Abstract
Placentation is a process that establishes the maternal-fetal interface and is required for successful pregnancy. The epithelial component of the placenta consists of trophoblast cells, which possess the capacity for multilineage differentiation and are responsible for placenta-specific functions. FOS-like antigen 1 (FOSL1), a component of AP-1 transcription factor complexes, contributes to the regulation of placental development. FOSL1 expression is restricted to trophoblast giant cells and invasive trophoblast cells. In the present study, we characterized the FOSL1 regulatory pathway in rat trophoblast cells. Transcriptome profiling in control and FOSL1 knockdown cells identified FOSL1-dependent gene sets linked to endocrine and invasive functions. FOSL1 was shown to occupy AP-1 binding sites within these gene loci, as determined by chromatin immunoprecipitation (ChIP). Complementary in vivo experiments using trophoblast-specific lentiviral delivery of FOSL1 short hairpin RNAs (shRNAs) provided in vivo validation of FOSL1 targets. FOSL1 actions require a dimerization partner. Coimmunoprecipitation, coimmunolocalization, and ChIP analyses showed that FOSL1 interacts with JUNB and, to a lesser extent, JUN in differentiating trophoblast cells. Knockdown of FOSL1 and JUNB expression inhibited both endocrine and invasive properties of trophoblast cells. In summary, FOSL1 recruits JUNB to form AP-1 transcriptional complexes that specifically regulate the endocrine and invasive trophoblast phenotypes.
INTRODUCTION
The placenta is a specialized tissue of pregnancy that permits development of the embryo within the female reproductive tract and effectively facilitates the redirection of resources from the mother to the fetus (1). Placentation is categorized based on the connectivity between maternal and embryonic tissues. In hemochorial placentation, as seen in rodents and most primate species, maternal blood directly bathes specialized extraembryonic cells referred to as trophoblasts (2). The trophoblast lineage arises early in embryonic development. As the embryo grows, a subset of totipotent stem cells becomes committed to the trophoblast cell lineage (3, 4). These cells are situated on the surface of the blastocyst and are called the trophectoderm. They give rise to a trophoblast stem (TS) cell population initially apposed to the inner cell mass of the blastocyst and expand into the extraembryonic ectoderm (5–7). TS cells differentiate into multiple specialized trophoblast cell types. In rat, TS cells differentiate into syncytial trophoblast cells, spongiotrophoblast cells, glycogen cells, trophoblast giant cells, and invasive trophoblast cells (8, 9). Each differentiated cell type contributes to a core function of the placenta. Syncytial trophoblast cells specialize in transport, spongiotrophoblast and trophoblast giant cells synthesize and secrete peptides and steroid hormones, glycogen cells are an energy reservoir, and invasive trophoblast cells penetrate the uterus and modify the uterine vasculature. Regulatory mechanisms controlling the trophoblast lineage have been investigated (10–13).
Activator protein 1 (AP-1) consists of a family of basic leucine zipper transcription factors induced in response to a variety of extracellular stimuli (14). The composition of the AP-1 family is best characterized as heterodimers of FOS family (FOS, FOSB, FOS-like antigen 1 [FOSL1], and FOSL2) and JUN family (JUN, JUNB, and JUND) proteins or as JUN family homodimers (15, 16). The AP-1 family plays an important role in the regulation of fundamental cellular processes, including cell proliferation, differentiation, motility, and invasion (14–16). There is a remarkable specificity of the actions of AP-1, which is determined by the composition of its constituent proteins (15, 16).
FOS and JUN family transcription factors are expressed in rodent and human trophoblast cells (17–21) and have been implicated in the regulation of transcription of an assortment of genes expressed in trophoblast cells (22–28). Mouse mutagenesis studies have demonstrated roles for FOSL1 and JUNB in placental development (29, 30). Null mutations at either Fosl1 or Junb loci result in early embryonic death. Initial phenotypic descriptions suggested that FOSL1 and JUNB contributed to the regulation of vascularization of the labyrinth zone of the mouse placenta (20, 29). FOSL1 is prominently expressed in trophoblast giant cells and in endovascular invasive trophoblast cells, placing it in a position to potentially regulate the transcription of genes involved in hormone biosynthesis and in vascular remodeling, respectively (20). In rat TS cells, FOSL1 expression is prominently increased during trophoblast differentiation correlated with the acquisition of both endocrine and invasive properties (20, 31). Furthermore, FOSL1 was identified as a downstream mediator of a phosphatidylinositol 3-kinase/AKT signaling pathway promoting trophoblast invasion and vascular remodeling (20). In vivo disruption of FOSL1 by using trophoblast-specific lentiviral delivery of Fosl1 short hairpin RNAs (shRNAs) inhibited the depth of endovascular trophoblast cell invasion (20). These actions of FOSL1 on the invasive trophoblast cell phenotype are conserved in rat and human trophoblast cells (20, 21).
In this study, we delve deeper into the actions of FOSL1 on trophoblast cell differentiation. Targets for FOSL1 action and FOSL1 dimerization partners in differentiating trophoblast cells are identified. In vitro and in vivo research strategies were performed by utilizing TS cells and lentiviral trophoblast-specific gene manipulation, respectively. Our experimental findings demonstrate a cooperative role for FOSL1 and JUNB in regulating trophoblast cell invasive and endocrine phenotypes.
MATERIALS AND METHODS
Animals.
Holtzman Sprague-Dawley rats were obtained from Harlan Laboratories (Indianapolis, IN). Animals were housed in an environmentally controlled facility with lights on from 0600 to 2000 h and were allowed free access to food and water. The presence of a seminal plug or sperm in the vaginal smear was designated day 0.5 of pregnancy. Tissues for histological analysis were frozen in dry ice-cooled heptane and stored at −80°C until processed. Tissue samples for RNA or protein analysis were frozen in liquid nitrogen and stored at −80°C until they were processed. Female rats were made pseudopregnant by mating with vasectomized males. The presence of a seminal plug was designated day 0.5 of pseudopregnancy. The University of Kansas Animal Care and Use Committee approved protocols for the care and use of animals.
Rcho-1 TS cells and rat blastocyst-derived TS cells.
Rcho-1 TS cells were maintained in stem state medium (RPMI 1640 culture medium [Life Technologies, Grand Island, NY] supplemented with 20% fetal bovine serum [FBS; Sigma-Aldrich, St. Louis, MO], 50 μM 2-mercaptoethanol [Sigma-Aldrich], 1 mM sodium pyruvate [Life Technologies], 100 μM penicillin, and 100 U/ml streptomycin [Life Technologies]) and passaged with trypsin-EDTA (Fisher, Pittsburgh, PA) (32, 33). Differentiation was induced by replacing stem state medium with differentiation-promoting medium (NCTC-135 medium [Sigma-Aldrich] supplemented with 1% horse serum [Life Technologies], 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate, 10 mM HEPES, 38 mM sodium bicarbonate, 100 μM penicillin, and 100 U/ml streptomycin [Fisher]). Rat blastocyst-derived TS cells were grown in stem state medium supplemented with fibroblast growth factor 4 (FGF4) (37.5 ng/ml; Sigma-Aldrich), heparin (1.5 μg/ml; Sigma-Aldrich), and rat embryonic fibroblast-conditioned medium (70% of the final volume), as described above. Differentiation was induced by the removal of FGF4, heparin, and rat embryonic fibroblast-conditioned medium (34). Stem cell state cells were collected within 48 h of subculture, and differentiated cells were maintained for 8 days in differentiation medium prior to harvesting.
shRNA constructs and production of lentiviral particles.
Two Fosl1 shRNAs were designed and subcloned into pLKO.1 by using AgeI and EcoRI restriction sites (20). Jun, Junb, Jund, and Grhl1 shRNA constructs subcloned into pLKO.1 were obtained from Sigma-Aldrich (St. Louis, MO). A control shRNA that does not target any known mammalian gene, pLKO.1-shSCR (plasmid 1864), was obtained from Addgene (Cambridge, MA). Sequences representing the sense target site for each of the shRNAs used for analyses are provided in Table 1. Lentiviral packaging vectors (Addgene) were used to produce lentiviral particles. Culture supernatants containing lentiviral particles were harvested every 24 h for 2 to 3 days, centrifuged to remove cell debris, filter sterilized, concentrated by ultracentrifugation, and stored at −80°C until use. Lentiviral vector titers were determined by measurement of p24 Gag antigen by an enzyme-linked immunosorbent assay (Advanced Bioscience Laboratories, Kensington, MD).
TABLE 1.
shRNA sequences
In vitro lentiviral transduction.
Rcho-1 TS cells and rat blastocyst-derived TS cells were exposed to lentiviral particles, selected with puromycin dihydrochloride (2 μg/μl; Sigma-Aldrich) for 2 days, and then maintained with a lower concentration of the antibiotic (1 μg/μl). Puromycin selective pressure was removed during in vitro differentiation.
In vivo lentiviral transduction.
Rat embryos were transduced with lentiviral particles as previously described (20, 35). Briefly, blastocysts collected on gestation day 4.5 were treated with pronase (10 mg/ml for 10 min; Sigma-Aldrich) to remove zonae pellucidae and incubated with concentrated lentiviral particles (750 ng of p24/ml) for 4.5 h. Transduced blastocysts were transferred to uteri of day 3.5 pseudopregnant rats for subsequent evaluation of gene knockdown and placentation site phenotypes on gestation day 11.5.
DNA microarray analysis.
DNA microarray analysis was performed by the University of Kansas Medical Center Biotechnology Support Facility, as previously described (31). Briefly, Affymetrix 230 2.0 DNA microarray chips (Affymetrix, Santa Clara, CA) were probed with cDNAs generated from differentiated Rcho-1 TS cells expressing control or Fosl1 shRNAs. Each experimental group was analyzed in triplicate. Hybridization signals were normalized to signals for the internal controls by using the MAS5 algorithm with Expression Console software (Affymetrix), and fold changes were computed. Significant differences were determined by paired two-tailed Student t tests. Microarray data were processed for functional analysis by using Ingenuity Pathway Analysis (Qiagen, Redwood City, CA). Probe sets included in the analysis were restricted to those with which we detected changes in gene expression of at least 2-fold between group comparisons with signal strengths of ≥500 for the maximal value.
Quantitative reverse transcription-PCR (qRT-PCR).
RNA was extracted by using Tri reagent (Sigma-Aldrich) according to the manufacturer's instructions. cDNAs were reverse transcribed from RNA by using cDNA synthesis reagents from Applied Biosystems (Foster City, CA), according to the manufacturer's instructions. Power SYBR green PCR master mix (Applied Biosystems) was used for PCR. Reaction mixtures were processed by using a 7500 real-time PCR system (Applied Biosystems). Conditions included an initial holding stage (50°C for 2 min and 95°C for 10 min) and 40 cycles (95°C for 15 s and 60°C for 1 min), followed by a dissociation stage (95°C for 15 s, 60°C for 1 min, and then 95°C for 15 s). Primers used for analyses are shown in Table 2. The comparative cycle threshold (ΔΔCT) method was used for relative quantification of the amount of mRNA normalized to the amount of 18S RNA. Values are presented relative to the values for the controls for each gene.
TABLE 2.
