Abstract
Aromatic amines are an important group of industrial chemicals, which are widely used for manufacturing of dyes, pesticides, drugs, pigments, and other industrial products. These compounds have been considered highly toxic to human beings due to their carcinogenic nature. Three groups of aromatic amines have been recognized: monocyclic, polycyclic, and heterocyclic aromatic amines. Bacterial degradation of several monocyclic aromatic amines has been studied in a variety of bacteria, which utilizes monocyclic aromatic amines as their sole source of carbon and energy. Several degradation pathways have been proposed and the related enzymes and genes have also been characterized. Many reviews have been reviewed toxicity of monocyclic aromatic amines; however, there is lack of review on biodegradation of monocyclic aromatic amines. The aim of this review is to summarize bacterial degradation of monocyclic aromatic amines. This review will increase our current understanding of biochemical and molecular basis of bacterial degradation of monocyclic aromatic amines.
Keywords: aniline, anthranilic acid, aminophenols, biodegradation, bioremediation
Introduction
Aromatic amines are derivatives of aromatic hydrocarbons containing an amino group, (-NH2) or an amine group (-NH), or a nitrogen (-N) atom in their structures. There are three types of aromatic amines: monocyclic, polycyclic, and heterocyclic, which have been observed in tobacco smoke, diesel exhaust, dyes, pesticides, pharmaceuticals, and polyurethane foams (Stellman, 1998; DeBruin and Josephy, 2002; DeBruin et al., 2002).
Many aromatic amines are recognized as known or suspect human carcinogens, and mutagenicity of aromatic amines has been demonstrated in many test systems, including Big Blue transgenic mice (Layton et al., 1995; Suter et al., 1996; Stellman, 1998; Chung, 2000; DeBruin and Josephy, 2002; DeBruin et al., 2002; Pira et al., 2010). Furthermore, they are potent inducer of the formation of methemoglobinemia in animals and humans (Ohta et al., 1983). Occupational exposure to aromatic amines causes an increased risk of bladder cancer in workers even 30 years after exposure (Pira et al., 2010). Several pesticides including diuron, metobromuron, linuron, isoproturon, chlorotoluron, acetochlor, bentazon, butachlor, metolachlor, amitraz, and vinclozolin may release several monocyclic aromatic amines in soil because of their microbial transformation (Dupret et al., 2011). Cigarette smoke releases several carcinogenic aromatic amines including p-toluidine, 2-naphthylamine, and 4-aminobiphenyl into the ambient air (Stabbert et al., 2003). Bladder cancer is strongly associated with cigarette smoking, probably due to exposure to aromatic amines in tobacco smoke (Pfeifer et al., 2002). Heterocyclic aromatic amines are generally produced in meats or fish when grilled or cooked at high temperatures (Steck et al., 2007). An epidemiological study showed that people who lifelong consume grilled meats and fish have a risk of post-menopausal breast cancer (Steck et al., 2007). Furthermore, higher exposures to heterocyclic aromatic amines may cause presence of DNA adducts, which are associated with carcinogenesis (Turesky, 2007).
Several reviews have been published dealing with toxicity of aromatic amines (Chung et al., 1997; Skipper et al., 2010; Besaratinia and Tommasi, 2013). Despite the fact, monocyclic aromatic amines are distributed throughout the environment including soil and groundwater, there is no review dealing with bacterial degradation of monocyclic aromatic amines. The aim of this review is to summarize bacterial degradation of monocyclic aromatic amines.
Bacterial Degradation of Monocyclic Aromatic Amines
Many bacteria have been isolated and characterized with their ability to mineralize or transform various monocyclic aromatic amines. Table 1 summarizes the role of various monocyclic aromatic amine-degrading bacteria. Bacterial degradation of monocyclic aromatic amines proceeds generally with release of ammonia. Ammonium ions may release either after the ring cleavage (Takenaka et al., 1997) or prior to the ring cleavage (Chang et al., 2003). Several mechanisms have been proposed for mineralization of monocyclic aromatic amines. Bacterial aerobic degradation of monocyclic aromatic amines may be initiated via one of the following mechanisms: (i) A dioxygenase may catalyze ring cleavage of aromatic amine (Takenaka et al., 1997), (ii) Dioxygenation of aromatic amine (Chang et al., 2003), (iii) Deamination of aromatic amine (Qu and Spain, 2011), (iv) Hydroxylation of aromatic amine (Takenaka et al., 2003), (v) Co-ligase mediated activation of aromatic amines to coenzyme A (CoA) thioesters (Schühle et al., 2001), and (vi) Dehalogenation of chlorinated aromatic amine (Hongsawat and Vangnai, 2011). In this section, the bacterial degradation of well-studied monocyclic aromatic amines including aniline, aminophenols, chloroaminophenols, anthranilate, 5-nitroanthranilate, 4-amino-3-hydroxybenzoate, methylanilines, and chloroanilines are discussed.
