Abstract
Long noncoding RNAs (lncRNAs) are emerging as important regulators in mammalian development, but little is known about their roles in monocyte/macrophage differentiation. Here we identified a long noncoding monocytic RNA (lnc-MC) that exhibits increased expression during monocyte/macrophage differentiation of THP-1 and HL-60 cells as well as CD34+ hematopoietic stem/progenitor cells (HSPCs) and is transcriptionally activated by PU.1. Gain- and loss-of-function assays demonstrate that lnc-MC promotes monocyte/macrophage differentiation of THP-1 cells and CD34+ HSPCs. Mechanistic investigation reveals that lnc-MC acts as a competing endogenous RNA to sequester microRNA 199a-5p (miR-199a-5p) and alleviate repression on the expression of activin A receptor type 1B (ACVR1B), an important regulator of monocyte/macrophage differentiation. We also noted a repressive effect of miR-199a-5p on lnc-MC expression and function, but PU.1-dominant downregulation of miR-199a-5p weakens the role of miR-199a-5p in the reciprocal regulation between miR-199a-5p and lnc-MC. Altogether, our work demonstrates that two PU.1-regulated noncoding RNAs, lnc-MC and miR-199a-5p, have opposing roles in monocyte/macrophage differentiation and that lnc-MC facilitates the differentiation process, enhancing the effect of PU.1, by soaking up miR-199a-5p and releasing ACVR1B expression. Thus, we reveal a novel regulatory mechanism, comprising PU.1, lnc-MC, miR-199a-5p, and ACVR1B, in monocyte/macrophage differentiation.
INTRODUCTION
Hematopoiesis is a highly orchestrated process wherein the pluripotent self-renewing hematopoietic stem cells (HSCs) give rise to all blood cell lineages, including monocytes/macrophages (1). Monocytes/macrophages are mononuclear phagocytes that play crucial roles in innate immunity and the inflammatory response, and defects in their biogenesis and function can contribute to a broad spectrum of pathologies (2, 3). Control of monocyte/macrophage differentiation is a complex process requiring the coordinated expression of stage-specific transcription factors, cytokines, and noncoding RNAs (4, 5).
PU.1 is a hematopoiesis-specific transcription factor that binds to a purine-rich sequence (GAGGAA) and regulates lineage-specific gene expression (6). Homozygous PU.1-deficient mice died at a late gestational stage, and PU.1 mutant embryos exhibited a defect in the generation of progenitors for monocytes and granulocytes (7). High expression of PU.1 in granulocyte-macrophage progenitors (GMPs) antagonizes C/EBPα function and favors monocyte development. Conversely, GMPs with low expression of PU.1 commit to granulocyte differentiation (8). In addition, transcription factors RUNX1, KLF4, and MafB are important regulators in monocyte/macrophage development (9–11). Colony-stimulating factors (CSF), including granulocyte-macrophage CSF, granulocyte CSF, and CSF-1, also play fundamental roles in the early and late stages of the monocyte/macrophage differentiation process (12).
MicroRNAs (miRNAs) are short (20- to 24-nucleotide [nt]) noncoding RNAs that are involved in posttranscriptional regulation of gene expression in multicellular organisms by affecting the stability or translation of mRNAs (13). A number of miRNAs have been reported to play crucial roles in hematopoietic lineage differentiation, including monocytopoiesis (14). miRNA 142-3p (miR-142-3p) and miR-29a, targeting TAB2 and CDK6, respectively, and both targeting CCNT2, are important regulators in monocytic differentiation (15). Downregulation of miR-17-5p, miR-20a, miR-106a, and miR-26a is required for monocytopoiesis (16, 17). During the induction of THP-1 cells by phorbol myristate acetate (PMA), increased expression of miR-155, miR-222, miR-424, and miR-503 was observed, and the combinatorial regulation of the four miRNAs was demonstrated to influence monocytic differentiation (18). miR-424 has been reported to be actively transcribed by PU.1 and to stimulate monocytic differentiation through translational repression of the transcriptional factor NFI-A (19). Our previous work showed that PU.1, as a transcriptional repressor, negatively regulates miR-199a-5p expression and that the downregulation of miR-199a-5p alleviates the repressive effect on the activin A receptor type 1B gene (ACVR1B), which consequently activates the transforming growth factor β (TGF-β) signal pathway and promotes monocyte/macrophage differentiation (20).
Long noncoding RNAs (lncRNAs) (>200 nucleotides) lack protein-coding potential and function as versatile regulators through interaction with DNA, RNA, and proteins to modulate gene expression on many levels (21). The mammalian genome transcribes thousands of lncRNAs, only some of which have been functionally characterized (22, 23). Although a number of well-defined lncRNAs have been reported to play crucial roles in many biological processes and pathological diseases (24, 25), only a few examples of lncRNAs that regulate hematopoiesis have been reported (26–29). In this study, we screened the potential lncRNAs involved in myeloid differentiation through combined analysis of the transcriptome-sequencing (RNA-Seq) data for white blood cells published by Cabili et al. (30) and the PU.1 chromatin immunoprecipitation sequencing (ChIP-seq) data in ChIPBase (31), and we identified a long noncoding monocytic RNA (lnc-MC) that is upregulated during the monocyte/macrophage differentiation of THP-1 and HL-60 cells and of CD34+ hematopoietic stem/progenitor cells (HSPCs) and that could be regulated by PU.1. We also demonstrated that during monocyte/macrophage differentiation, lnc-MC can be negatively regulated by miR-199a-5p and that lnc-MC can also soak up miR-199a-5p to alleviate the repression of ACVR1B, suggesting that lnc-MC may act as a competing endogenous RNA (ceRNA) to modulate monocytopoiesis.
MATERIALS AND METHODS
Cell culture and induction of differentiation.
The human acute monocytic leukemia cell line THP-1 was grown in RPMI 1640 medium (Gibco, BRL, United Kingdom) supplemented with 10% fetal bovine serum (FBS) (HyClone). The human acute promyelocytic leukemia cell line HL-60 was maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS. Monocyte/macrophage differentiation of THP-1 and HL-60 cells was induced with PMA at a final concentration of 10 nM. 293TN cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS.
Separation and induction culture of CD34+ HSPCs.
Human umbilical cord blood (UCB) was obtained from normal full-term deliveries at Beijing Hospital and the First Hospital of Hebei Medical University. Informed consent to the performance of the biological studies was obtained from the individuals examined, and the related study was approved by the Ethics Committees of the hospitals and the Institutional Review Board of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. Mononuclear cell (MNC) fractions were isolated from the samples by Percoll density (d) gradient [d = 1.077 g/ml] centrifugation (Amersham Biotech, Germany), and CD34+ cells were enriched from MNCs through positive immunomagnetic selection (CD34 MultiSort kit; Miltenyi Biotec, Bergisch Gladbach, Germany). Monocyte/macrophage differentiation culture of CD34+ HSPCs was performed as described previously (15).
RNA extraction and qRT-PCR.
Total RNA was extracted from cell samples by using the TRIzol reagent (Invitrogen) and was quantified using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Bremen, Germany). The first strand of cDNA was synthesized by using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Oligo(dT) was used as the primer for reverse transcription of mRNA. Stem-loop reverse transcription primers were used for the reverse transcription of miRNA. U6 and lnc-MC were reverse transcribed using strand-specific primers. Quantitative real-time PCR (qRT-PCR) was performed in a Bio-Rad CFX96 system (Bio-Rad, Foster City, CA) using SYBR premix (TransGen Biotech). The primers used for reverse transcription and qRT-PCR are listed in Table S1 in the supplemental material.