Specific primer sequences used for qRT-PCR analysis
| Target gene | Forward primer | Reverse primer | GenBank accession no. |
|---|---|---|---|
| 18s | GCAATTATTCCCCATGAACG | GGCCTCACTAAACCATCCAA | NR_046237.1 |
| Fosl1 | ATCCCCGACCTCTGACCTAT | CAAGGCGTTCCTTCTGCTT | NM_012953.1 |
| Prl3d1 | TTCGGGCTCTGGTATGCAAC | TGGACACAATGGCAGTTGGTTTGG | NM_017363.3 |
| Prl5a1 | TCCACACCAGACATTCCAGA | TTTCCAGGAAGCCAACATTC | NM_138527.1 |
| Gcm1 | CCCCAACAGGTTCCACTAGA | AGGGGAGTGGTACGTGACAG | NM_017186.2 |
| Pcdh12 | GAGCCTGGTTCGACTCTCTG | GGGCTTGGCCGAGTATTTAT | NM_053944.1 |
| Crispld2 | GTATCCCCCTGCCTCCAAC | CAGTGCACAGCCAGGTTCT | NM_138518.2 |
| Grhl1 | AAGGGATCCACCACCCTATC | ATGAAGACCTTCGCCTCATC | XM_234006.8 |
| Cgm4 | GTTCCCTGTGTATCCGCAAT | GTGAGCTTGGCAGGGTTAAA | NM_012525.1 |
| Cyp11a1 | ACAAGCTGCCCTTCAAGAAC | CGCAGCATCTCCTGTACCTT | NM_017286.2 |
| Ppap2b | GCCTCCTTCTCCATGTTCAC | AAGGCCATCATGAGCAAAGT | NM_138905.2 |
| Mmp9 | AACTTCGACGCTGACAAGAA | TTTAGAGCCACGACCATACA | NM_031055.1 |
| Jun | TAACAGTGGGTGCCAACTCA | CGCAACCAGTCAAGTTCTCA | NM_021835.3 |
| Junb | CAGTTACTCCCCAGCCTCTG | ATGTGGGAGGTAGCTGATGG | NM_021836.2 |
| Jund | TGTTGCGTGTGTGTTTCCTT | CCAAGGATTACGGAACAGGA | NM_138875.4 |
| Ddx60 | GTCCTAAGCCAAAGAAGGAT | TGCCCTTCTTTTTGTTCTTCC | XM_008771220.1 |
| Mfap5 | AGAAGGCCTTGCTGCTTGT | CCGCCAGAACTGTATCGTCT | NM_001108644.1 |
| Rdh12 | CTCTTCTCGCCCTTCTTCAA | GCGCTCAGCTGTTTTCTTGT | NM_001108037.1 |
| Micall2 | GACGTGAGCATCACCAACAT | AGCTGTTCCTCAGCCACTTG | XM_006248975.2 |
| Il1r2 | CTCCCCTGGAGACAATACCA | TGGTGTTGGAAGATGTACCAGT | NM_053953.1 |
| Mmd | GATTTTCTCCAGCCTTGGTG | AATGATGCCATCGCTCTTG | NM_001007673.1 |
| LOC171573 | TGAGGCTAAAGAGGACCAGG | CTGACATGCTCCATTGCCAG | NM_138537.2 |
| Flt1 | GGGTGTCTGCTTCTCACAGG | TTTTTGTCCTCCTGGCTCAC | NM_019306.1 |
| Hspb8 | GAAGGTGGGATTGTCTCCAA | CCTGGTTGTCTTGAGGAAGC | NM_053612.2 |
| Ccdc125 | AAAGCCACAAGCCACACAC | ATTCCATTTCCCACTGCAAG | NM_001134761.1 |
| Fdxr | AGATCTGTGTGGTCGGCAGT | AGCGGGCTGTCTGTGTAAAT | NM_024153.1 |
| Rilpl1 | AGACCAAGGAGCAGGAGATG | GGTTGGGGTCCTTGAGGT | NM_001191665.1 |
| Ceacam9 | CCTGCTGATCAGAAACGTCA | AGGTGAGGGTCACAGAACCA | NM_053919.2 |
| Chdh | GTTGGGAATGCAGATGACCT | ACCTCCAGAGCCACTCCAG | NM_198731.2 |
| Ceacam3 | TGGTACAAAGGGCTGACAAA | TCCACAGGTCAAAGTGGAGAA | NM_012702.2 |
| Sema6d | GGCCAGTGATGCTGTCATTT | TATGTTCCACGGCGATTTCT | NM_001107768.1 |
| Cd47 | GAAAAACCGCCCAGTTTCGTGGT | CGGCAACCAGCAGCAGAATGA | NM_019195.2 |
| Cd9 | GCATGCTGGGATTGTTCTTCGGAT | ATGGATGGCTTTGAGTGTTTCCCG | NM_053018.1 |
| Il17f | CAAAACCAGGGCATTTCTGT | GACCAGGATTTCTTGCTGGA | NM_001015011.1 |
| Prl4a1 | TCTGTCTTTGTCTCACCAATGG | GTTCAAACCCTCGGCCTTG | NM_017036.2 |
| Dio3 | CCGCATATGGTGCCTATTTT | GCCCACCAATTCAGTCACTT | NM_017210.3 |
| Mt1a | CACCAGATCTCGGAATGGAC | GTTCGTCACTTCAGGCACAG | NM_138826.4 |
| Adm | TCATCGCAGTCAGTCTTGGA | ACGACTTAGCGCCCACTTAT | NM_012715.1 |
| Zfp36l2 | TTGCAGTTCCGACCATTACA | AACCCCGGAGTGAAGCTC | NM_001036626.1 |
| Ptprk | TTTGAGTGGGTCCATGTCAG | CCTTCTGGCTGTACAACAGGT | NM_001029902.2 |
| Tex19.2 | TTTTGGCCTCATGGGAGATA | CAGCTTCACACTTGCTCCAA | NM_001109622.1 |
| Ppap2a | ACCATTTACAAAGCCGTTGG | AGTGCCCCGAGTAGAAGGAC | NM_022538.2 |
| Ceacam1 | ACCCAATCAAGCTGGACGTA | TTGAGGGTTTGTGCTCTGTG | NM_031755.2 |
| Igdcc3 | TGAGGGCGACTATGAATGTG | TGGGGACTCTGTTCTTCTCC | XM_006243296.2 |
| Akap12 | GCAGGAGGAGACATCAGAAA | CTTCGCCTTCATCCTTCTTG | NM_057103.2 |
| Icam2 | TTTCACTTGTTCGGGAAAGC | TCTGCCACAGAGCAGAGAGA | NM_001007725.1 |
| Rbks | GCTTGCCAAAAACTGGAGAA | GGAGGCTGTTCCTGTAGCTG | NM_001108703.1 |
| Myo1f | CCAAGAACCCTCTGAACCTG | GATGGCGTCCTCAGTGATCT | NM_001108076.1 |
| Chchd10 | CTCAGCTGTAGGGCATGTCA | GAAGCCCTCACACAGGGTTA | NM_001007008.1 |
| Gbx2 | GACGGCAAAGCCTTCTTG | CGCTCTCCAGAGAGAAGCTC | NM_053708.2 |
| H19 | AGCTCGGACTGGAGACTAGG | GGCAAAGGAAAGAACAGACG | NR_027324.1 |
| Bex1 | AGCAGGAGGAGGAGGAAGAG | TCCTCCTTTTTCTGATGGTCA | NM_001037365.1 |
| Serpinh1 | CCAGAGGTCACCAAGGATGT | GGTGCATCATCGTAACACCC | NM_017173.1 |
| Mmp12 | GCACATTTTGATGAGGCAGA | TTGATTTTGGATTATTGGAATGC | NM_053963.2 |
| Il11ra1 | CAAGTTCCGGTTGCAATACC | TAGGAGTACCCCAGGCCTCT | NM_139116.1 |
| Grhl2 | TGGAAGCCACCAAATCTCTC | GGTTTATTGCTGCGGTTGTT | NM_001134527.1 |
| Grhl3 | GAGACCCAGCCTGTGTTGTT | CCTCGCTTGCATTTCTTGTA | NM_001106690.1 |
Primary antibodies.
Antibodies to FOSL1 (SC-605) and JUNB (SC-8051 for all applications except chromatin immunoprecipitation [ChIP], for which SC-73 was used) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). We obtained antibodies to GRHL1 (HPA005798) and pancytokeratin (F3418) from Sigma-Aldrich. Antibodies to JUN (610326), JUND (5226-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AB2302), and histone H3 (ab1791) were obtained from BD Biosciences (Franklin Lakes, NJ), Epitomics (Burlingame, CA), Millipore (Billerica, MA), and Abcam (Cambridge, MA), respectively. Antibodies to PRL3D1 and CYP11A1 were previously generated and characterized by our laboratory (36, 37).
Western blotting.
Whole-cell lysates were prepared in lysate buffer (Cell Signaling Technology, Danvers, MA). Nuclear lysates were prepared with an NE-PER nuclear protein extraction kit (Thermo Fisher Scientific, Rockford, IL). Lysates were fractionated by SDS-PAGE. Separated proteins were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes. Blots were probed with the indicated antibodies overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies to rabbit IgG (Cell Signaling Technology) or mouse IgG (Sigma-Aldrich) for 1 h at room temperature. Reaction products were visualized by incubation with Luminata Western HRP substrates according to the manufacturer's instructions (Millipore). Lysates from 293FT cells (Invitrogen, Eugene, OR) transfected with rat Jund cDNA subcloned into pCMV SPORT6 (GE Healthcare, Lafayette, CO) were used as a positive control for JUND immunoblots.
Immunolocalization.
Ten-micrometer cryosections of placentation sites were prepared and stored at −80°C until use. Tissue sections mounted onto glass slides or Rcho-1 TS cells plated onto chamber slides were fixed in 4% paraformaldehyde, washed with phosphate-buffered saline (PBS), and permeabilized in PBS containing 0.25% Triton X-100. Following a blocking step with 10% normal goat serum for 30 min, slides containing the specimens were incubated with the indicated primary antibodies overnight at 4°C and subsequently incubated with secondary antibodies for an additional 30 min at room temperature. We used a secondary goat anti-rabbit antibody conjugated with cyanine 3 bis-N-hydroxysuccinimide (NHS) ester (Cy3; Jackson ImmunoResearch Laboratories Inc., West Grove, PA), a secondary goat anti-rabbit antibody conjugated with Alexa Fluor 488 (Invitrogen), or a secondary goat anti-mouse antibody conjugated with fluorescein isothiocyanate (FITC; Sigma-Aldrich). Nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Carlsbad, CA). Trophoblast cells were visualized by immunostaining with pancytokeratin antibodies. Fluorescence images were captured by using a Leica DMI 4000 microscope equipped with a charge-coupled-device camera (Leica Microsystems GmbH, Welzlar, Germany).
Analysis of DNA content.
DNA content was estimated by flow cytometry, as previously described (31). Cells were trypsinized, fixed in 70% ethanol, and then stained with propidium iodine.
Chromatin immunoprecipitation.