Table 1.
A list of bacteria involved in degradation of monocyclic aromatic amines.
| Bacteria | Aromatic amine(s) | Mode of action | Reference |
|---|---|---|---|
| Acinetobacter sp. YAA | Aniline | Aerobic, degraded via catechol and its meta-cleavage pathway | Fujii et al. (1997) |
| Delftia tsuruhatensis AD9 | Aniline | Aerobic, degraded via catechol and its meta-cleavage pathway | Liang et al. (2005) |
| Delftia sp. XYJ6 | Aniline | Aerobic, degraded via catechol and its ortho-cleavage pathway | Chengbin et al. (2009) |
| Delftia sp. HY99 | Aniline | Both aerobic and anaerobic, Aerobically degraded via catechol, and anaerobically transformed to 4-aminobenzoic acid | Kahng et al. (2000) |
| Desulfobacterium aniline | Aniline | Anaerobic, degraded via 4-aminobenzoic acid and 4-aminobenzoyl-CoA | Schnell and Schink (1991) |
| Frateuria sp. ANA-18 | Aniline | Aerobic, degraded via catechol and its ortho-cleavage pathway | Murakami et al. (2003) |
| Pseudomonas sp. AW-2 | Aniline | Aerobic, degraded via catechol and its meta-cleavage pathway | Murakami et al. (1998) |
| Burkholderia xenovorans LB400 | 2-Aminophenol | Aerobic, degraded via direct ring cleavage to 2-aminomuconic-6-semialdehyde | Chirino et al. (2013) |
| Pseudomonas sp. AP-3 | 2-Aminophenol | Aerobic, degraded via direct ring cleavage to 2-aminomuconic-6-semialdehyde | Takenaka et al. (2005) |
| Pseudomonas pseudoalcaligenes JS45 | 2-Aminophenol | Aerobic, degraded via direct ring cleavage to 2-aminomuconic-6-semialdehyde | Nishino and Spain (1993) |
| Burkholderia sp. AK-5 | 4-Aminophenol | Aerobic, degraded via 1,4-benzenediol and 1,2,4-benzenetriol | Takenaka et al. (2003) |
| Arthrobacter sp. SPG | 2-Chloro-4-aminophenol | Aerobic, degraded via chlorohydroquinone and hydroquinone | Arora et al. (2014a) |
| Burkholderia sp. RKJ 800 | 4-Chloro-2-aminophenol | Aerobic, degraded via 4-chlorocatechol | Arora et al. (2014b) |
| Acinetobacter sp. ADP1 | Anthranilate | Aerobic, degraded via the catechol pathway | Eby et al. (2001) |
| Azoarcus evansii | Anthranilate | Both aerobic and anaerobic, Aerobically degraded via 2-aminobenzoyl-CoA and anaerobically degraded via benzoyl CoA. | Schühle et al. (2001) |
| Burkholderia cepacia DBO1 | Anthranilate | Aerobic, degraded via the catechol pathway | Chang et al. (2003) |
| Geobacillus thermodenitrificans NG80-2 | Anthranilate | Aerobic, via 3-hydroxyanthranilate | Liu et al. (2010) |
| Nocardia opaca | Anthranilate | Aerobic, degraded via the catechol pathway and the gentisate pathway | Cain (1968) |
| Pseudomonas aeruginosa PAO1 | Anthranilate | Aerobic, degraded via the catechol pathway | Costaglioli et al. (2012) |
| Pseudomonas sp. PAMC 2593 | Anthranilate | Aerobic, degraded via the catechol pathway | Kim et al. (2015) |
| Bradyrhizobium sp. JS329 | 5-Nitroanthranilate | Aerobic, degraded via 5-nitrosalicylic acid | Qu and Spain (2010) |
| Bordetella sp. 10d | 4-Amino-3-hydroxybenzoate | Aerobic, degraded via direct ring cleavage | Orii et al. (2006) |
