol Cells 29:93–98. doi: 10.1007/s10059-010-0040-0. [DOI] [PubMed] [Google Scholar]
- 35.Toba G, Yamamoto D, White K. 2010. Life-span phenotypes of elav and Rbp9 in Drosophila suggest functional cooperation of the two ELAV-family protein genes. Arch Insect Biochem Physiol 74:261–265. doi: 10.1002/arch.20377. [DOI] [PubMed] [Google Scholar]
- 36.Soller M, White K. 2005. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol Cell Biol 25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 38.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet 36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 39.Stowers RS, Schwarz TL. 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haussmann IU, Li M, Soller M. 2011. ELAV-mediated 3′-end processing of ewg transcripts is evolutionarily conserved despite sequence degeneration of the ELAV-binding site. Genetics 189:97–107. doi: 10.1534/genetics.111.131383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraus ME, Lis JT. 1994. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol Cell Biol 14:5360–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinn LM, Dickins RA, Coombe M, Hime GR, Bowtell DD, Richardson H. 2004. Drosophila Hfp negatively regulates dmyc and stg to inhibit cell proliferation. Development 131:1411–1423. doi: 10.1242/dev.01019. [DOI] [PubMed] [Google Scholar]
- 43.Samuels ME, Bopp D, Colvin RA, Roscigno RF, Garcia-Blanco MA, Schedl P. 1994. RNA binding by Sxl proteins in vitro and in vivo. Mol Cell Biol 14:4975–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yannoni YM, White K. 1999. Domain necessary for Drosophila ELAV nuclear localization: function requires nuclear ELAV. J Cell Sci 112:4501–4512. [DOI] [PubMed] [Google Scholar]
- 45.Koushika SP, Soller M, DeSimone SM, Daub DM, White K. 1999. Differential and inefficient splicing of a broadly expressed Drosophila erect wing transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol Cell Biol 19:3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lisbin MJ, Gordon M, Yannoni YM, White K. 2000. Function of RRM domains of Drosophila melanogaster ELAV: RNP1 mutations and RRM domain replacements with ELAV family proteins and SXL. Genetics 155:1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soller M, Haussmann IU, Hollmann M, Choffat Y, White K, Kubli E, Schafer MA. 2006. Sex-peptide-regulated female sexual behavior requires a subset of ascending ventral nerve cord neurons. Curr Biol 16:1771–1782. doi: 10.1016/j.cub.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 48.Toba G, White K. 2008. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res 36:1390–1399. doi: 10.1093/nar/gkm1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preibisch S, Saalfeld S, Tomancak P. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coulom H, Birman S. 2004. Chronic exposure to rotenone models sporadic Parkinson's disease in Drosophila melanogaster. J Neurosci 24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim-Ha J, Kim J, Kim YJ. 1999. Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol Cell Biol 19:2505–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haussmann IU, Soller M. 2010. Differential activity of EWG transcription factor isoforms identifies a subset of differentially regulated genes important for synaptic growth regulation. Dev Biol 348:224–230. doi: 10.1016/j.ydbio.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Soller M, Li M, Haussmann IU. 2008. Regulation of the ELAV target ewg: insights from an evolutionary perspective. Biochem Soc Trans 36:502–504. doi: 10.1042/BST0360502. [DOI] [PubMed] [Google Scholar]
- 54.Bopp D, Saccone G, Beye M. 2014. Sex determination in insects: variations on a common theme. Sex Dev 8:20–28. doi: 10.1159/000356458. [DOI] [PubMed] [Google Scholar]
- 55.Salz HK. 2011. Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev 21:395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Bell LR. 1994. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev 8:2072–2085. doi: 10.1101/gad.8.17.2072. [DOI] [PubMed] [Google Scholar]
- 57.Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. 2010. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci 13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labourier E, Bourbon HM, Gallouzi IE, Fostier M, Allemand E, Tazi J. 1999. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev 13:740–753. doi: 10.1101/gad.13.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrmann I, Dalgliesh C, Liu Y, Danilenko M, Crosier M, Overman L, Arthur HM, Lindsay S, Clowry GJ, Venables JP, Fort P, Elliott DJ. 2013. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet 9:e1003474. doi: 10.1371/journal.pgen.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi J, Su S, McGuffin ME, Mattox W. 2006. Concentration dependent selection of targets by an SR splicing regulator results in tissue-specific RNA processing. Nucleic Acids Res 34:6256–6263. doi: 10.1093/nar/gkl755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venables JP, Bourgeois CF, Dalgliesh C, Kister L, Stevenin J, Elliott DJ. 2005. Up-regulation of the ubiquitous alternative splicing factor Tra2β causes inclusion of a germ cell-specific exon. Hum Mol Genet 14:2289–2303. doi: 10.1093/hmg/ddi233. [DOI] [PubMed] [Google Scholar]
- 62.Caceres JF, Stamm S, Helfman DM, Krainer AR. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 63.Chen M, David CJ, Manley JL. 2012. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol 19:346–354. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci U S A 108:15864–15869. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samson ML. 1998. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics 150:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bronicki LM, Jasmin BJ. 2013. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. RNA 19:1019–1037. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilgers V, Lemke SB, Levine M. 2012. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev 26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, Molfenter J, Zhu H, Lou H. 2010. Promotion of exon 6 inclusion in HuD pre-mRNA by Hu protein family members. Nucleic Acids Res 38:3760–3770. doi: 10.1093/nar/gkq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brauer U, Zaharieva E, Soller M. 2014. Regulation of ELAV/Hu RNA-binding proteins by phosphorylation. Biochem Soc Trans 42:1147–1151. doi: 10.1042/BST20140103. [DOI] [PubMed] [Google Scholar]
- 70.Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. 2005. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci U S A 102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 72.Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. 2004. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci U S A 101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kornblihtt AR. 2005. Promoter usage and alternative splicing. Curr Opin Cell Biol 17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Oktaba K, Zhang W, Lotz TS, Jun DJ, Lemke SB, Ng SP, Esposito E, Levine M, Hilgers V. 2015. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell 57:341–348. doi: 10.1016/j.molcel.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.David CJ, Manley JL. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.