ChIP analysis was performed according to a previously reported procedure (20). Briefly, Rcho-1 TS cells stably transduced with control, Fosl1, or Junb shRNA-expressing lentiviruses were grown to confluence in 150-mm dishes and differentiated for 8 days. Cells were then fixed with 1% formaldehyde, and purified nuclear lysates were sonicated on ice to prepare DNA fragments at a size of ∼500 bp. Lysates were immunoprecipitated with 5 μg of FOSL1 or JUNB antibodies and collected on protein A-agarose beads (Sigma-Aldrich). Rabbit IgG (BD Biosciences) was used as a nonspecific control. Immunoprecipitated chromatin fragments were washed and eluted from protein A-agarose beads. DNA-protein complexes were reverse cross-linked and purified by using a QIAquick PCR purification kit (Qiagen). Purified DNA fragments were characterized by quantitative PCR (qPCR) using primers listed in Table 3. Putative AP-1 response elements (TGAGTCA) were targeted in the ChIP analysis and identified by searching specific gene loci, including 10 kbp flanking the locus at the 5′ and 3′ ends, using rVista 2.0 (http://rvista.dcode.org/) (38). Occupancy/enrichment was normalized to values for input samples by use of the ΔΔCT method and presented relative to values for IgG controls.
TABLE 3.
Specific primer sequences used for ChIP analysis
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| Prl3d1 | ATCATATGGGGGAAATGCAG | TCATTCAACTTTCTGCCTCCT |
| Crispld2 | TCGAGTGACCAGGTGTGTGT | CCCTCCCAAAAGTGACTCAA |
| Grhl1 | CCTTGGGGCTTCAGCCATTT | ATAAGTGGGCAAGGGTCCTG |
| Cgm4 | GGGAACAGTGCTTTTGAGGA | TAACTTAGTGGCCGGGACAT |
| Cyp11a1 | GAGTCCCAGACCCAGAGAGG | CCACTGATTCCACAGCAATG |
| Ppap2b | CCGACTTCTGGTTTCTGGTC | CGCTACCATGAGGAAAGGAG |
| Mmp9 | GAGTCAGCGTAAGCCTGGAG | AGCAGAATTTGCGGAGGTTT |
| Fosl1 | AGTCACTGAGGCTGAGTCAC | TATGTCCCCAGCCCAATACT |
Coimmunoprecipitation.
Rcho-1 TS cells or rat blastocyst-derived TS cells were washed with ice-cold PBS and extracted in cell lysis buffer (Epitomics). Lysates were immunoprecipitated by using the indicated antibodies and collected on protein A-agarose beads. Rabbit IgG or mouse IgG (BD Biosciences) was used as a nonspecific control. Immunoprecipitated protein complexes were washed with PBS, eluted with cell lysate buffer, separated by SDS-PAGE, and processed for Western blot analysis using the Clean Blot IP detection reagent (Thermo).
Matrigel invasion assay.
In vitro cellular invasive activities were measured as previously described (39). Rcho-1 TS cells were differentiated for 8 days, trypsinized, and placed onto transwell inserts (6.5-mm diameter and 8-μm pore size; BD Biosciences) coated with 400 μg/ml phenol red-free Matrigel (BD Biosciences) at a density of 3 × 104 cells per insert. Cells were initially plated in stem state medium overnight and then switched to differentiation medium and allowed to invade for 72 h. Invaded cells situated on the undersurface of the membrane were stained with Diff-Quik (Dade Behring, Newark, DE), visualized by stereomicroscopy, and counted.
Progesterone radioimmunoassay.
Measurements of progesterone concentrations in medium conditioned by Rcho-1 TS cells were performed by using a radioimmunoassay (RIA), as previously reported (40). Progesterone concentrations were normalized to cellular DNA content with an E.Z.N.A. tissue DNA kit (Omega Bio-Tek, Norcross, GA).
Statistical analyses.
Values are expressed as means ± standard errors of the means (SEM). All experiments were conducted at least in triplicate and were replicated two to three times. Statistical comparisons between two means were evaluated with Student's t test. Analysis of variance and Tukey's post hoc tests were used for assessing differences among three or more means by using GraphPad Prism (GraphPad Software Inc., La Jolla, CA).
Microarray data accession number.
The array results have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE68272.
RESULTS
FOSL1 and the trophoblast lineage.
The hemochorial placenta consists of multiple trophoblast lineages. These cell types differentiate from TS cells and include trophoblast giant cells, spongiotrophoblast cells, glycogen cells, invasive trophoblast cells, and syncytial trophoblasts (8, 9). The activation of specific sets of genes defines each lineage. Monitoring the expression of Prl3d1, Tpbpa, Prl5a1, Gcm1, and Pcdh12 provides insights into the development of trophoblast giant cells, spongiotrophoblast cells, invasive trophoblast cells, syncytial trophoblasts, and glycogen trophoblast cells (32, 34, 41). To evaluate the mechanisms of trophoblast cell differentiation, we utilized Rcho-1 TS cells, which can be induced to differentiate (32).
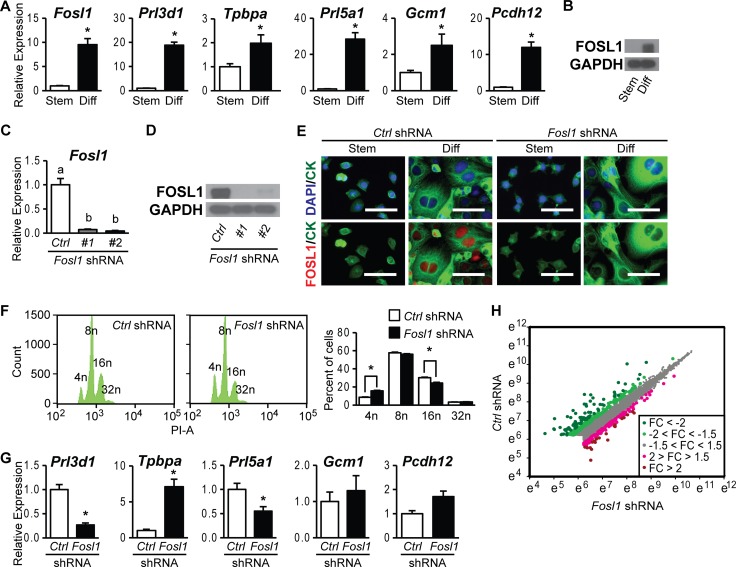
The expression level of FOSL1 was low in stem state Rcho-1 TS cells, as we reported previously (Fig. 1A and B) (20, 31). After differentiation, FOSL1 expression was dramatically increased, localized to nuclei, and correlated with elevated expression levels of the trophoblast cell lineage-specific markers Prl3d1, Tpbpa, Prl5a1, Gcm1, and Pcdh12 (Fig. 1A) (20, 31). We next evaluated the role of FOSL1 in trophoblast differentiation following genetic manipulation by lentiviral vector-transduced Fosl1 shRNAs. FOSL1 expression was efficiently inhibited in differentiated trophoblast cells expressing Fosl1 shRNAs (Fig. 1C to E). Knockdown of FOSL1 did not adversely affect trophoblast giant cell formation but significantly decreased trophoblast giant cell DNA content (Fig. 1F). Similar effects on ploidy have been observed for Rcho-1 TS cells following inhibition of phosphatidylinositol 3-kinase (31). Note that Rcho-1 TS cells are tetraploid (33). Disruption of FOSL1 also compromised the expression of the trophoblast cell differentiation-associated transcripts Prl3d1 and Prl5a1 but not that of Gcm1 or Pcdh12 (Fig. 1G). Tpbpa transcript levels were significantly increased following FOSL1 knockdown, suggesting that FOSL1 represses Tpbpa expression and/or the presence of Tpbpa-expressing trophoblast cells. These findings further support the involvement of FOSL1 as a regulator of the differentiation of specific trophoblast cell lineages.
FIG 1.
FOSL1 regulation of the trophoblast cell phenotype. (A) qRT-PCR analysis of Fosl1, Prl3d1, Tpbpa, Prl5a1, Gcm1, and Pcdh12 expression in stem (Stem) and differentiated (Diff) Rcho-1 TS cells. (B) Western blot analysis of FOSL1 and GAPDH expression in stem and differentiated Rcho-1 TS cells. (C and D) FOSL knockdown efficiency in differentiated Rcho-1 TS cells expressing control (Ctrl) or Fosl1 shRNAs was validated by qRT-PCR (C) and Western blot (D) analyses. GAPDH was used as an internal control for Western blot analysis. (E) FOSL1 localization and knockdown efficiency in stem and differentiated Rcho-1 TS cells expressing control or Fosl1 shRNAs were assessed by immunocytochemistry. FOSL1 was localized to nuclei. Cells were counterstained with cytokeratin (CK) and/or DAPI. Bar = 100 μm. (F) DNA content was estimated by propidium iodine staining followed by flow cytometry. PI-A, propidium iodide staining area. (G) qRT-PCR analysis of Prl3d1, Tpbpa, Prl5a1, Gcm1, and Pcdh12 transcripts in differentiated Rcho-1 TS cells expressing control or Fosl1 shRNAs. (H) Scatter plot of data from microarray analysis shown as the natural logarithm of raw signal values for differentiated Rcho-1 TS cells expressing control (x axis) or Fosl1 (y axis) shRNAs. The magnitudes of fold changes (FC) are depicted as different-colored dots. Bars in the histograms in panels A, C, F, and G represent means ± SEM. Asterisks or different letters above each bar indicate significant differences (P < 0.05).
FOSL1 targets in differentiating trophoblast cells.
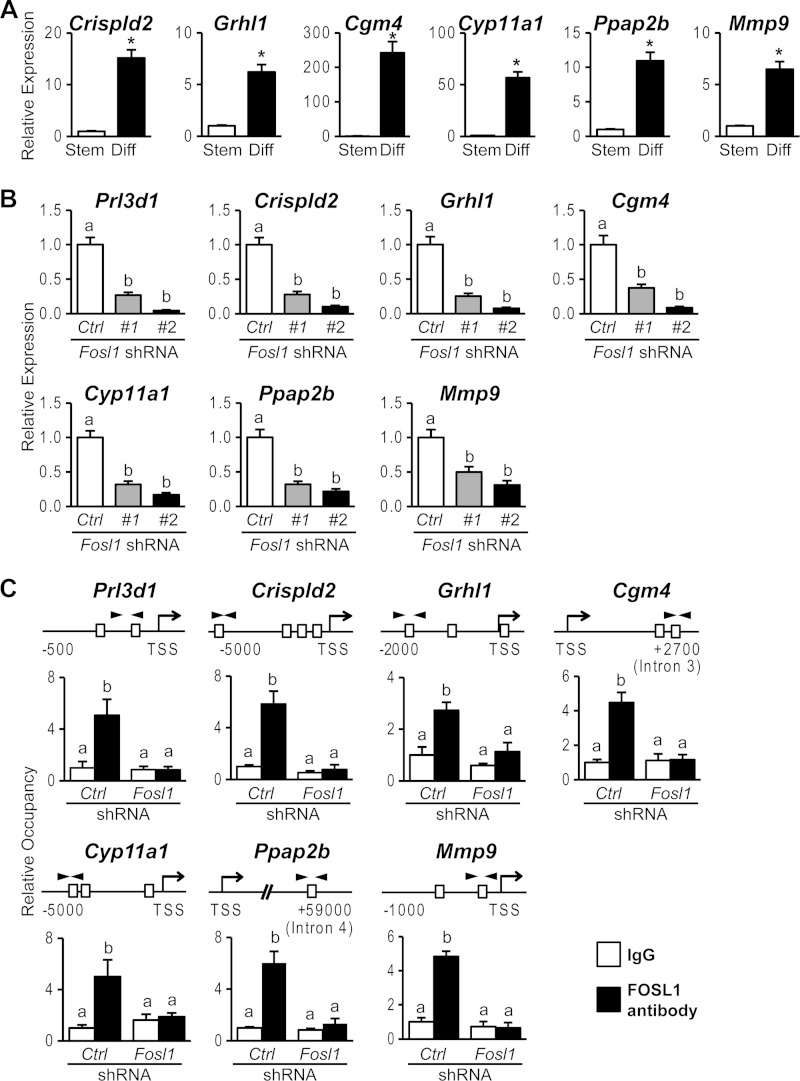
Since FOSL1 is known to interact with DNA and regulate transcription, we next sought to identify components of the transcriptome affected by FOSL1. DNA microarray analysis was performed by using differentiated trophoblast cells under control conditions and following FOSL1 knockdown. Transcripts regulated by FOSL1 were functionally categorized into pathways regulating specific cellular functions (Fig. 1H and Tables 4 to 6). A subset of the FOSL1-dependent transcripts was selected for further analysis by qRT-PCR. This subset of transcripts exhibited increased expression during trophoblast cell differentiation and a dependence on FOSL1 (Fig. 2A and B). We next determined whether FOSL1 interacted with putative AP-1 binding motifs located in the vicinity of these target genes. FOSL1 ChIP analysis of the differentiated trophoblast cells was performed, which demonstrated FOSL1 occupancy at consensus AP-1 binding sites within these target gene loci (Fig. 2C). Collectively, these observations are consistent with a role for FOSL1 in the transcriptional regulation of trophoblast differentiation.