Immunoblot analysis.
Cell lysates were subjected to SDS-PAGE (10% separation gel) and were transferred to a polyvinylidene difluoride (PVDF) membrane. Primary antibodies against the following proteins were used: PU.1 (antibody 2258; Cell Signaling Technology), Ago2 (antibody 2897; Cell Signaling Technology), ACVR1B (ab109300; Abcam), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog no. 10494-1-AP; Proteintech). Horseradish peroxidase-conjugated secondary antibodies were used. Signals were detected using an ECL (enhanced chemiluminescence) kit (Millipore).
Plasmid constructs.
The cDNAs of PU.1, pri-miR-199a, and lnc-MC were amplified and inserted into pmiRNA1 (System Biosciences [SBI], Mountain View, CA) and pcDNA6 (Invitrogen) in order to obtain their expression plasmids. The fragments of lnc-MC containing the miR-199a-5p binding site and its corresponding mutants were inserted into the pMIR-Report luciferase reporter vector (Ambion, Austin, TX). The short hairpin RNA (shRNA) sequences for PU.1 and lnc-MC were synthesized, annealed, and inserted into pll3.7 (Addgene). Sequences containing two consecutive miR-199a-5p complementary sequences, used to construct the miR-199a-5p sensor, were synthesized, annealed, and inserted into the pGL3-Control luciferase reporter vector (Promega). All the primers and oligonucleotides used for plasmid construction are listed in Table S2 in the supplemental material.
Transient transfection and luciferase reporter assay.
293TN cells were plated into 24-well plates to reach approximately 50% confluence on the following day. The cells were cotransfected with the pGL3-Control/pMIR-Report-based constructs, pRL-TK (Promega), and pcDNA6-based expression plasmids using Lipofectamine Plus (Invitrogen). The transfection medium was replaced with complete medium after 5 to 6 h. The cells were cultured at 37°C under 5% CO2 for an additional 24 to 48 h. Then the cells were lysed with passive lysis buffer (Promega), and reporter gene expression was assessed using the dual-luciferase reporter assay system (Promega).
Flow cytometry analysis.
The infected THP-1 cells and CD34+ HSPCs were induced toward monocyte/macrophage differentiation and harvested at different time points of differentiation. The cells were rinsed twice with phosphate-buffered saline (PBS) and were resuspended in 100 μl PBS. Then the cells were incubated with phycoerythrin (PE)- and allophycocyanin (APC)-conjugated anti-CD14 (eBioscience) at 4°C for 30 min. The cells were washed with 1 ml PBS, resuspended in 200 μl PBS, and analyzed immediately using an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA).
May-Grünwald-Giemsa staining.
The infected THP-1 cells and CD34+ HSPCs were induced toward monocyte/macrophage differentiation and were harvested at different time points of differentiation. The cells were rinsed twice with PBS and resuspended in FBS, and then the freshly prepared and air-dried cell smears were fixed in methanol for 10 min. The slides were stained in pure May-Grünwald solution for 5 min, washed in distilled water for 2 min, and incubated in a 10% Giemsa-water solution for 20 min. The slides were then washed in distilled water, air dried, and observed under a BX51 optical microscope (Olympus, Tokyo, Japan).
Lentivirus production and cell infection.
The recombinant lentiviruses for overexpression and knockdown were produced using pmiRNA1- and pll3.7-based constructs. Lentivirus packaging was performed by using the pPACKH1 lentiviral vector packaging kit (LV500A-1; SBI, Mountain View, CA) according to the manufacturer's instructions. The virus particles were condensed using the PEG-it virus precipitation solution (SBI, Mountain View, CA). The THP-1 cells and CD34+ HSPCs were infected with lentivirus in 6-well plates containing 5 μg/ml Polybrene (Sigma-Aldrich). After 24 h of infection, the cells were placed in fresh medium and induced toward monocyte/macrophage differentiation.
ChIP and RIP.
THP-1 and HL-60 cells were plated onto 10-cm plates and grown to approximately 90% confluence. Then the cells were collected, and chromatin immunoprecipitation (ChIP) was performed as described previously (20). The precipitated DNA was amplified by PCR. All the primers used for ChIP-PCR are listed in Table S3 in the supplemental material. THP-1 cells were plated onto 10-cm plates and grown to approximately 80% confluence. Then the cells were induced toward monocyte/macrophage differentiation for 48 h, and RNA immunoprecipitation (RIP) was performed by using the Magna RIP RNA-binding protein immunoprecipitation kit (product code 17-700; Millipore) according to the manufacturer's instructions. Primary antibodies against the following proteins were used: PU.1 (antibody 2258; Cell Signaling Technology), Ago2 (antibody 2897; Cell Signaling Technology), and rabbit IgG (catalog no. PP64B; Millipore).
Pulldown assay with biotinylated DNA probe.
The pulldown assay was performed as described previously (32) with minor modifications. The biotinylated DNA probe complementary to lnc-MC was synthesized and dissolved in 400 μl of wash/binding buffer (0.5 M NaCl, 20 mM Tris-HCl [pH 7.5], and 1 mM EDTA). The probes were incubated with streptavidin-coated agarose beads (product no. S1638-1ML; Sigma) at 4°C for 10 h to generate probe-coated agarose beads. The THP-1 cell lysate was precleared by incubation with 40 μl of avidin-agarose beads (catalog no. 20219; Pierce) at 4°C for 1 h and was then incubated with probe-coated beads. After washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for quantitative real-time PCR. The following primer sequences were used: lnc-MC pulldown probe, 5′-GCCTGTAATTCCAATGTGATACCC-3′; random pulldown probe, 5′-GTGATGTCTAGCGCTTGGGCTTTG-3′.
RNA fluorescence in situ hybridization (FISH).
THP-1 cells were induced with PMA toward monocyte/macrophage differentiation and were harvested at 50 h of induction. The cells were rinsed briefly in PBS and were resuspended in FBS. Then the freshly prepared and air-dried cell smears were fixed in 4% formaldehyde plus 10% acetic acid in PBS (pH 7.4) for 15 min at 20°C. The cell smears were permeated with PBS containing 0.2% Triton X-100 for 5 min at 20°C and were then washed in PBS (3 times, for 10 min each time). Hybridization was carried out using a biotin-labeled DNA probe (Tianyi Biotech) in a moist chamber at 37°C for 12 to 16 h. After being washed in PBS (5 times, for 10 min each time), the cell smears were incubated with fluorescein isothiocyanate (FITC)-conjugated streptavidin (catalog no. SA1001; Life Technologies) for 2 h at 20°C. Then the cell smears were washed in PBS (5 times, for 10 min each time), air dried, and covered with glass slides using DAPI (4′,6-diamidino-2-phenylindole) Fluoromount-G mounting medium (SouthernBiotech). Fluorescence images were acquired with a BX51 optical microscope (Olympus, Tokyo, Japan).
Statistical analysis.
Student's t test (two-tailed) was performed to analyze the data. Statistical significance was set at a P value of <0.05.
RESULTS
lnc-MC is identified as a potential lncRNA involved in myeloid differentiation.