| Desulfobacula toluolica Tol2 | 4-Methylaniline | Anaerobic, transformed into p-aminophenylacetic acid and phenylacetic acid | Raber et al. (1998) |
| Pseudomonas testosterone | 4-Methylaniline | Aerobic, degraded via 4-methyl-catechol | Raabe et al. (1984) |
| Pseudomonas cepacia CMA1 | 3-Chloro-4-methylaniline | Aerobic, degraded via release of ammonium and chloride ions | Stockinger et al. (1992) |
| Acinetobacter baylyi GFJ2 | 3,4-Dichloroaniline | Aerobic, degraded via 4-chloroaniline | Hongsawat and Vangnai (2011) |
| Alcaligenes faecalis H1 | 3,4-Dichloroaniline | Aerobic, degraded via 4,5-dichloropyrocatechol | Surovtseva et al. (1993) |
| Pseudomonas fluorescens 26-K | 3,4- Dichloroaniline | Aerobic, degraded via 4-amino-2-chlorophenol | Travkin et al. (2003) |
| Rhodococcus rhodochrous CTM | 2-Methylaniline, 4-Chloro-2-methylaniline, and 3-Chloro-2-methylaniline | Aerobic, degraded via corresponding methylcatechols | Fuchs et al. (1991) |
| Brevundimonas diminuta INMI KS-7 | 3-Chloroaniline, 4-Chloroaniline, and 3,4-Dichloroaniline | Aerobic, degraded via corresponding chloropyrocatechols | Surovtseva et al. (1986) |
| Diaphorobacter PCA039 | 4-Chloroaniline | Aerobic, degraded via 4-chlorocatechol | Zhang et al. (2010b) |
| Bacillus megaterium IMT21 | 2,3-Dichloroaniline (2,3-DCA), 2,4-Dichloroaniline (2,4-DCA), 2,5-Dichloroaniline (2,5-DCA), 3,4-Dichloroaniline (3,4-DCA), and 3,5-Dichloroaniline (3,5-DCA) | Aerobic, 2,3-DCA, 2,4-DCA, and 2,5-DCA degraded via dichloroaminophenols whereas 3,4-DCA and 3,5-DCA degraded via dichloroacetanilides | Yao et al. (2011) |
Bacterial Degradation of Aniline
Aniline, which is the simplest aromatic amine, is mainly used for synthesis of dyes, antioxidants, rubbers, pharmaceuticals, and herbicides (Stellman, 1998). In this subsection, pathways for aerobic and anaerobic bacterial degradation of aniline are summarized. Several aerobic bacteria have been isolated and characterized for mineralization of aniline (Fujii et al., 1997; Fukumori and Saint, 1997; Murakami et al., 1998; Liang et al., 2005; Chengbin et al., 2009). These bacteria initiate aniline degradation with formation of catechol that is degraded further via the ortho-cleavage or the meta-cleavage pathway. Pseudomonas putida UCC22 (Fukumori and Saint, 1997), Acinetobacter sp. YAA (Fujii et al., 1997), Pseudomonas sp. AW-2 (Murakami et al., 1998), and Delftia tsuruhatensis AD9 (Liang et al., 2005) metabolize aniline via the meta-cleavage pathway whereas, Frateuria sp. ANA-18 (Murakami et al., 2003) and Delftia sp. XYJ6 (Chengbin et al., 2009) degrade aniline via the ortho-cleavage pathway.
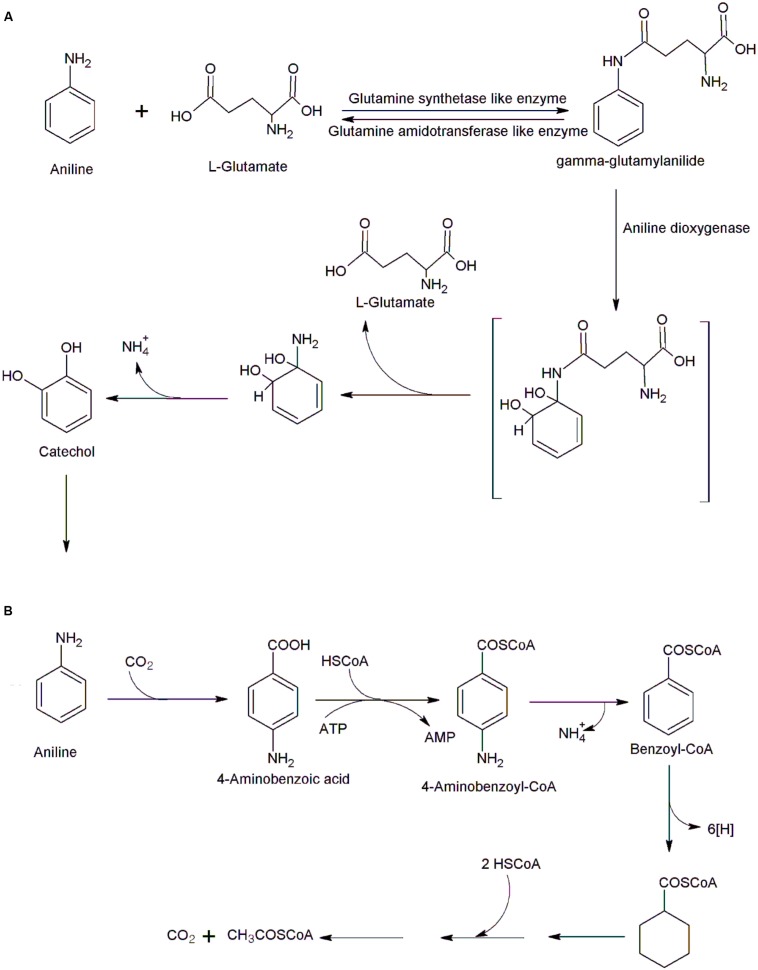