TABLE 4.
Functional analysis of selected genes regulated by FOSL1
| Molecule(s) in network | Score | Top disease(s) and/or function(s) |
|---|---|---|
| ADM, AKAP12, BCAR3, Cgm4, CSRP1, CYP11A1, DIO3, DUSP14, FDXR, FOSL1, IGFBP3, IL1R2, LY6E, MMD, MMP9, MT1, NOV, SEMA6D, TXNIP | 41 | Cardiovascular system development/function, organ morphology, organismal development |
| CHDH, CHSY1, DENND2C, EIF4E3, FLYWCH2, GNPTAB, HECA, MOSPD1, MYO1F, PARVG, PHLDB2, PPAP2A, RASL10B, RDH12, RILPL1, SALL1, TSPAN33 | 26 | Cellular compromise, cell-to-cell signaling and interaction, cellular movement |
| CD47, GNE, HSPB1, HSPB8, IGDCC3, KRT15, NEDD4L, PBX3, Prl3d1, PTPRK, RDM1, ZAK, ZFP36L2 | 26 | Hereditary disorder, neurological disease, organismal injury and abnormalities |
| ARHGDIB, CEACAM1, CLDN4, CRISPLD2, F3, ICAM2, LAMC2, PDGFA, PLSCR1, PPAP2B, STIM1, TIMP3 | 26 | Cellular movement, cancer, tumor morphology |
| CLDN4, CPEB2, FAM110B, FDXR, FOSL1, FXYD6, GRHL1, HNF4A, ITIH3, RBKS, ROBO3, TIMP3 | 14 | Cellular movement, cellular development, reproductive system development and function |
| CHCHD10 | 2 | Cancer, cell cycle, cell death and survival |
| MLLT3 | 2 | Cancer, cardiovascular disease, cardiovascular system development and function |
TABLE 5.
Selected genes downregulated by Fosl1 shRNA in differentiated Rcho-1 TS cells identified by microarray analysis and validated by qRT-PCR analysis
| Protein | Gene symbol | Functiona |
Fosl1 knockdown/control knockdown transcript expression ratio by: |
|
|---|---|---|---|---|
| Microarray | qRT-PCR (±SEM) | |||
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | Ddx60 | RNA helicase | 0.04 | 0.17 ± 0.02 |
| Microfibrillar-associated protein 5 | Mfap5 | Cell adhesion/ECM | 0.07 | 0.37 ± 0.11 |
| Retinol dehydrogenase 12 | Rdh12 | Retinoid metabolism | 0.12 | 0.17 ± 0.02 |
| Prolactin family 3, subfamily d, member 1 | Prl3d1 | Hormone/cytokine | 0.13 | 0.27 ± 0.04 |
| Cys-rich secretory protein LCCL domain 2 | Crispld2 | ECM assembly | 0.17 | 0.28 ± 0.04 |
| Mical-like 2 | Micall2 | Cytoskeleton organizer | 0.20 | 0.17 ± 0.02 |
| Interleukin 1 receptor type II | Il1r2 | Decoy cytokine receptor | 0.20 | 0.09 ± 0.02 |
| Monocyte-to-macrophage differentiated | Mmd | Progestin/adipoq receptor family, unknown function | 0.20 | 0.20 ± 0.03 |
| Grainyhead-like 1 | Grhl1 | Transcription factor | 0.21 | 0.25 ± 0.04 |
| Carcinoembryonic antigen gene family 4 | Cgm4 | Ligand; immune/vasculature | 0.21 | 0.37 ± 0.05 |
| Cytochrome P450, family 11, subfamily a, polypeptide 1 | Cyp11a1 | Steroid biosynthesis | 0.25 | 0.32 ± 0.05 |
| Spleen protein 1 precursor | LOC171573 | Unknown function | 0.27 | 0.17 ± 0.03 |
| Fms-related tyrosine kinase 1 | Flt1 | Growth factor receptor; vasculature | 0.28 | 0.29 ± 0.04 |
| Heat shock protein B8 | Hspb8 | Chaperone; stress response | 0.30 | 0.36 ± 0.05 |
| Coiled-coil domain-containing 125 | Ccdc125 | Cell motility | 0.32 | 0.52 ± 0.10 |
| Phosphatidic acid phosphatase 2B (LPP3) | Ppap2b | Phospholipid phosphatase; signal transduction | 0.34 | 0.32 ± 0.05 |
| Ferredoxin reductase | Fdxr | Mitochondrial flavoprotein; electron transport | 0.34 | 0.66 ± 0.05 |
| Rab-interacting lysosomal protein-like 1 | Rilpl1 | Regulation of cilium function | 0.36 | 0.57 ± 0.09 |
| Carcinoembryonic antigen-cell adhesion molecule 9 | Ceacam9 | Cell adhesion | 0.36 | 0.34 ± 0.04 |
| Choline dehydrogenase | Chdh | Mitochondrial phospholipid and 1-carbon metabolism | 0.36 | 0.36 ± 0.05 |
| Matrix metalloproteinase 9 | Mmp9 | ECM remodeling | 0.38 | 0.50 ± 0.08 |
| Carcinoembryonic antigen-related cell adhesion molecule 3 | Ceacam3 | Cell adhesion | 0.40 | 0.13 ± 0.03 |
| Semaphorin 6D | Sema6d | Ligand; cell guidance | 0.41 | 0.71 ± 0.14 |
| Cd47 molecule | Cd47 | Cell adhesion/thrombospondin receptor | 0.44 | 0.55 ± 0.07 |
| Cd9 molecule | Cd9 | Tetraspanin; cell adhesion/signal transduction | 0.55 | 0.59 ± 0.08 |
| Interleukin 17F | Il17f | Cytokine | 0.66 | 0.64 ± 0.17 |
| Prolactin family 4, subfamily a, member 1 | Prl4a1 | Hormone/cytokine | 0.66 | 0.34 ± 0.06 |
ECM, extracellular matrix.
TABLE 6.
Selected genes upregulated by Fosl1 shRNA in differentiated Rcho-1 TS cells identified by microarray and validated by qRT-PCR analysis
| Protein | Gene symbol | Functiona |
Fosl1 knockdown/control knockdown transcript expression ratio by: |
|
|---|---|---|---|---|
| Microarray | qRT-PCR (±SEM) | |||
| Deiodinase, iodothyronine, type iii | Dio3 | Thyroid hormone inactivation | 3.52 | 2.86 ± 0.23 |
| Metallothionein 1a | Mt1a | Metal ion homeostasis | 2.97 | 5.85 ± 0.42 |
| Adrenomedullin | Adm | Ligand; vasculature | 2.58 | 2.20 ± 0.43 |
| Zinc finger protein 36, c3h-type-like 2 | Zfp36l2 | Putative transcription factor | 2.45 | 2.46 ± 0.27 |
| Protein tyrosine phosphatase, receptor type, k | Ptprk | Protein tyrosine phosphatase; signal transduction | 2.36 | 2.77 ± 0.25 |
| Testis-expressed gene 19.2 protein | Tex19.2 | Unknown function | 2.30 | 1.72 ± 0.54 |
| Phosphatidic acid phosphatase type 2a | Ppap2a | Phospholipid phosphatase; signal transduction | 2.21 | 3.17 ± 0.09 |
| Carcinoembryonic antigen-related cell adhesion molecule 1 | Ceacam1 | Cell adhesion | 2.16 | 4.36 ± 0.37 |
| Immunoglobulin superfamily, dcc subclass, member 3 | Igdcc3 | Unknown function | 2.15 | 2.52 ± 0.13 |
| A kinase (Prka) anchor protein 12 | Akap12 | Scaffold protein for protein kinase A/C; signal transduction | 2.13 | 1.92 ± 0.20 |
| Intercellular adhesion molecule 2 | Icam2 | Cell adhesion; leukocytes, immune function | 2.11 | 1.85 ± 0.19 |
| Ribokinase | Rbks | Ribose metabolism; energy production, nucleotide and amino acid biosynthesis | 2.09 | 3.32 ± 0.05 |
| Myosin IF | Myo1f | Unconventional myosin; intracellular movement | 2.08 | 3.35 ± 0.12 |
| Coiled-coil-helix-coiled-coil-helix domain-containing 10 | Chchd10 | Mitochondrial protein, possibly oxidative phosphorylation | 2.08 | 2.30 ± 0.09 |
| Gastrulation brain homeobox 2 | Gbx2 | Transcription factor | 1.97 | 2.89 ± 0.62 |
| H19, imprinted maternally expressed transcript | H19 | Noncoding RNA; imprinting | 1.94 | 2.25 ± 0.09 |
| Brain-expressed gene 1 protein | Bex1 | Adaptor; signal transduction | 1.92 | 1.99 ± 0.21 |
| Serine (or cysteine) peptidase inhibitor, clade h, member 1 | Serpinh1 | Collagen-specific chaperone; collagen biosynthesis | 1.89 | 1.73 ± 0.29 |
| Matrix metallopeptidase 12 | Mmp12 | ECM remodeling | 1.76 | 1.89 ± 0.64 |
| Interleukin 11 receptor, alpha chain 1 | Il11ra1 | Cytokine receptor-associated signal transducer | 1.73 | 1.85 ± 0.26 |
ECM, extracellular matrix.
FIG 2.
FOSL1-dependent genes activated in trophoblast cells differentiating in vitro. (A) qRT-PCR analyses of Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in stem (Stem) or differentiated (Diff) Rcho-1 TS cells. (B) qRT-PCR analyses of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control (Ctrl) or Fosl1 shRNAs. (C) FOSL1 occupancy at DNA regions possessing putative AP-1 binding elements within Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 loci. ChIP was used for determination of FOSL1 occupancy in differentiated Rcho-1 TS cells expressing control or Fosl1 shRNAs. Schematic representations of putative AP-1 elements (boxes) at each gene locus and the location of primers used to amplify regions containing the putative AP-1 elements (arrowheads) are shown at the top. IgG was included as a negative control. Bars represent means ± SEM. Values with an asterisk or different characters indicate significant differences (P < 0.05). TSS, transcription start site.