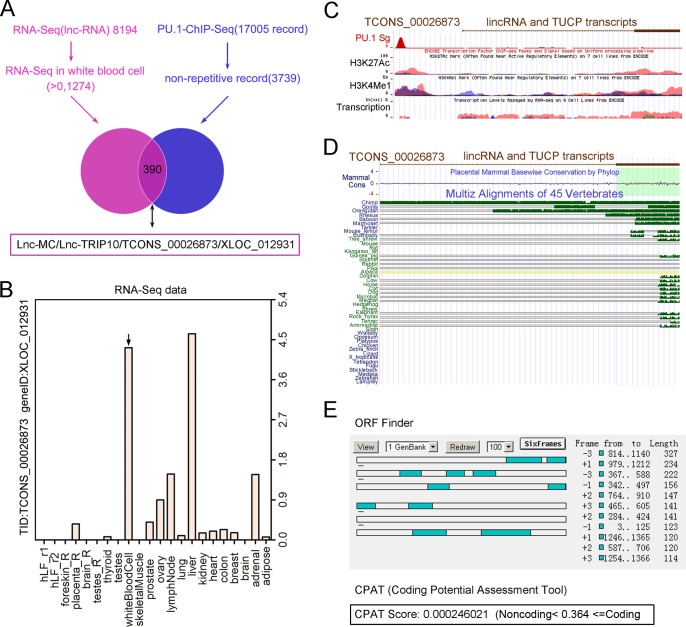
To identify lncRNAs involved in myeloid differentiation, we comprehensively analyzed the RNA-Seq data for white blood cells (30) and the PU.1 ChIP-seq data (31), and we finally obtained 390 candidate lncRNAs (Fig. 1A). lnc-MC was chosen for further investigation on the basis of its relatively high abundance in 22 tissues and cell lines (Fig. 1B) and the PU.1, H3K27Ac (histone H3 acetylated on Lys27), and H3K4Me1 (histone H3 monomethylated on Lys4) ChIP-seq signals on the lnc-MC locus (Fig. 1C). lnc-MC is also known as lnc-TRIP10/TCONS_00026873/XLOC_012931; it is annotated in LNCipedia (33), and its gene is located at chromosome 19, positions 6656385 to 6662832. Its conservation was analyzed using the UCSC Genome Browser (http://genome.ucsc.edu/index.html) and is presented in Fig. 1D, which shows that lnc-MC is conserved only among some primates (such as the chimpanzee). The coding potential of lnc-MC was analyzed using ORF Finder and the Coding Potential Assessment Tool (CPAT), which suggested that lnc-MC tends to be a noncoding RNA (Fig. 1E).
FIG 1.
Bioinformatic analysis of lnc-MC to infer its potential role in myeloid differentiation. (A) The long noncoding RNA-Seq data for white blood cells are drawn from the supplemental file of the paper of Cabili et al. (30). The PU.1 ChIP-seq data pertaining to lncRNA were downloaded from ChIPBase. The data common to the two data sets were determined with a tool developed by a member of our lab, and the results are depicted as the intersection of two circles. (B) Expression spectrum of lnc-MC in 22 tissues and cell lines based on RNA-Seq data. lnc-MC expression in white blood cells is indicated by a vertical arrow. (C) UCSC Genome Browser screenshot showing the locations of PU.1 ChIP-seq, H3K27Ac ChIP-Seq, H3K4Me1 ChIP-seq, and RNA-Seq signals on the lnc-MC locus. (D) lnc-MC conservation among vertebrates was mapped using the UCSC Genome Browser. (E) The protein-coding potential of lnc-MC was analyzed using ORF Finder and the Coding Potential Assessment Tool.
lnc-MC mediates PMA-induced monocyte/macrophage differentiation.
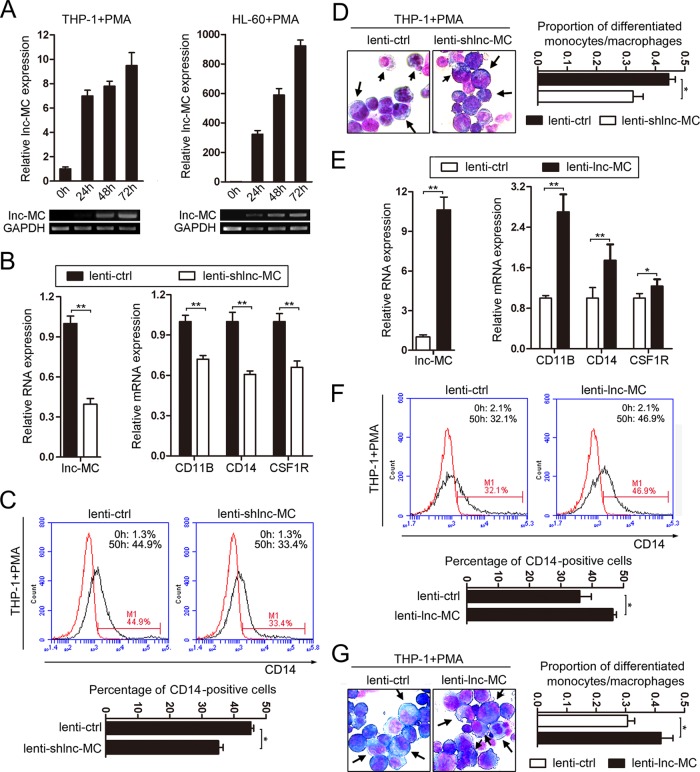
Next, we performed qRT-PCR and semiquantitative PCR to detect lnc-MC expression during the PMA-induced monocyte/macrophage differentiation of THP-1 and HL-60 cells and during the all-trans-retinoic acid (ATRA)-induced granulocyte differentiation of HL-60 and NB-4 cells. The results showed that lnc-MC was upregulated significantly during the PMA-induced time course of THP-1 and HL-60 cell differentiation (Fig. 2A). No significant changes were seen in lnc-MC expression over the time course of the ATRA-induced granulocyte differentiation of HL-60 and NB-4 cells (data not shown). The expression spectrum of lnc-MC suggests that it may play a positive role in monocyte/macrophage differentiation. To investigate the effect of lnc-MC on monocyte/macrophage differentiation, THP-1 cells were infected with a recombinant lentivirus expressing either short hairpin RNAs specific for lnc-MC (lenti-shlnc-MC) or lnc-MC (lenti-lnc-MC), followed by PMA induction for 50 h. Real-time PCR analysis revealed that lenti-shlnc-MC infection significantly decreased the levels of lnc-MC and the monocyte/macrophage differentiation markers (CD11B, CD14, and CSF1R) (Fig. 2B) from levels with the lentivirus control (lenti-ctrl) infection. Flow cytometry analysis also showed decreased CD14 expression in lenti-shlnc-MC-infected THP-1 cells (Fig. 2C). In addition, May-Grünwald-Giemsa staining demonstrated that upon PMA induction, lenti-shlnc-MC-infected THP-1 cells exhibited a lower proportion of differentiated monocytes/macrophages (with clear cytoplasm and a bean-shaped nucleus) than lenti-ctrl-infected cells (Fig. 2D). These results suggested that knockdown of lnc-MC in THP-1 cells impaired PMA-induced monocyte/macrophage differentiation.