The initial conversion of aniline to catechol is a multistep reaction catalyzed by three enzymes, a glutamine synthetase (GS)-like enzyme, glutamine amidotransferase like enzyme, and an aniline dioxygenase (a large and small subunits of an oxygenase component and a ferredoxin-reductase component; Fujii et al., 1997; Fukumori and Saint, 1997; Murakami et al., 1998; Liang et al., 2005). In the first step, GS like enzyme catalyzed ATP-dependent ligation of L-glutamate to aniline to form gamma-glutamylanilide (Figure 1A; Takeo et al., 2013). The next step, catalyzed by aniline dioxygenase involves conversion of gamma-glutamylanilide into catechol (Takeo et al., 2013). High concentrations of gamma-glutamylanilide are cytotoxic, but the action of another enzyme, glutamine amidotransferase, prevents its accumulation by converting it to aniline (Takeo et al., 2013). Five genes encoding these three enzymes involved in the conversion of aniline to catechol have been identified in a number of bacteria including P. putida UCC22 (Fukumori and Saint, 1997), Acinetobacter sp. YAA (Fujii et al., 1997), Frateuria sp. ANA-18 (Murakami et al., 2003), Delftia acidovorans 7N (Urata et al., 2004), Delftia tsuruhatensis AD9 (Liang et al., 2005) and Delftia sp. AN3 (Zhang et al., 2008). These genes are located on either plasmid or chromosomal DNA. The plasmids of P. putida UCC22 (pTDN1,) and Acinetobacter sp. YAA (pYA1) contain aniline oxidation genes (tdnQTA1A2B or atdA1A2A3A4; Fujii et al., 1997; Fukumori and Saint, 1997). Murakami et al. (2003) expressed all five genes from Frateuria sp. ANA-18 in Escherichia coli and the recombinant bacteria exhibited the aniline oxidation activities. They demonstrated that deletion of tdnA1A2 or tdnQ genes resulted in loss of aniline oxidation activity. Apart of a tdn gene cluster, Frateuria sp. contain two catechol catabolic gene clusters cat1 and cat2 (Murakami et al., 2003). The gene cluster cat1 may involve in the ortho-cleavage pathway of aniline degradation (Murakami et al., 2003). Takeo et al. (2013) reported characterization of the atdA1 gene (encoding the enzyme similar to GS) from Acinetobacter sp. YAA and confirmed that the AtdA1 catalyzes conversion of aniline to gamma-glutamylanilide.
FIGURE 1.
Bacterial degradation of aniline. (A) Initial steps of aniline degradation in Acinetobacter sp. YAA (Takeo et al., 2013), and (B) anaerobic degradation of aniline in Desulfobacterium aniline (Schnell and Schink, 1991).
Anaerobic degradation of aniline was studied in sulfate-reducing bacterium Desulfobacterium aniline (Schnell and Schink, 1991). Initially, aniline is carboxylated to 4-aminobenzoic acid that is transformed to 4-aminobenzoyl-CoA (Figure 1B). The 4-aminobenzoyl-CoA undergoes reductive deamination to form benzoyl-CoA which enters the normal benzoate pathway, to form three acetyl-CoA. Few bacteria are able to degrade aniline under either aerobic or anaerobic conditions; Delftia sp. HY99 is one example of this capability (Kahng et al., 2000). Strain HY99 mineralized aniline acerbically via catechol and transformed aniline to 4-aminobenzoic acid under anaerobic conditions (Kahng et al., 2000).