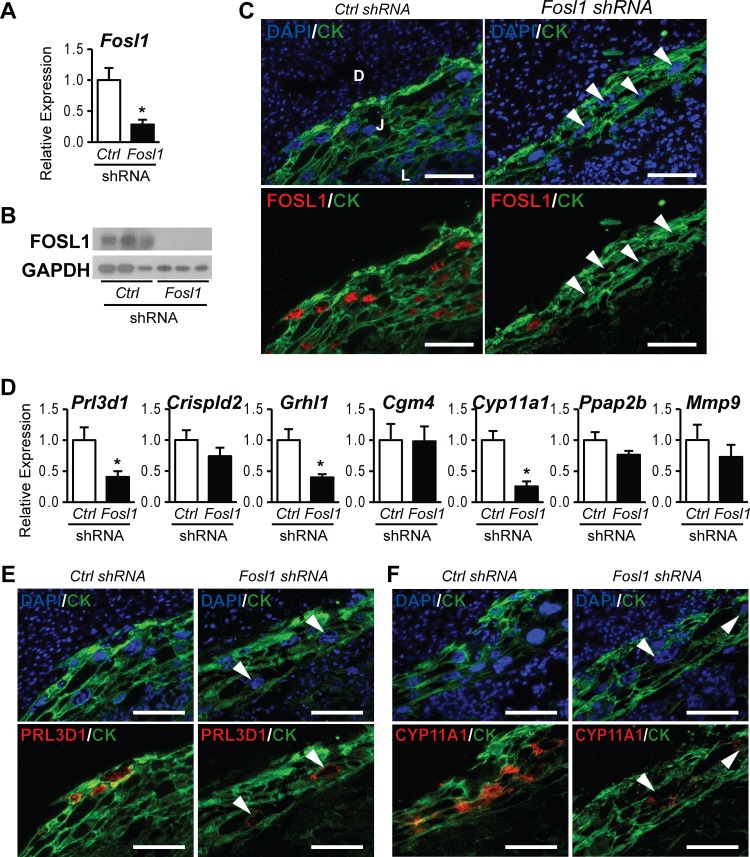
We subsequently evaluated the in vivo role of FOSL1 using a trophoblast lineage-specific gene knockdown strategy (20, 35). FOSL1 was strongly expressed in nuclei of trophoblast giant cells within the uteroplacental interface at embryonic day 11.5 (E11.5), and its expression significantly decreased following FOSL1 knockdown (Fig. 3A to C), similar to our previously reported observations (20). In vivo FOSL1 knockdown significantly attenuated the expression of Prl3d1, Grhl1, and Cyp11a1 (Fig. 3D). Immunohistochemical analysis further confirmed the dependence of PRL3D1 and CYP11A1 expression on FOSL1 (Fig. 3E and F). Discrepancies in FOSL1 targets identified by using in vitro versus in vivo models may be linked to several factors, including (i) the impact of distinct environmental factors in the uterine decidua and/or the embryo on the in vivo behavior of trophoblast cells, which are absent in cell culture; (ii) the presence of unique factors under in vitro culture conditions, which are absent in the in vivo model; and (iii) distinct compositions of trophoblast lineages in the dissected in vivo specimens versus the Rcho-1 TS cell cultures.
FIG 3.
FOSL1-dependent genes activated in trophoblast cells differentiating in vivo. Blastocysts were transduced with a lentivirus expressing control (Ctrl) or Fosl1 shRNAs and were transferred to day 3.5 pseudopregnant rats. (A to C) Trophoblasts dissected from placentation sites were evaluated on gestation day 11.5. FOSL1 knockdown efficiency was assessed by qRT-PCR (A), Western blot (B), and immunohistochemistry (C) analyses. (D) qRT-PCR analyses of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in control or Fosl1 knockdown placentation sites. (E and F) Immunohistochemical analyses of PRL3D1 (E) and CYP11A1 (F) expression in control or Fosl1 knockdown placentation sites. Bars in the histograms in panels A and D represent means ± SEM. Values significantly different from those of controls are indicated with an asterisk (P < 0.05). Tissue sections were immunostained for FOSL1, PRL3D1, or CYP11A1 and counterstained with cytokeratin (CK) and/or DAPI. Arrowheads indicate trophoblast giant cells, which show minimal expression of FOSL1, PRL3D1, or CYP11A1 (C, E, and F). D, uterine decidua; J, junctional zone of the placenta; L, labyrinth zone of the placenta.
The findings of these experiments indicate that FOSL1 targets a defined set of genes that are activated during trophoblast cell differentiation.
GRHL1 regulation of trophoblast cell differentiation.
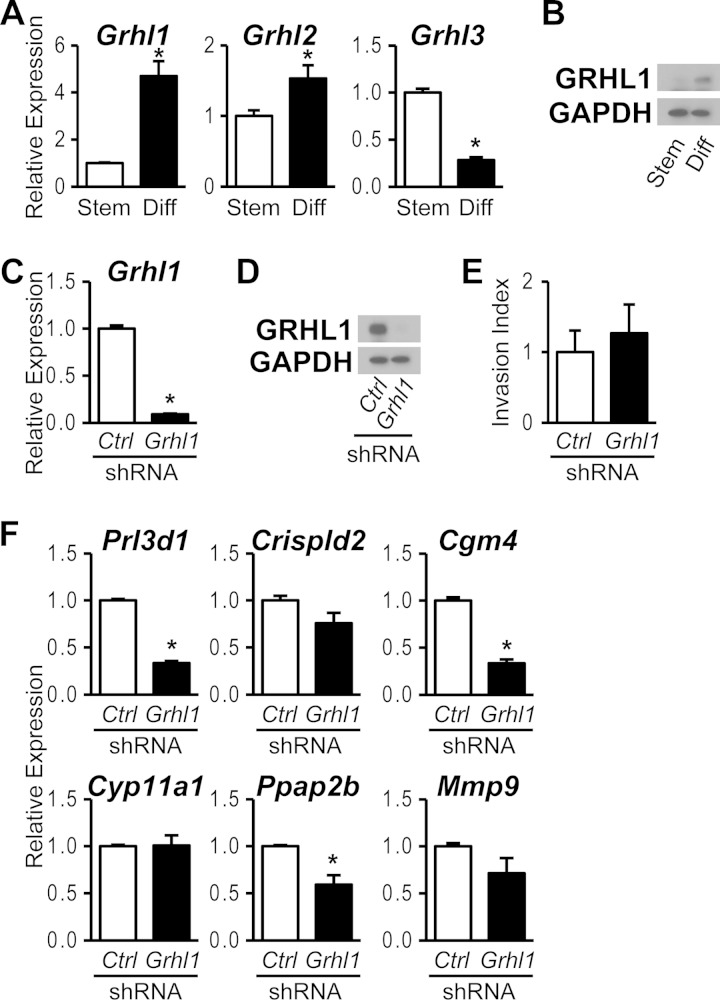
Among the FOSL1 target genes was the transcription factor GRHL1, an evolutionarily conserved regulator of epithelial cell differentiation (42, 43) and a component of the human trophoblast transcriptome (44, 45). GRHL1 belongs to the mammalian GRHL transcription factor family consisting of GRHL1, GRHL2, and GRHL3 (42, 43). The expression of Grhl1 was robustly upregulated during trophoblast cell differentiation, whereas the expression level of Grhl2 exhibited only a modest increase accompanying differentiation (Fig. 4A and B). In contrast, Grhl3 was highly expressed in the stem state, and expression declined following differentiation (Fig. 4A). The differentiation-dependent increase in GRHL1 expression and its dependence on FOSL1 led us to evaluate a possible role for GRHL1 as a mediator of FOSL1 actions on the differentiated trophoblast cell phenotype. GRHL1 expression was successfully inhibited with Grhl1 shRNAs (Fig. 4C and D). Disruption of GRHL1 expression did not significantly affect trophoblast cell migration through a Matrigel extracellular matrix but inhibited a subset of FOSL1 targets, including Prl3d1, Cgm4, and Ppap2b (Fig. 4E and F). These findings suggest that GRHL1 contributes to the downstream actions of FOSL1 on trophoblast cell differentiation.
FIG 4.
GRHL1-dependent genes activated in trophoblast cells differentiating in vitro. (A) qRT-PCR analyses of Grhl1, Grhl2, and Grhl3 transcripts in stem (Stem) and differentiated (Diff) Rcho-1 TS cells. (B) Western blot analysis of GRHL1 and GAPDH expression in stem and differentiated Rcho-1 TS cells. (C and D) GRHL1 knockdown efficiency in differentiated Rcho-1 TS cells expressing control (Ctrl) or Grhl1 shRNA was validated by qRT-PCR (C) and Western blot (D) analyses. (E) Analysis of trophoblast invasive abilities in differentiated Rcho-1 TS cells expressing control or Grhl1 shRNAs by Matrigel transwell chamber assays. (F) qRT-PCR analysis of Prl3d1, Crispld2, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control or Grhl1 shRNAs. Bars in the histograms in panels A, C, E, and F represent means ± SEM. Values significantly different from those of controls are indicated with an asterisk (P < 0.05).
FOSL1 interactions with the JUN family.
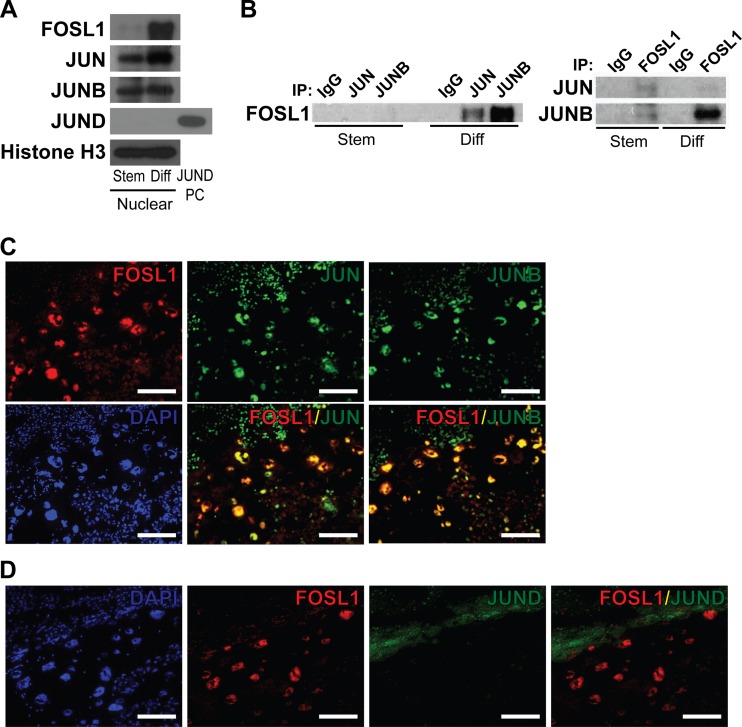
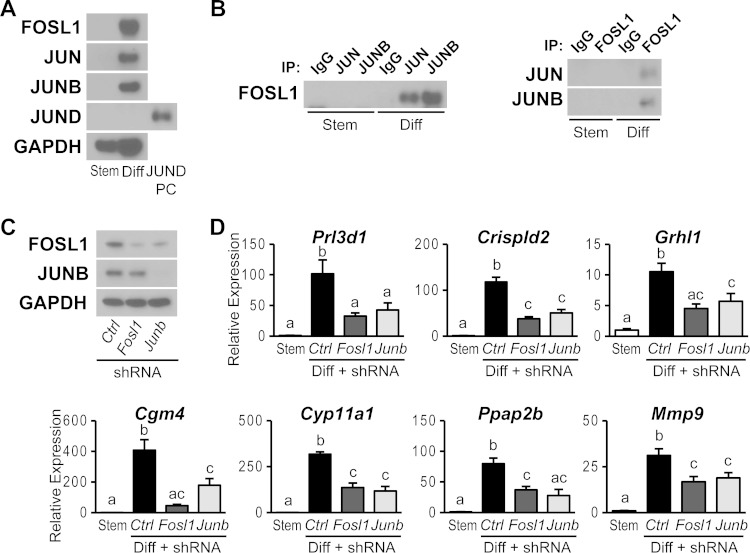
FOSL1 requires a dimerization partner to regulate gene transcription (14–16). This partner is often a member of the JUN family (14–16). This led to an evaluation of potential FOSL1 partners in trophoblast cells. JUN and JUNB were readily detected in nuclei of differentiating trophoblast cells, whereas JUND expression was below the level of sensitivity of the assay (Fig. 5A). Coimmunoprecipitation experiments demonstrated that FOSL1 interacts with JUN and JUNB (Fig. 5B). The relative abundance of the interacting proteins was most impressive for interactions between FOSL1 and JUNB. Dual-fluorescence immunohistochemistry analysis demonstrated colocalization of FOSL1 with JUN and JUNB in trophoblast cells from midgestation placentation sites (Fig. 5C and D). These findings suggest that FOSL1 heterodimerizes with JUNB and, to a lesser extent, with JUN in differentiating trophoblast cells.