FIG 2.
lnc-MC mediates PMA-induced monocyte/macrophage differentiation. (A) qRT-PCR and semiquantitative PCR analyses of lnc-MC expression during PMA-induced monocyte/macrophage differentiation of THP-1 and HL-60 cells. GAPDH mRNA was used as an internal control. (B) qRT-PCR detection of lnc-MC and monocyte/macrophage differentiation marker CD11B, CD14, and CSF1R mRNAs in THP-1 cells infected with lenti-shlnc-MC or lenti-ctrl, followed by PMA induction for 50 h. Three independent experiments were performed, and data are means ± standard deviations. Asterisks indicate significant differences by Student's t test (*, P < 0.05; **, P < 0.01). (C) CD14 expression was evaluated by cytometric analysis in infected and PMA-induced cells. (Top) Results of a representative experiment. Red and black curves show results for untreated cells and anti-CD14 antibody-stained cells, respectively. (Bottom) Statistical analysis of three experiments. (D) May-Grünwald-Giemsa staining analysis of infected and PMA-induced cells. (Left) Results of a representative experiment. The arrows point to differentiated monocytes/macrophages. (Right) Statistical analysis of counts of differentiated monocytes/macrophages in five fields. (E) qRT-PCR detection of lnc-MC and CD11B, CD14, and CSF1R mRNAs in THP-1 cells infected with lenti-lnc-MC or lenti-ctrl, followed by PMA induction for 50 h. Three independent experiments were performed, and data are means ± standard deviations. (F) CD14 expression was evaluated by cytometric analysis in infected and PMA-induced cells. (Top) Results of a representative experiment. Red and black curves show results for untreated cells and anti-CD14 antibody-stained cells, respectively. (Bottom) Statistical analysis of three experiments. (G) May-Grünwald-Giemsa staining analysis of infected and PMA-induced cells. (Left) Results of a representative experiment. (Right) Statistical analysis of counts of differentiated monocytes/macrophages in five fields.
In contrast, enhanced expression of lnc-MC in THP-1 cells promoted PMA-induced monocyte/macrophage differentiation. Compared with the lenti-ctrl-infected THP-1 cells, the lenti-lnc-MC-infected cells exhibited significant expression of lnc-MC, which increased the mRNA levels of the monocyte/macrophage differentiation markers (CD11B, CD14, and CSF1R) (Fig. 2E). The overexpression of lnc-MC also resulted in increased CD14 expression (Fig. 2F) and a higher proportion of differentiated monocytes/macrophages (Fig. 2G).
Taken together, both knockdown and overexpression of lnc-MC have significant effects on PMA-induced monocyte/macrophage differentiation, suggesting that lnc-MC functions as a crucial regulator in monocyte/macrophage differentiation.
PU.1 modulates lnc-MC expression during monocyte/macrophage differentiation.
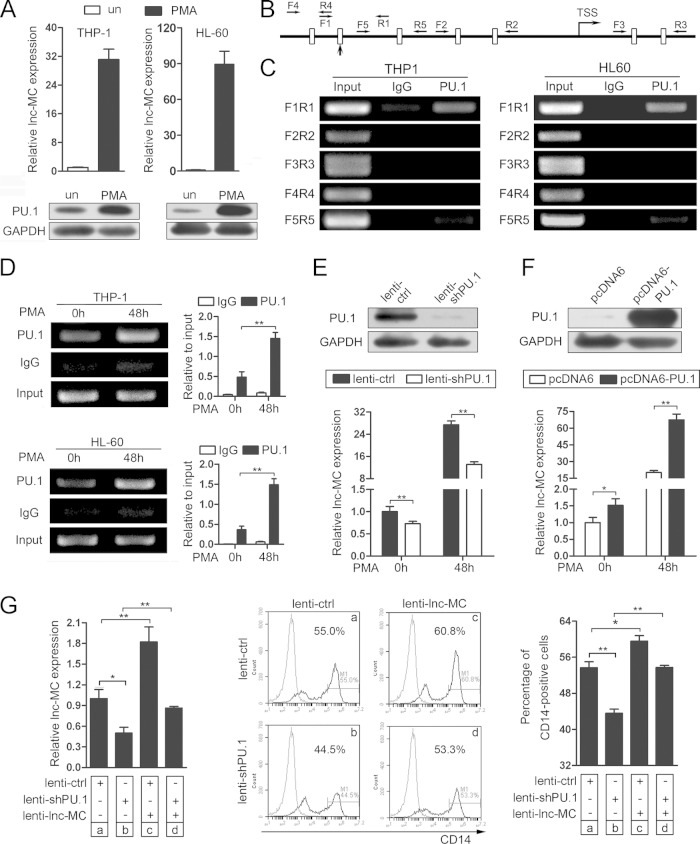
PU.1 is a master transcriptional factor that is essential for monocyte progenitor commitment from GMPs and for monocyte/macrophage maturation (4) and that regulates gene transcription both positively (19) and negatively (34). To evaluate whether PU.1 modulates lnc-MC expression during monocyte/macrophage development, we first detected the expression of PU.1 and lnc-MC during PMA-induced monocyte/macrophage differentiation of THP-1 and HL-60 cells. The results showed that both lnc-MC and PU.1 were upregulated (Fig. 3A), suggesting that PU.1 may positively regulate lnc-MC expression during monocyte/macrophage development.
FIG 3.
PU.1 modulates lnc-MC expression during monocyte/macrophage differentiation. (A) qRT-PCR analysis of lnc-MC expression and immunoblot analysis of PU.1 expression in THP-1 and HL-60 cells without (un) or with PMA induction for 48 h. The level of lnc-MC was normalized to the GAPDH mRNA level and is shown as fold expression relative to the levels in untreated cells. (B) Sketch map showing the putative PU.1 binding sites in lnc-MC gene loci. Boxes represent the putative binding sites of PU.1, and horizontal arrows flanking the boxes indicate the primers used for ChIP-PCR. The vertical arrow indicates the box confirmed by ChIP-PCR. The right-angle arrow indicates the transcriptional start site (TSS) and the direction of transcription. (C) ChIP-PCR assay of PU.1 binding. Only the box indicated by the vertical arrow in panel B was confirmed to be bound by PU.1. (D) Enhanced PU.1 binding to the site upstream of lnc-MC was detected in PMA-induced THP-1 and HL-60 cells. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by Student's t test. (E) Immunoblot analysis of PU.1 and qRT-PCR analysis of lnc-MC expression in THP-1 cells infected with lenti-shPU.1 or lenti-ctrl, followed by PMA induction. (F) Immunoblot analysis of PU.1 and qRT-PCR analysis of lnc-MC expression in THP-1 cells transfected with pcDNA6-PU.1 or pcDNA6, followed by PMA induction. (G) Rescue assays by infection with a combination of lenti-sh-PU.1 (or lenti-ctrl) and lenti-lnc-MC in CD34+ HSPCs. (Left) The expression levels of lnc-MC after infection were determined by qRT-PCR. (Center) CD14 expression was evaluated through by cytometry analysis. The gray and black curves indicate untreated cells and anti-CD14 antibody-stained cells, respectively. (Right) Statistical analysis of two independent CD14 expression experiments.
We identified a potential PU.1 binding signal upstream of lnc-MC gene loci using the UCSC Genome Browser (Fig. 1C). In addition, we predicted all potential PU.1 binding sites in a 3-kb region near lnc-MC gene loci according to the PU.1 binding motif, as depicted in Fig. 3B. Next, a PU.1 ChIP assay was performed in THP-1 and HL-60 cells to validate the predictions. As shown in Fig. 3C, only one site (indicated by a vertical arrow in Fig. 3B) upstream of lnc-MC gene loci was verified as the true binding site of PU.1; the others were not bound by PU.1, in agreement with the result from the UCSC Genome Browser. Furthermore, enhanced PU.1 binding to this site was noted after PMA induction of THP-1 or HL-60 cells (Fig. 3D). Then we infected THP-1 cells with lenti-shPU.1, which significantly downregulated PU.1 expression (Fig. 3E, top). PU.1 knockdown also decreased lnc-MC expression during PMA-induced monocyte/macrophage differentiation (Fig. 3E, bottom). On the other hand, PU.1 overexpression by pcDNA6-PU.1 transfection in THP-1 cells (Fig. 3F, top) remarkably increased lnc-MC expression during PMA-induced monocyte/macrophage differentiation (Fig. 3F, bottom). These results demonstrated that upregulation of lnc-MC during PMA-induced monocyte/macrophage differentiation was at least partially due to transcriptional activation of lnc-MC by PU.1.