FIG 5.
Association of FOSL1 with JUN family members in trophoblast cells. (A) Western blot analysis of AP-1 family proteins using nuclear extracts from stem (Stem) or differentiated (Diff) Rcho-1 TS cells. Lysates from 293FT cells overexpressing JUND were used as a positive control (PC) for JUND expression. (B) Coimmunoprecipitation analysis for detection of FOSL1 binding partners. (Left) Immunoprecipitation (IP) using IgG or antibodies to JUN or JUNB and Western blotting for FOSL1. (Right) Immunoprecipitation using IgG or antibodies to FOSL1 and Western blotting for JUN or JUNB. (C) Immunohistochemical characterization of FOSL1 and JUN or of FOSL1 and JUNB on the same sections of gestation day 13.5 rat placenta. (D) Immunohistochemical characterization of FOSL1 and JUND on serial sections of gestation day 13.5 rat placenta. Nuclei were visualized with DAPI. Bar = 100 μm.
JUN family regulation of trophoblast cell differentiation.
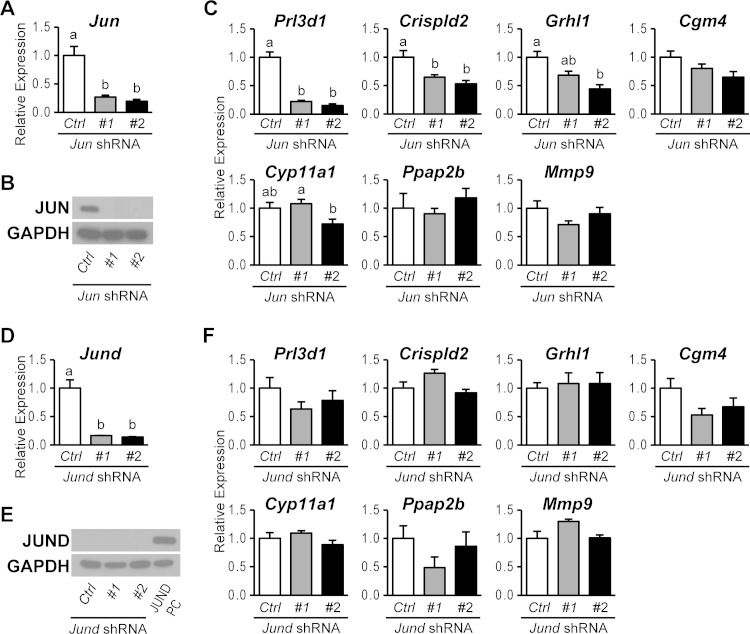
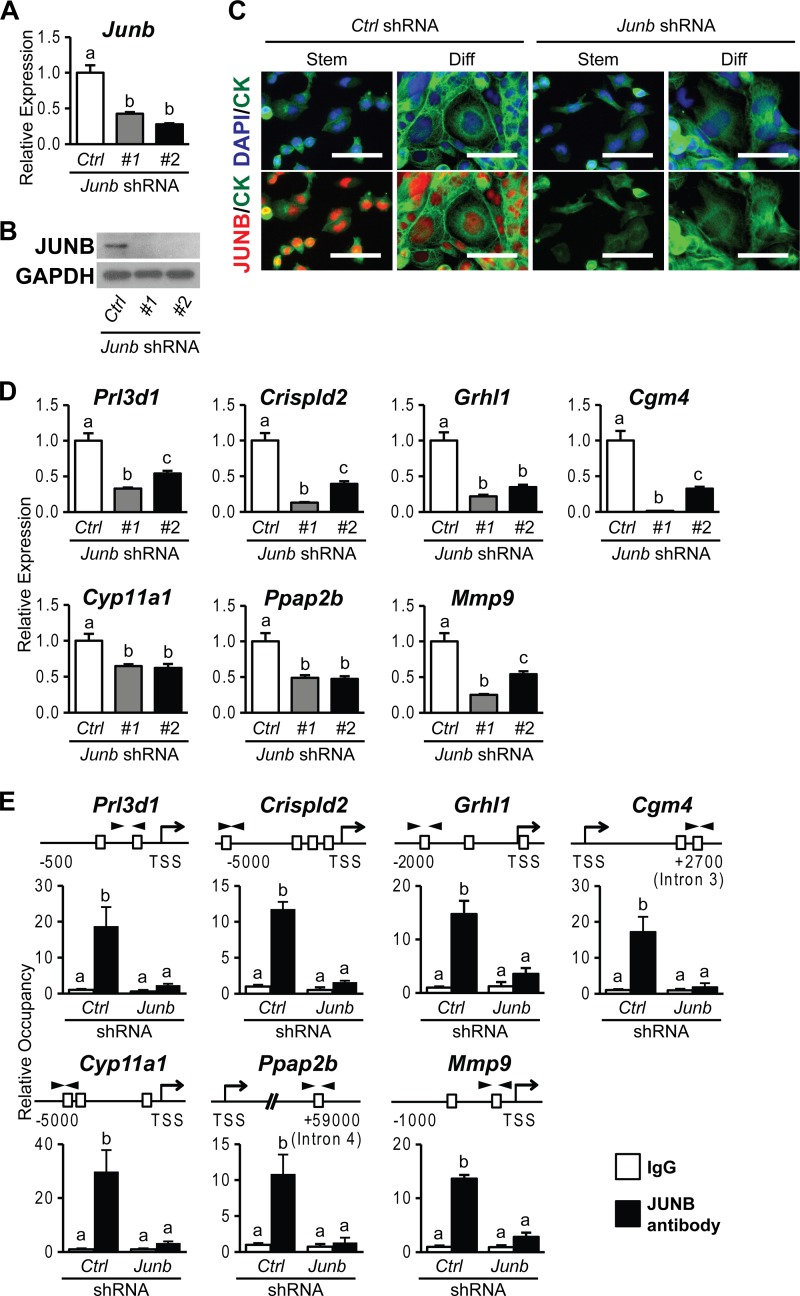
Physical interactions of FOSL1 with JUN family members prompted an investigation of the potential involvement of each JUN family member in FOSL1-guided trophoblast cell differentiation. JUN, JUND, and JUNB expression was silenced in differentiating trophoblast cells by using specific sets of shRNAs. FOSL1 targets were evaluated following knockdown of JUN family members. The expression of each JUN family member was successfully inhibited by using two different shRNAs (Fig. 6A, B, D, and E and 7A to C). JUN knockdown showed strong inhibition of Prl3d1 expression and moderate but significant inhibition of Crispld2 and Grhl1 expression (Fig. 6C). The expression of other FOSL1 target genes (Cgm4, Cyp11a1, Ppap2b, and Mmp9) was not significantly affected by JUN knockdown. JUND knockdown did not affect the expression of any of the previously identified FOSL1 target genes (Fig. 6F). In contrast, JUNB knockdown phenocopied FOSL1 knockdown. Each of the FOSL1 target genes investigated was significantly inhibited by JUNB knockdown (Fig. 7D). JUNB ChIP analysis further demonstrated that JUNB occupancy was increased at the target gene loci (Fig. 7E). The findings mirror the coimmunoprecipitation results for interactions between FOSL1 and JUN family members in differentiated trophoblast cells. In summary, JUN represents a redundant contributor to the activation of a subset of FOSL1 targets, whereas the evidence indicates that FOSL1 and JUNB fully cooperate in the regulation of gene expression during trophoblast cell differentiation.
FIG 6.
JUN- and JUND-dependent genes activated in trophoblast cells differentiating in vitro. (A and B) JUN knockdown efficiency in differentiated Rcho-1 TS cells expressing control (Ctrl) or Jun shRNAs was validated by qRT-PCR (A) and Western blot (B) analyses. (C) qRT-PCR analysis of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control or Jun shRNAs. (D and E) JUND knockdown efficiency in differentiated Rcho-1 TS cells expressing control or Jund shRNAs was validated by qRT-PCR (D) and Western blot (E) analyses. Lysates from 293FT cells overexpressing JUND were used as a positive control (PC) for JUND expression. (F) qRT-PCR analysis of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control or Jund shRNAs. Bars in the histograms in panels A, C, D, and F represent means ± SEM. Different letters above each bar indicate significant differences (P < 0.05).
FIG 7.
JUNB-dependent genes activated in trophoblast cells differentiating in vitro. (A and B) JUNB knockdown efficiency in differentiated Rcho-1 TS cells expressing control (Ctrl) or Junb shRNAs was validated by qRT-PCR (A) and Western blot (B) analyses. GAPDH was used as an internal control. (C) JUNB localization and knockdown efficiency in stem (Stem) and differentiated (Diff) Rcho-1 TS cells expressing control or Junb shRNAs were assessed by immunocytochemistry. Cells were immunostained for JUNB and counterstained with cytokeratin (CK) and/or DAPI. Bar = 100 μm. (D) qRT-PCR analysis of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in differentiated Rcho-1 TS cells expressing control or Junb shRNAs. (E) ChIP analysis of JUNB occupancy in differentiated Rcho-1 TS cells expressing control or Junb shRNAs. Schematic representations of putative AP-1 elements (boxes) at each gene locus and the location of primers used to amplify regions containing the putative AP-1 elements (arrowheads) are shown at the top. IgG was included as a negative control. Bars in the histograms represent means ± SEM. Different letters above each bar indicate significant differences (P < 0.05).
FOSL1 and JUNB regulation of the trophoblast phenotype.