To further confirm that lnc-MC is regulated by PU.1 during monocyte/macrophage differentiation, we performed rescue assays. CD34+ HSPCs were infected with lenti-shPU.1 or lenti-ctrl. Twenty-four hours later, the cells were reinfected with lenti-lnc-MC or lenti-ctrl, followed by induction of monocyte/macrophage differentiation. As expected, reinfection with lenti-lnc-MC alleviated the downregulation of lnc-MC resulting from lenti-shPU.1 treatment (Fig. 3G, left, d versus b). In agreement with the expression of lnc-MC, reinfection with lenti-lnc-MC also ameliorated the blockade of monocyte/macrophage differentiation caused by lenti-shPU.1 infection, a finding presented as CD14 expression detected through flow cytometry (Fig. 3G, center and right, d versus b). These results demonstrate that the master transcription factor PU.1 controls monocyte/macrophage differentiation partially by modulating lnc-MC expression.
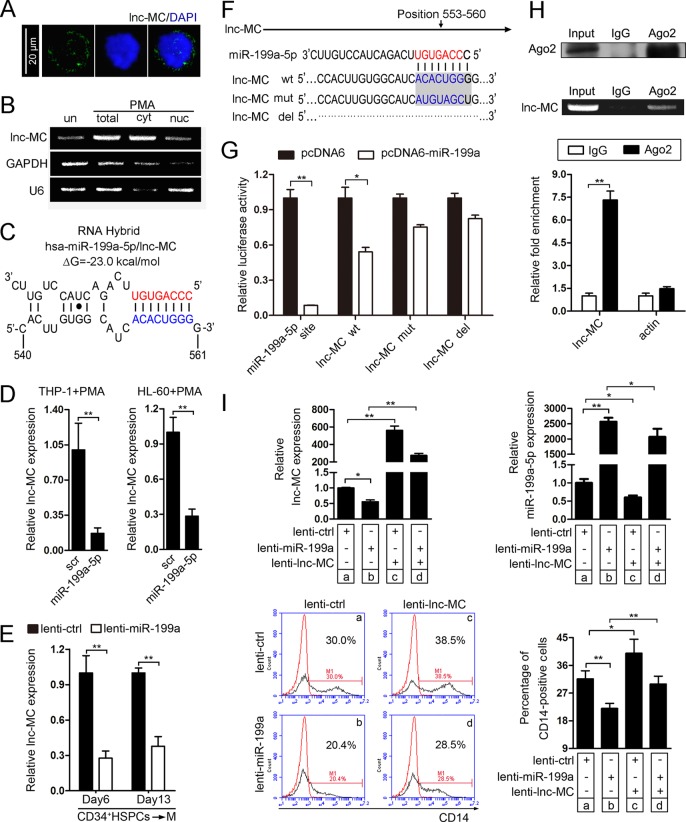
lnc-MC can be negatively regulated and antagonized by miR-199a-5p.
To investigate the molecular mechanism of lnc-MC involvement in monocyte/macrophage differentiation, we first determined the subcellular location of lnc-MC. RNA FISH (Fig. 4A) and semiquantitative PCR of nuclear and cytoplasmic fractions (Fig. 4B) suggested that lnc-MC was distributed mainly in the cytoplasm. Some lncRNAs have been reported to interact with miRNAs and regulate each other reciprocally (35). Since miRNAs are a class of very important posttranscriptional regulators, we sought to determine whether lnc-MC can regulate monocyte/macrophage differentiation through association with miRNAs. We next analyzed the miRNAs downregulated during monocyte/macrophage differentiation by using miRNA array data together with the data for published miRNAs that have been demonstrated to negatively regulate monocyte/macrophage differentiation (20). Finally, we focused on miR-199a-5p, which was predicted, by use of the RNAhybrid Web tool, to bind to lnc-MC stably (Fig. 4C). To investigate the effect of miR-199a-5p on lnc-MC, miR-199a-5p was overexpressed in THP-1 and HL-60 cells by using a miR-199a-5p mimic and in CD34+ HSPCs by using lenti-miR-199a. The enhanced expression of miR-199a-5p decreased lnc-MC expression significantly (Fig. 4D and E). Then we produced a luciferase construct of lnc-MC (lnc-MC wt), a mutated form (lnc-MC mut), and a deleted form (lnc-MC del) (Fig. 4F). A luciferase reporter assay revealed that miR-199a-5p could suppress the luciferase activity of lnc-MC wt but had less effect on lnc-MC mut and lnc-MC del (Fig. 4G).
FIG 4.
lnc-MC is negatively regulated by miR-199a-5p during monocyte/macrophage differentiation. (A) RNA FISH assay of lnc-MC in PMA-induced THP-1 cells. More than 20 cells were examined, and similar results were obtained. (B) Semiquantitative PCR detection of lnc-MC in the cytoplasmic (cyt) and nuclear (nuc) fractions of untreated (un) and PMA-induced THP-1 cells. GAPDH and U6 served as cytoplasmic and nuclear localization controls, respectively. (C) The interaction between lnc-MC and miR-199a-5p was predicted using the RNAhybrid Web tool. Partial sequences of lnc-MC (bottom) and miR-199a-5p (top) are shown. Numbers below the sequences indicate the positions of nucleotides relative to the transcriptional start site of lnc-MC. (D) qRT-PCR analysis of lnc-MC expression in THP-1 and HL-60 cells transfected with a miR-199a-5p mimic or a scrambled control (scr), followed by PMA induction for 48 h. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by Student's t test. (E) qRT-PCR analysis of lnc-MC expression at day 6 and day 13 of monocyte/macrophage induction culture of CD34+ HSPCs infected with lenti-miR-199a or lenti-ctrl. (F) miR-199a-5p sequence showing the predicted binding site (red) and lnc-MC sequences showing mutated or deleted nucleotides. wt, wild type; mut, mutant; del, deletant. (G) Luciferase reporter assays of 293TN cells cotransfected with each pMIR-Report-based construct and pcDNA6-miR-199a (or pcDNA6). Three independent experiments were performed, and data are means ± standard deviations. (H) Immunoprecipitation using an anti-Ago2 or anti-IgG antibody and extracts of PMA-induced THP-1 cells. (Top) Ago2 in immunoprecipitates was analyzed by immunoblotting. (Center) RNA levels in immunoprecipitates were determined by semiquantitative PCR. (Bottom) RNA levels in immunoprecipitates were determined by qRT-PCR. The levels of lnc-MC and β-actin are presented as fold enrichment in anti-Ago2 relative to anti-IgG immunoprecipitates. (I) Rescue assays in THP-1 cells. THP-1 cells were infected with lenti-miR-199a or lenti-ctrl. Twenty-four hours later, the cells were reinfected with lenti-lnc-MC or lenti-ctrl and were exposed to a medium with PMA for another 48 h. (Top) The expression levels of lnc-MC and miR-199a-5p were determined by qRT-PCR. (Bottom) (Left) CD14 expression was evaluated by flow cytometry analysis. The red and black curves indicate untreated cells and anti-CD14 antibody-stained cells, respectively. (Right) Statistical analysis of four independent experiments.
Ago2, the core component of the RNA-induced silencing complex (RISC), associates with miRNAs to form miRNA ribonucleoprotein complexes (miRNPs) and is indispensable for miRNA function (36, 37). To test whether lnc-MC could associate with Ago2, RIP was carried out using an antibody against Ago2 and extracts of PMA-induced THP-1 cells. The specificity of the anti-Ago2 antibody was confirmed by immunoprecipitation (IP) and immunoblotting (Fig. 4H, top). As revealed by the RIP semiquantitative PCR (Fig. 4H, center) and RIP-qPCR (Fig. 4H, bottom), lnc-MC was preferentially enriched in Ago2-containing miRNPs relative to control IgG immunoprecipitates.
To further investigate the effect of lnc-MC modulation by miR-199a-5p on monocyte/macrophage differentiation, we designed rescue assays. THP-1 cells were infected with lenti-miR-199a or lenti-ctrl. Twenty-four hours later, the cells were reinfected with lenti-lnc-MC or lenti-ctrl and were exposed to a medium with PMA. As expected, infection with lenti-miR-199a significantly increased miR-199a-5p expression (Fig. 4I, top right, b versus a), decreased lnc-MC expression (Fig. 4I, top left, b versus a), and impaired monocyte/macrophage differentiation as revealed by CD14 expression (Fig. 4I, bottom). However, reinfection with lenti-lnc-MC not only recovered the expression of lnc-MC (Fig. 4I, top left, d versus b) but also ameliorated the blockade of monocyte/macrophage differentiation resulting from lenti-miR-199a infection, as revealed by CD14 expression detected through flow cytometry (Fig. 4I, bottom). A lower lnc-MC level was also observed in cells coinfected with lenti-miR-199a and lenti-lnc-MC than in cells coinfected with lenti-ctrl and lenti-lnc-MC (Fig. 4I, top left, d versus c), again demonstrating the inhibitory effect of miR-199-5p on lnc-MC expression. Collectively, these results demonstrated that lnc-MC could be negatively regulated and its function disturbed by miR-199a-5p during monocyte/macrophage differentiation.
lnc-MC negatively regulates miR-199a-5p expression and activity.
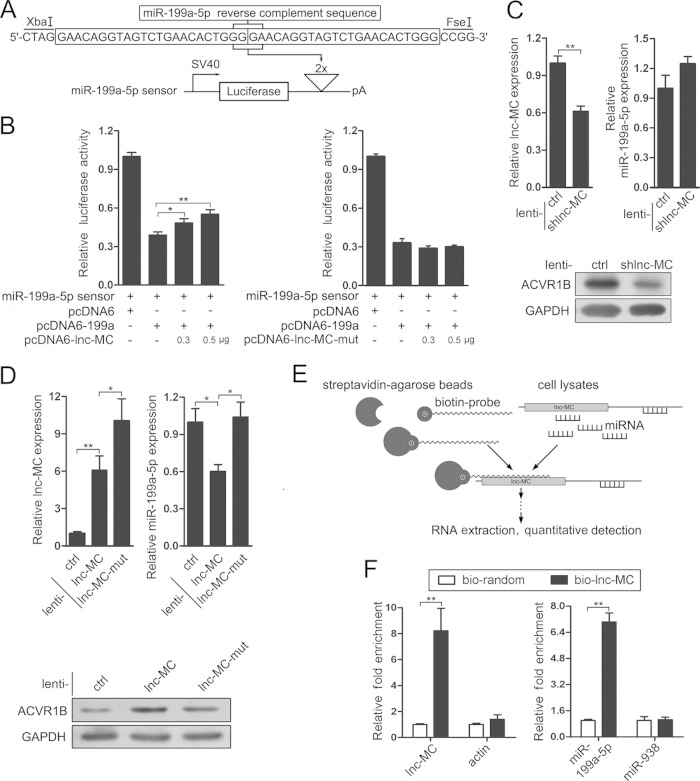
Having demonstrated that lnc-MC is a target of miR-199a-5p, we wanted to determine whether lnc-MC might also act as a sponge to sequester miR-199a-5p. For this purpose, we performed luciferase reporter assays using a miR-199a-5p sensor (Fig. 5A). The repressive effect of miR-199a-5p on the luciferase activity of the miR-199a-5p sensor was ameliorated by the cotransfection of increasing amounts of pcDNA6-lnc-MC (Fig. 5B, left) but was not influenced by increasing amounts of pcDNA6-lnc-MC-mut (containing lnc-MC with the miR-199a-5p binding site mutated) (Fig. 5B, right). In addition, inhibition of lnc-MC by lenti-shlnc-MC infection in THP-1 cells (Fig. 5C, top left) had less effect on miR-199a-5p expression (Fig. 5C, top right), which may be due to the gradual decrease in endogenous miR-199a-5p levels during differentiation and the further knockdown of lnc-MC on the basis of the low abundance of endogenous lnc-MC. Infection of THP-1 cells by lenti-shlnc-MC significantly decreased the protein level of ACVR1B (the target of miR-199a-5p) (Fig. 5C, bottom). However, overexpression of lnc-MC by lenti-lnc-MC infection (Fig. 5D, top left) decreased miR-199a-5p expression significantly (Fig. 5D, top right), leading to the upregulation of ACVR1B (Fig. 5D, bottom), while infection with lenti-lnc-MC-mut had less effect on miR-199a-5p expression (Fig. 5D, top right) and the ACVR1B level (Fig. 5D, bottom), suggesting that lnc-MC influences ACVR1B expression by soaking up miR-199a-5p. In addition, the inhibitory effect of lnc-MC on miR-199a-5p expression was also observed after coinfection with lenti-miR-199a and lenti-lnc-MC (Fig. 4I, top right). A decrease in endogenous miR-199a-5p levels was detected in cells infected with lenti-ctrl and lenti-lnc-MC compared with cells infected with lenti-ctrl only (Fig. 4I, top right, c versus a), and a decreased miR-199a-5p level was also observed in cells coinfected with lenti-miR-199a and lenti-lnc-MC compared with cells coinfected with lenti-miR-199a (d versus b). To further demonstrate the physical interaction between lnc-MC and miR-199a-5p, RNA pulldown was carried out using a biotin-labeled DNA probe (Fig. 5E). lnc-MC and miR-199a-5p were preferentially enriched in the lnc-MC-probe pulldown components versus random-probe pulldown components, whereas the enrichment of actin and miR-938 (used as negative controls) showed no difference between the two groups (Fig. 5F). Taken together, these results strongly suggest that lnc-MC interacts physically with miR-199a-5p and regulates its expression and activity.
FIG 5.
lnc-MC regulates miR-199a-5p expression and activity negatively. (A) miR-199a-5p sensor construct. (B) Luciferase reporter assays. 293TN cells were cotransfected with the miR-199a-5p sensor construct and either pcDNA6 or pcDNA6-199a alone or together with different doses of pcDNA6-lnc-MC or pcDNA6-lnc-MC-mut. Three independent experiments were performed, and data are means ± standard deviations. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by Student's t test. (C) qRT-PCR analysis of lnc-MC (top left) and miR-199a-5p (top right), and immunoblot analysis of ACVR1B expression (bottom), in THP-1 cells infected with lenti-shlnc-MC or lenti-ctrl, followed by PMA induction. (D) qRT-PCR analysis of lnc-MC (top left) and miR-199a-5p (top right), and immunoblot analysis of ACVR1B expression (bottom), in THP-1 cells infected with either lenti-lnc-MC, lenti-lnc-MC-mut, or lenti-ctrl, followed by PMA induction. (E) Schematic outline for identification of lnc-MC-associated RNA components by using the biotin-streptavidin system. (F) Relative abundances of lnc-MC and actin RNAs, as well as relative abundances of miR-199a-5p and miR-938, in lnc-MC probe and random probe pulldown assays are plotted as relative fold enrichment after normalization against GAPDH and U6 snRNA, respectively. Data are means ± standard deviations (n = 3).