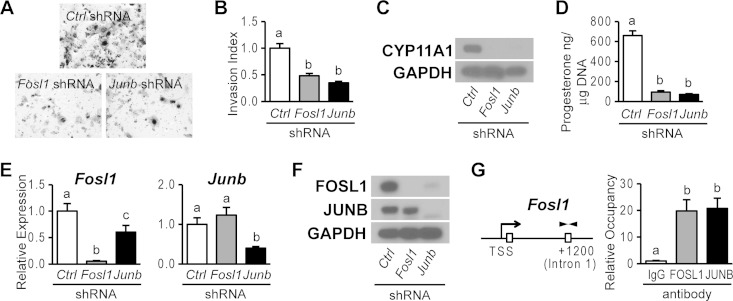
Trophoblast cells exhibit a number of specialized functions, including their capacity to invade extracellular matrices and produce steroid hormones. Trophoblast cell invasion is critical for remodeling the uterine interface (46) and steroid hormone production, especially progesterone, a hormone indispensable for the maintenance of pregnancy (47). In the experiments described above (Fig. 2 and 7), FOSL1 and JUNB were directly linked to the regulation of expression of Mmp9, which encodes a matrix metalloproteinase implicated in trophoblast cell invasion (48, 49), and Cyp11a1, which encodes the rate-limiting enzyme controlling progesterone biosynthesis (50). Consequently, the effects of FOSL1 and JUNB knockdowns on trophoblast cell invasion and progesterone production were investigated. Disruption of either FOSL1 or JUNB similarly inhibited the movement of trophoblast cells through Matrigel (Fig. 8A and B), an in vitro measure of their invasive potential. Additionally, knockdown of either FOSL1 or JUNB inhibited CYP11A1 protein concentrations and progesterone production in trophoblast cells (Fig. 8C and D). Since we obtained similar phenotypes when either FOSL1 or JUNB was disrupted, we examined the potential roles of each transcription factor in the expression of its partner. FOSL1 knockdown inhibited FOSL1 expression but not JUNB expression, whereas JUNB knockdown inhibited both JUNB and FOSL1 expressions (Fig. 8E and F). ChIP analysis further demonstrated that both FOSL1 and JUNB occupy the Fosl1 gene locus (Fig. 8G). Thus, FOSL1 and JUNB also contribute to positive feedback regulation of FOSL1 expression during trophoblast differentiation.
FIG 8.
FOSL1 and JUNB regulation of the differentiated trophoblast phenotype. (A and B) Analysis of trophoblast invasive abilities in differentiated Rcho-1 TS cells expressing control, Fosl1, or Junb shRNAs by Matrigel chamber assays. (A) Representative filters showing trophoblast invasion through Matrigel. (B) Graphic representation of results from the Matrigel invasion chamber assays. Cells from eight replicates were counted, and results were normalized to values for control samples. (C) Western blot analysis of CYP11A1 expression in differentiated Rcho-1 TS cells expressing control, Fosl1, or Junb shRNAs. (D) RIA of progesterone in medium conditioned by differentiated Rcho-1 TS cells expressing control, Fosl1, or Junb shRNAs. Progesterone concentrations are normalized to cell DNA content. (E and F) qRT-PCR (E) and Western blot (F) analyses of FOSL1 and JUNB expression in differentiated Rcho-1 TS cells expressing control, Fosl1, or Junb shRNAs. (G) ChIP analysis of FOSL1 and JUNB occupancy at the Fosl1 locus in differentiating Rcho-1 TS cells. A schematic representation of putative AP-1 elements (boxes) at the Fosl1 gene locus and the location of primers used to amplify the region containing the putative AP-1 elements (arrowheads) is shown on the left. IgG was included as a negative control. Bars in the histograms represent means ± SEM. Different letters above each bar indicate significant differences (P < 0.05).
Partnership of FOSL1 and JUNB in blastocyst-derived rat TS cell differentiation.
The roles of FOSL1 and JUNB were next investigated by using blastocyst-derived rat TS cells (Fig. 9A to D). Blastocyst-derived rat TS cells exhibit many similarities to Rcho-1 TS cells but also exhibit some differences, including their dependence on FGF4 for proliferation and their capacity for differentiation (34). Similar to Rcho-1 TS cells, rat blastocyst-derived TS cells prominently expressed JUN and JUNB, whereas JUND expression was below the level of detection (Fig. 9A). Additionally, FOSL1 was shown to specifically interact with JUN and JUNB (Fig. 9B), as was demonstrated in Rcho-1 TS cells (Fig. 4B). Each of the FOSL1 and JUNB targets identified in Rcho-1 TS cells was also validated for blastocyst-derived rat TS cells (Fig. 9C and D). Some differences in the expression of JUN and JUNB were noted for rat TS cells, which included differentiation-dependent increases in both JUN and JUNB expression levels (Fig. 9A). These trophoblast cell-associated JUN and JUNB expression patterns may reflect the disparate conditions used to culture Rcho-1 TS cells versus rat blastocyst-derived TS cells or, alternatively, intrinsic differences in their developmental states (34).
FIG 9.
FOSL1 and JUNB regulation of the differentiated trophoblast phenotype in blastocyst-derived rat TS cells. (A) Western blot analysis of AP-1 family proteins using cell lysates from stem and differentiated (Diff) blastocyst-derived rat TS cells. Lysates from 293FT cells overexpressing JUND were used as a positive control (PC) for JUND expression. (B) Coimmunoprecipitation analysis for detection of FOSL1 binding partners. (Left) Immunoprecipitation (IP) using IgG or antibodies to JUN or JUNB and Western blot analysis of FOSL1. (Right) Immunoprecipitation using IgG or antibodies to FOSL1 and Western blot analysis of JUN or JUNB. (C) FOSL1 and JUNB knockdown efficiencies in differentiated rat TS cells expressing control (Ctrl), Fosl1, or Junb shRNAs were validated by Western blot analysis. GAPDH was used as an internal control. (D) qRT-PCR analysis of Prl3d1, Crispld2, Grhl1, Cgm4, Cyp11a1, Ppap2b, and Mmp9 transcripts in stem or differentiated blastocyst-derived rat TS cells expressing control, Fosl1, or Junb shRNAs. Bars in the histograms represent means ± SEM. Different letters above each bar indicate significant differences (P < 0.05).
Collectively, our findings indicate that FOSL1 and JUNB cooperate to control elements of trophoblast cell differentiation, including the acquisition of invasive and endocrine properties.
DISCUSSION
Elucidation of signaling pathways controlling trophoblast cell differentiation is key to understanding the process of placentation. In the present study, we utilized Rcho-1 TS cells, rat blastocyst-derived TS cells, and developing rat placentation sites to characterize the involvement of FOSL1 and JUNB in the regulation of trophoblast cell differentiation. FOSL1 and JUNB cooperate in differentiating trophoblast cells to regulate target genes dictating endocrine and invasive trophoblast lineages.
The expression pattern of FOSL1 was helpful in identifying it as a candidate regulator of trophoblast cell differentiation. In the TS cell stem state, FOSL1 is expressed at very low levels and does not possess a recognized function. Thus, stable TS cells expressing shRNAs against Fosl1 were readily established. These Fosl1 shRNAs effectively inhibited the differentiation-dependent upregulation of FOSL1, altering the differentiated trophoblast cell transcriptome and the ability of trophoblast cells to invade extracellular matrices and produce peptides and steroid hormones. FOSL1 targets such as GRHL1 may in turn regulate elements of the FOSL1-directed differentiation pathway. The actions of FOSL1 could be tracked to its occupancy at regions containing AP-1 binding motifs for several target genes identified by transcriptome profiling. This does not represent the full breadth of the involvement of FOSL1 in regulating the differentiated trophoblast cell genome. FOSL1 can be viewed as an entry point into a regulatory network controlling trophoblast cell differentiation. Identification of its genome-wide binding sites via ChIP sequencing and its binding partners will provide additional insight into the regulation of trophoblast cell differentiation.
We utilized a candidate approach to identify FOSL1-interacting proteins. FOSL1 is known to bind to JUN family proteins (14–16). Both JUN and JUNB were shown to bind to FOSL1 and participate in some of its actions. Among the FOSL1 targets and cellular responses examined in this report, JUNB knockdown phenocopied FOSL1 knockdown. FOSL1 and JUNB have been independently implicated as regulators of placental development through mouse mutagenesis experiments (29, 30). Furthermore, disruption of the Ppap2b gene, a target for FOSL1 action (this study), also leads to abnormalities in placentation resembling the phenotypes of Fosl1- and Junb-null mouse models (51). Mutagenesis of the Jun gene results in embryonic death due to liver pathologies and no overt placental abnormalities (52), implying that its role in the placenta may overlap those of other genes. JUN and JUNB exhibit increased abundances accompanying the differentiation of rat blastocyst-derived TS cells, whereas Rcho-1 TS cells express JUN and JUNB similarly in the stem and differentiated states. Disruption of JUN or JUNB expression in the Rcho-1 TS cell stem state slowed cell proliferation, suggesting that JUN and JUNB could be acting as homodimers or interacting with other JUN or FOS family members or some other protein partner(s). Furthermore, the implication is that the differentiation-dependent increase in FOSL1 expression redirects JUN and JUNB to a new set of targets characteristic of the differentiated trophoblast cell phenotype. JUND was not identified as a participant in the actions of FOSL1. Nonetheless, JUND contributes to the regulation of placentation. JUND is a positive regulator of glial cell missing 1 (GCM1) in developing trophoblast cells (28). GCM1 is a transcription factor and is critical for the development of the syncytial trophoblast lineage and the bidirectional transport of nutrients and wastes between maternal and fetal compartments (53, 54). Thus, at least in part, FOSL1 and JUND expression patterns are segregated and contribute to distinct interfaces: trophoblast-maternal and trophoblast-fetal, respectively. Rcho-1 TS cells and rat blastocyst-derived TS cells do not readily differentiate into syncytial trophoblasts (32–34), which is consistent with our inability to demonstrate the involvement of JUND in the regulation of trophoblast differentiation. In addition to the JUN family, FOSL1 is known to interact with several other regulatory proteins (55); however, the nature of these and other potential protein interactions in FOSL1 regulation of trophoblast differentiation remains to be determined.
In summary, FOSL1 and JUNB play a fundamental role in regulating trophoblast differentiation. They exhibit overlapping expression patterns in trophoblast cells, interact with each other, and share target genes. Target genes for FOSL1 and JUNB include genes encoding proteins contributing to cell motility, invasion, and endocrine functions of trophoblast cells. We conclude that FOSL1 recruitment of JUNB to specific targets within the genome during differentiation is a key regulatory step in the acquisition of trophoblast cell invasive and endocrine phenotypes.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants HD020676, HD079363, and GM103418). K.K. is supported by American Heart Association and Japan Society for the Promotion of Science postdoctoral fellowships.
We thank Dong-soo Lee and Adam Krieg (University of Kansas Medical Center, Kansas City, KS) for assistance with and advice on the in vivo lentiviral knockdown and ChIP analyses, respectively. We also appreciate the comments and advice of Michael W. Wolfe, KUMC, during various stages of this project, and we thank Stacy McClure, Lesley Shriver, and Martin Graham for administrative assistance.