Role and mechanism of lnc-MC in monocyte/macrophage differentiation of CD34+ HSPCs.
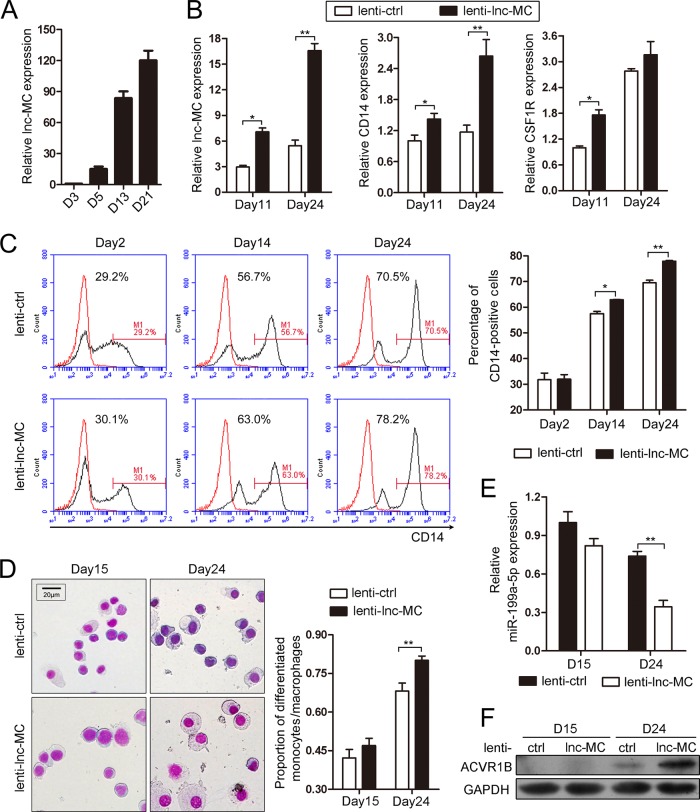
To confirm the function of lnc-MC in monocyte/macrophage differentiation of HSPCs, we first determined lnc-MC expression during monocyte/macrophage induction culture of CD34+ HSPCs derived from UCB. The results showed a time course increase in lnc-MC expression during differentiation (Fig. 6A), in agreement with the results in THP-1 and HL-60 cell lines. Next, CD34+ HSPCs were infected with lenti-ctrl or lenti-lnc-MC and were then induced toward monocyte/macrophage differentiation for 24 days. The lenti-lnc-MC infection increased lnc-MC expression significantly over that with the lenti-ctrl infection (Fig. 6B, left). Overexpression of lnc-MC promoted monocyte/macrophage differentiation, as revealed by increased mRNA expression of the differentiation markers CD14 and CSF1R (Fig. 6B, center and right), elevated CD14 expression as evaluated by flow cytometry (Fig. 6C), and a higher proportion of differentiated cells (monocyte/macrophage-like cells with clear cytoplasm and the nucleus located on one side) (Fig. 6D). Since we have demonstrated that lnc-MC can interact directly with miR-199a-5p, which can suppress monocyte/macrophage differentiation through its repressive effect on ACVR1B, we determined the expression of miR-199a-5p and ACVR1B at day 15 and day 24 after lenti-lnc-MC and lenti-ctrl infection and induction culture. The results revealed that enhanced expression of lnc-MC decreased miR-199a-5p expression remarkably at day 24 of differentiation (Fig. 6E); meanwhile, the expression of ACVR1B was increased (Fig. 6F). These results demonstrated that lnc-MC can promote monocyte/macrophage differentiation of HSPCs by decreasing the expression and disturbing the function of endogenous miR-199a-5p, leading to the release of ACVR1B expression.
FIG 6.
Role and mechanism of lnc-MC in monocyte/macrophage differentiation of CD34+ HSPCs. (A) qRT-PCR analysis of lnc-MC expression during monocyte/macrophage differentiation of CD34+ HSPCs. D, day. (B) qRT-PCR analysis of lnc-MC and monocyte/macrophage differentiation marker CD14 and CSF1R mRNAs during induction culture of CD34+ HSPCs infected with lenti-lnc-MC or lenti-ctrl. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) by Student's t test. (C) CD14 expression during induction culture of infected CD34+ HSPCs was evaluated by flow cytometry analysis. (Left) Histograms showing results of a representative experiment. The red and black curves indicate untreated cells and anti-CD14 antibody-stained cells, respectively. (Right) The results from two independent experiments were statistically analyzed and are presented as means ± standard deviations. (D) May-Grünwald-Giemsa staining analysis during induction culture of infected CD34+ HSPCs. (Left) Results of a representative experiment. (Right) Statistical analysis of counts of differentiated monocytes/macrophages in five fields. (E) qRT-PCR analysis of miR-199a-5p expression during induction culture of infected CD34+ HSPCs. (F) Immunoblot analysis of ACVR1B expression during induction culture of infected CD34+ HSPCs.
DISCUSSION
Monocytes/macrophages are critical effectors and regulators of inflammation and the innate immune response (38). Abnormal biogenesis and function of monocytes/macrophages can lead to a broad spectrum of pathological diseases, such as leukemia, immunological disorders, and atherosclerosis (39, 40). Monocyte/macrophage differentiation, an important branch of hematopoiesis, involves a number of important regulators. In the past few years, lncRNAs have attracted much attention for their versatile regulatory functions. lncRNAs can contribute to protein-DNA interaction (24), organize nuclear architecture (41), regulate miRNA biogenesis (42), influence mRNA stability and translation (43), and directly alter protein function (44), and they participate in various biological and pathological processes, such as the maintenance of pluripotency (45), regulation of neurogenesis (24), muscle differentiation (22), and cancer development (46). However, only a few lncRNAs that are involved in hematopoietic lineage differentiation have been reported. Murine linc-EPS inhibits apoptosis by repressing Pycard expression and confers erythroid differentiation (28). The lncRNA EGO regulates eosinophil granule protein transcript expression (26), and HOTAIRM1 shows specific expression in the myeloid lineage and regulates retinoic acid (RA)-driven granulocytic differentiation (27). Wang et al. reported that during dendritic cell differentiation, lnc-DC promotes STAT3 signaling by interacting with the C terminus of STAT3 to prevent the dephosphorylation of STAT3 Y705 by SHP1 (47). To date, few lncRNAs have been reported to participate in monocyte/macrophage differentiation. In this study, we demonstrated that lnc-MC facilitates monocyte/macrophage differentiation in both PMA-induced THP-1 cells and normal CD34+ HSPCs.
PU.1, a master transcription factor in hematopoietic lineage differentiation, plays a critical role in monocytic lineage commitment and monocyte/macrophage maturation (8). PU.1 has been reported to direct a broad spectrum of gene expression during monocyte/macrophage differentiation, including the expression of protein-coding genes, such as the macrophage colony-stimulating factor receptor gene (CSF1R) (48), and noncoding genes, such as miR-424 (19). Our previous work has demonstrated that miR-199a-5p negatively regulates monocyte/macrophage differentiation by targeting ACVR1B and that miR-199a-2 is transcriptionally repressed by PU.1 (20). Here we revealed that lnc-MC could also be directly activated by PU.1 during monocyte/macrophage differentiation. In addition, lnc-MC could antagonize miR-199a-5p and further enhance the role of PU.1 during differentiation.
ceRNAs were recently introduced as RNA transcripts that affect each other's expression levels through competition for their miRNA coregulators. Salmena et al. proposed a ceRNA hypothesis, that transcripts “talk” to each other using miRNA response elements (MREs) as letters of new languages (49). In the past few years, further publications on ceRNAs have extended ceRNA categories into pseudogenes, lncRNAs, circular RNAs (circRNAs), small noncoding RNAs, and even mRNAs (50). For the hematopoietic system, Guo et al. reported that lncRNA-BGL3, which regulates Bcr-Abl-mediated chronic myeloid leukemia (CML), acts as a ceRNA to soak up miR-17, miR-20a, miR-20b, miR-93, miR-106a, and miR-106b and alleviate their repression of PTEN (52). Here we demonstrated that lnc-MC could interact directly and physically with miR-199a-5p, functioning as a ceRNA to release ACVR1B expression. We have shown previously that ACVR1B promotes the activation of the TGF-β signaling pathway by increasing the levels of phosphorylated Smad2 and Smad3, which increases C/EBPα expression and finally facilitates monocyte/macrophage differentiation (20).
On the basis of our results, we constructed a model for the involvement of lnc-MC in the regulation of monocyte/macrophage differentiation (Fig. 7). Following monocytic/macrophagic induction of HL-60 and THP-1 cells as well as HSPCs, the increased PU.1 expression, on one side, represses miR-199a-5p expression (20) and, on the other side, activates lnc-MC expression. The upregulation of lnc-MC further strengthens the role of PU.1 by reducing miR-199a-5p expression and activity, which alleviates the repression of ACVR1B expression. The increased ACVR1B expression promotes monocyte/macrophage differentiation. Taking the findings together, our study demonstrated positive regulation of monocyte/macrophage differentiation by lnc-MC and uncovered an elaborate regulation mechanism composed of PU.1, lnc-MC, miR-199a-5p, and ACVR1B.
FIG 7.
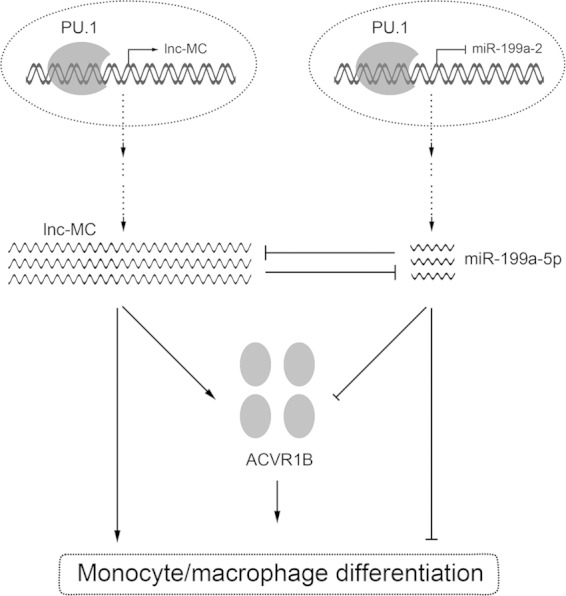
Schematic representation of the involvement of lnc-MC in the regulation of monocyte/macrophage differentiation. lnc-MC, transcriptionally activated by PU.1, facilitates monocyte/macrophage differentiation by acting as a ceRNA to sequester miR-199a-5p and release ACVR1B expression.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shao-Wei Wang of Beijing Hospital and Li-Xia Huang of the First Hospital of Hebei Medical University for providing the UCB samples.
This work was supported by the National Natural Science Foundation of China (grants 31171311 and 30970616, to J.-W.Z.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00429-15.
REFERENCES
- 1.Zhu J, Emerson SG. 2002. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene 21:3295–3313. doi: 10.1038/sj.onc.1205318. [DOI] [PubMed] [Google Scholar]
- 2.Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. 2011. Monocyte trafficking in acute and chronic inflammation. Trends Immunol 32:470–477. doi: 10.1016/j.it.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman AD. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 5.El Gazzar M, McCall CE. 2012. MicroRNAs regulatory networks in myeloid lineage development and differentiation: regulators of the regulators. Immunol Cell Biol 90:587–593. doi: 10.1038/icb.2011.74. [DOI] [PubMed] [Google Scholar]
- 6.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. 1990. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 7.Anderson KL, Smith KA, Conners K, McKercher SR, Maki RA, Torbett BE. 1998. Myeloid development is selectively disrupted in PU.1 null mice. Blood 91:3702–3710. [PubMed] [Google Scholar]
- 8.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. 2003. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat Immunol 4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbauer F, Tenen DG. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 10.Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. 2000. MafB is an inducer of monocytic differentiation. EMBO J 19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK. 2007. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J 26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geissler K, Harrington M, Srivastava C, Leemhuis T, Tricot G, Broxmeyer HE. 1989. Effects of recombinant human colony stimulating factors (CSF) (granulocyte-macrophage CSF, granulocyte CSF, and CSF-1) on human monocyte/macrophage differentiation. J Immunol 143:140–146. [PubMed] [Google Scholar]
- 13.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen CZ, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 15.Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, Tan ZQ, Luo ZM, Yang GH, Shen C, Zhang JW. 2012. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 119:4992–5004. doi: 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- 16.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. 2007. MicroRNAs 17-5p–20a–106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 17.Salvatori B, Iosue I, Mangiavacchi A, Loddo G, Padula F, Chiaretti S, Peragine N, Bozzoni I, Fazi F, Fatica A. 2012. The microRNA-26a target E2F7 sustains cell proliferation and inhibits monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis 3:e413. doi: 10.1038/cddis.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, Yasuda J, Kawai J, Hayashizaki Y, Hume DA, Suzuki H. 2010. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 19.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, Bozzoni I. 2007. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A 104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin HS, Gong JN, Su R, Chen MT, Song L, Shen C, Wang F, Ma YN, Zhao HL, Yu J, Li WW, Huang LX, Xu XH, Zhang JW. 2014. miR-199a-5p inhibits monocyte/macrophage differentiation by targeting the activin A type 1B receptor gene and finally reducing C/EBPα expression. J Leukoc Biol 96:1023–1035. doi: 10.1189/jlb.1A0514-240R. [DOI] [PubMed] [Google Scholar]
- 21.Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Zhang H, Mei Y, Wu M. 2014. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell 53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Ng SY, Bogu GK, Soh BS, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, Esplin MS, Weiss RB, Gleich GJ. 2007. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 109:5191–5198. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. 2009. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W, Yuan B, Flygare J, Lodish HF. 2011. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. 2014. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 123:570–581. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. 2013. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 41:D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. 2014. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 33.Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. 2013. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellon T, Perrotti D, Calabretta B. 1997. Granulocytic differentiation of normal hematopoietic precursor cells induced by transcription factor PU.1 correlates with negative regulation of the c-myb promoter. Blood 90:1828–1839. [PubMed] [Google Scholar]
- 35.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. 2013. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 37.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parks BW, Lusis AJ. 2013. Macrophage accumulation in atherosclerosis. N Engl J Med 369:2352–2353. doi: 10.1056/NEJMcibr1312709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yona S, Jung S. 2010. Monocytes: subsets, origins, fates and functions. Curr Opin Hematol 17:53–59. doi: 10.1097/MOH.0b013e3283324f80. [DOI] [PubMed] [Google Scholar]
- 41.Mao YS, Sunwoo H, Zhang B, Spector DL. 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liz J, Portela A, Soler M, Gomez A, Ling H, Michlewski G, Calin GA, Guil S, Esteller M. 2014. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell 55:138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. 2013. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. 2014. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. 2014. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 48.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. 1994. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol 14:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay Y, Rinn J, Pandolfi PP. 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, Ouyang J, Zhang L, Tan H, Chen R, Huang S, Chen JL. 19 May 2014. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.