REFERENCES
- 1.Amoroso EC. 1968. The evolution of viviparity. Proc R Soc Med 61:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wooding FBP, Burton GJ. 2008. Comparative placentation—structures, functions, and evolution. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Rossant J. 2001. Stem cells from the mammalian blastocyst. Stem Cells 19:477–482. doi: 10.1634/stemcells.19-6-477. [DOI] [PubMed] [Google Scholar]
- 4.Cockburn K, Rossant J. 2010. Making the blastocyst: lessons from the mouse. J Clin Invest 120:995–1003. doi: 10.1172/JCI41229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner RL, Beddington RS. 1988. Multi-lineage ‘stem' cells in the mammalian embryo. J Cell Sci 10(Suppl):11–27. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 7.Guzman-Ayala M, Ben-Haim N, Beck S, Constam DB. 2004. Nodal protein processing and fibroblast growth factor 4 synergize to maintain a trophoblast stem cell microenvironment. Proc Natl Acad Sci U S A 101:15656–15660. doi: 10.1073/pnas.0405429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares MJ, Chapman BM, Rasmussen CA, Dai G, Kamei T, Orwig KE. 1996. Differentiation of trophoblast endocrine cells. Placenta 17:277–289. doi: 10.1016/S0143-4004(96)90051-X. [DOI] [PubMed] [Google Scholar]
- 9.Soares MJ, Chakraborty D, Rumi MAK, Konno T, Renaud SJ. 2012. Rat placentation: an experimental model for investigating the hemochorial maternal-fetal interface. Placenta 33:233–243. doi: 10.1016/j.placenta.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons DG, Cross JC. 2005. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol 284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Roberts RM, Fisher SJ. 2011. Trophoblast stem cells. Biol Reprod 84:412–421. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer PL, Pearton DJ. 2012. Trophoblast development. Reproduction 143:231–246. doi: 10.1530/REP-11-0374. [DOI] [PubMed] [Google Scholar]
- 13.Latos PA, Hemberger M. 2014. Review: the transcriptional and signalling networks of mouse trophoblast stem cells. Placenta 35(Suppl):S81–S85. doi: 10.1016/j.placenta.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Shaulian E, Karin M. 2002. AP-1 as a regulator of cell life and death. Nat Cell Biol 4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 15.Hess J, Angel P, Schorpp-Kistner M. 2004. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 16.Eferl R, Wagner EF. 2003. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 17.Bamberger AM, Bamberger CM, Aupers S, Milde-Langosch K, Löning T, Makrigiannakis A. 2004. Expression pattern of the activating protein-1 family of transcription factors in the human placenta. Mol Hum Reprod 10:223–228. doi: 10.1093/molehr/gah011. [DOI] [PubMed] [Google Scholar]
- 18.Briese J, Sudahl S, Schulte HM, Löning T, Bamberger AM. 2005. Expression pattern of the activating protein-1 family of transcription factors in gestational trophoblastic lesions. Int J Gynecol Pathol 24:265–270. doi: 10.1097/01.pgp.0000163023.49965.10. [DOI] [PubMed] [Google Scholar]
- 19.Marzioni D, Todros T, Cardaropoli S, Rolfo A, Lorenzi T, Ciarmela P, Romagnoli R, Paulesu L, Castellucci M. 2010. Activating protein-1 family of transcription factors in the human placenta complicated by preeclampsia with and without fetal growth restriction. Placenta 31:919–927. doi: 10.1016/j.placenta.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Kent LN, Rumi MA, Kubota K, Lee DS, Soares MJ. 2011. FOSL1 is integral to establishing the maternal-fetal interface. Mol Cell Biol 31:4801–4813. doi: 10.1128/MCB.05780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renaud SJ, Kubota K, Rumi MA, Soares MJ. 2014. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem 289:5025–5039. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shida MM, Ng YK, Soares MJ, Linzer DI. 1993. Trophoblast-specific transcription from the mouse placental lactogen-I gene promoter. Mol Endocrinol 7:181–188. doi: 10.1210/mend.7.2.8469232. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, Duckworth ML. 1999. Identification of a placental-specific enhancer in the rat placental lactogen II gene that contains binding sites for members of the Ets and AP-1 (activator protein 1) families of transcription factors. Mol Endocrinol 13:385–399. doi: 10.1210/mend.13.3.0243. [DOI] [PubMed] [Google Scholar]
- 24.Orwig KE, Soares MJ. 1999. Transcriptional activation of the decidual/trophoblast prolactin-related protein gene. Endocrinology 140:4032–4039. doi: 10.1210/endo.140.9.6954. [DOI] [PubMed] [Google Scholar]
- 25.Peters TJ, Chapman BM, Wolfe MW, Soares MJ. 2000. Placental lactogen-I gene activation in differentiating trophoblast cells: extrinsic and intrinsic regulation involving mitogen-activated protein kinase signaling pathways. J Endocrinol 165:443–456. doi: 10.1677/joe.0.1650443. [DOI] [PubMed] [Google Scholar]
- 26.Bischof P, Truong K, Campana A. 2003. Regulation of trophoblastic gelatinases by proto-oncogenes. Placenta 24:155–163. doi: 10.1053/plac.2002.0890. [DOI] [PubMed] [Google Scholar]
- 27.Abell AN, Granger DA, Johnson NL, Vincent-Jordan N, Dibble CF, Johnson GL. 2009. Trophoblast stem cell maintenance by fibroblast growth factor 4 requires MEKK4 activation of Jun N-terminal kinase. Mol Cell Biol 29:2748–2761. doi: 10.1128/MCB.01391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashif M, Hellwig A, Hashemolhosseini S, Kumar V, Bock F, Wang H, Shahzad K, Ranjan S, Wolter J, Madhusudhan T, Bierhaus A, Nawroth P, Isermann B. 2012. Nuclear factor erythroid-derived 2 (Nfe2) regulates JunD DNA-binding activity via acetylation: a novel mechanism regulating trophoblast differentiation. J Biol Chem 287:5400–5411. doi: 10.1074/jbc.M111.289801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schorpp-Kistner M, Wang ZQ, Angel P, Wagner EF. 1999. JunB is essential for mammalian placentation. EMBO J 18:934–948. doi: 10.1093/emboj/18.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiber M, Wang ZQ, Jochum W, Fetka I, Elliott C, Wagner EF. 2000. Placental vascularisation requires the AP-1 component fra1. Development 127:4937–4948. [DOI] [PubMed] [Google Scholar]
- 31.Kent LN, Konno T, Soares MJ. 2010. Phosphatidylinositol 3 kinase modulation of trophoblast cell differentiation. BMC Dev Biol 10:97. doi: 10.1186/1471-213X-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faria TN, Soares MJ. 1991. Trophoblast cell differentiation: establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology 129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- 33.Sahgal N, Canham LN, Canham B, Soares MJ. 2006. Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol Med 121:159–178. [PubMed] [Google Scholar]
- 34.Asanoma K, Rumi MAK, Kent LN, Chakraborty D, Renaud SJ, Wake N, Lee D-S, Kubota K, Soares MJ. 2011. FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev Biol 351:110–119. doi: 10.1016/j.ydbio.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DS, Rumi MA, Konno T, Soares MJ. 2009. In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis 47:433–439. doi: 10.1002/dvg.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roby KF, Larsen D, Deb S, Soares MJ. 1991. Generation and characterization of antipeptide antibodies to rat cytochrome P-450 side-chain cleavage enzyme. Mol Cell Endocrinol 79:13–20. doi: 10.1016/0303-7207(91)90090-F. [DOI] [PubMed] [Google Scholar]
- 37.Hamlin GP, Lu XJ, Roby KF, Soares MJ. 1994. Recapitulation of the pathway for trophoblast giant cell differentiation in vitro: stage-specific expression of members of the prolactin gene family. Endocrinology 134:2390–2396. doi: 10.1210/endo.134.6.8194465. [DOI] [PubMed] [Google Scholar]
- 38.Loots GG, Ovcharenko I. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters TJ, Albieri A, Bevilacqua E, Chapman BM, Crane LH, Hamlin GP, Seiki M, Soares MJ. 1999. Differentiation-dependent expression of gelatinase B/matrix metalloproteinase-9 in trophoblast cells. Cell Tissue Res 295:287–296. doi: 10.1007/s004410051235. [DOI] [PubMed] [Google Scholar]
- 40.Terranova PF, Garza F. 1983. Relationship between the preovulatory luteinizing hormone (LH) surge and androstenedione synthesis of preantral follicles in the cyclic hamster: detection by in vitro responses to LH. Biol Reprod 29:630–636. doi: 10.1095/biolreprod29.3.630. [DOI] [PubMed] [Google Scholar]
- 41.Rampon C, Prandini M, Bouillot S, Pointu H, Tillet E, Frank R, Vernet M, Huber P. 2005. Protocadherin 12 (VE-cadherin 2) is expressed in endothelial, trophoblast, and mesangial cells. Exp Cell Res 302:48–60. doi: 10.1016/j.yexcr.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Moussian B, Uv AE. 2005. An ancient control of epithelial barrier formation and wound healing. Bioessays 27:987–990. doi: 10.1002/bies.20308. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Samakovlis C. 2012. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr Top Dev Biol 98:35–63. doi: 10.1016/B978-0-12-386499-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 44.Henderson YC, Frederick MJ, Jayakumar A, Choi Y, Wang MT, Kang Y, Evans R, Spring PM, Uesugi M, Clayman GL. 2007. Human LBP-32/MGR is a repressor of the P450scc in human choriocarcinoma cell line JEG-3. Placenta 28:152–160. doi: 10.1016/j.placenta.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Henderson YC, Frederick MJ, Wang MT, Hollier LM, Clayman GL. 2008. LBP-1b, LBP-9, and LBP-32/MGR detected in syncytiotrophoblasts from first-trimester human placental tissue and their transcriptional regulation. DNA Cell Biol 27:71–79. doi: 10.1089/dna.2007.0640. [DOI] [PubMed] [Google Scholar]
- 46.Knöfler M. 2010. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int J Dev Biol 54:269–280. doi: 10.1387/ijdb.082769mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer TE, Bazer FW. 2002. Biology of progesterone action during pregnancy recognition and maintenance of pregnancy. Front Biosci 7:d1879–d1898. doi: 10.2741/spencer. [DOI] [PubMed] [Google Scholar]
- 48.Librach CL, Werb Z, Fitzgerald ML, Chiu K, Corwin NM, Esteves RA, Grobelny D, Galardy R, Damsky CH, Fisher SJ. 1991. 92-kD type IV collagenase mediates invasion of human cytotrophoblasts. J Cell Biol 113:437–449. doi: 10.1083/jcb.113.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plaks V, Rinkenberger J, Dai J, Flannery M, Sund M, Kanasaki K, Ni W, Kalluri R, Werb Z. 2013. Matrix metalloproteinase-9 deficiency phenocopies features of preeclampsia and intrauterine growth restriction. Proc Natl Acad Sci U S A 110:11109–11114. doi: 10.1073/pnas.1309561110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller WL, Auchus RJ. 2011. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev 32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escalante-Alcalde D, Hernandez L, Le Stunff H, Maeda R, Lee HS, Jr-Gang-Cheng, Sciorra VA, Daar I, Spiegel S, Morris AJ, Stewart CL. 2003. The lipid phosphatase LPP3 regulates extra-embryonic vasculogenesis and axis patterning. Development 130:4623–4637. doi: 10.1242/dev.00635. [DOI] [PubMed] [Google Scholar]
- 52.Hilberg F, Aguzzi A, Howells N, Wagner EF. 1993. c-Jun is essential for normal mouse development and hepatogenesis. Nature 365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 53.Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet 25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- 54.Lu J, Zhang S, Nakano H, Simmons DG, Wang S, Kong S, Wang Q, Shen L, Tu Z, Wang W, Wang B, Wang H, Wang Y, van Es JH, Clevers H, Leone G, Cross JC, Wang H. 2013. A positive feedback loop involving Gcm1 and Fzd5 directs chorionic branching morphogenesis in the placenta. PLoS Biol 11:e1001536. doi: 10.1371/journal.pbio.1001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinke AW, Baek J, Ashenberg O, Keating AE. 2013. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]