Bacterial Degradation of Aminophenols
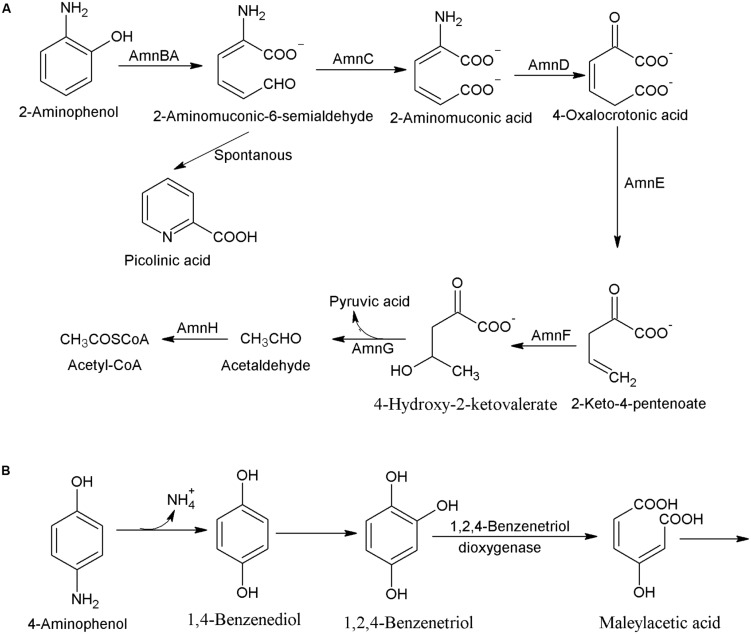
2-Aminophenol and 4-aminophenol are two major isomers of aminophenol, which are widely used for pharmaceuticals, drugs, and dyes. In this subsection, the pathways for bacterial degradation of 2-aminophenol and 4-aminophenol are summarized. The aerobic bacterial degradation of 2-aminophenol initiates with ring-cleavage of 2-aminophenol to 2-aminomuconic-6-semialdehyde by the 2-aminophenol-1,6-dioxygenase (AmnBA; Takenaka et al., 1997). The next step, catalyzed by 2-aminomuconic acid dehydrogenase (AmnC) involves conversion of 2-aminomuconic-6-semialdehyde to 2-aminomuconic acid that deaminates into 4-oxalocrotonic acid by the 2-aminomuconate deaminase (AmnD) with a concomitant release of ammonium (Figure 2A). In next step, 4-oxalocrotonic acid decarboxylase (AmnE) catalyzes conversion of 4-oxalocrotonic acid to 2-keto-4-pentenoate that is transformed to 4-hydroxy-2-ketovalerate by a hydratase (AmnF; Chirino et al., 2013). Next degradation step involves conversion of 4-hydroxy-2-ketovalerate to pyruvic acid and acetaldehyde by a 4-hydroxy-2-oxovalerate aldolase (AmnG; Chirino et al., 2013). Acetaldehyde is further converted to acetyl coenzyme by acetaldehyde dehydrogenase (AmnH). An amn gene cluster involved in the 2-aminophenol degradation has been observed in Pseudomonas sp. AP-3 (Takenaka et al., 2005), P. pseudoalcaligenes JS45 (Nishino and Spain, 1993), P. putida HS12 (Park and Kim, 2000), P. knackmussi B13 (Gaillard et al., 2006), and Burkholderia xenovorans LB400 (Chirino et al., 2013). In Burkholderia xenovorans LB400, the ring cleavage product, 2-aminomuconic-6-semialdehyde is partially converted to picolinic acid that reduced bacterial growth during the 2-aminophenol degradation (Chirino et al., 2013). Literature studies also show the non-enzymatic conversion of 2-aminomuconic-6-semialdehyde to picolinic acid in absence of nicotinamide adenine dinucleotide (Nishino and Spain, 1993).
FIGURE 2.
Bacterial degradation pathway of (A) 2-aminophenol in Pseudomonas sp. AP3 (Murakami et al., 1998), and Burkholderia xenovorans LB400 (Chirino et al., 2013), and (B) 4-aminophenol in Burkholderia sp. AK-5 (Takenaka et al., 2003).
The degradation pathway of 4-aminophenol was studied in Burkholderia sp. AK-5 that utilized it as its sole source of carbon, nitrogen, and energy (Takenaka et al., 2003). Initially, 4-aminophenol is hydroxylated to 1,4-benzenediol that is further hydroxylated to 1,2,4-benzenetriol (Takenaka et al., 2003). The next step, catalyzed by a 1,2,4-benzenetriol dioxygenase involves ring-cleavage of 1,2,4-benzenetriol to maleylacetic acid (Figure 2B). This bacterium expresses a Fe-containing superoxide dismutase and a 2-hydroxy-1,4-benzoquinone reductase that prevents the autoxidation of the labile intermediate 1,2,4-benzenetriol to 2-hydroxy-1,4-benzoquinone (Takenaka et al., 2011).
Bacterial Degradation of Chloroaminophenols
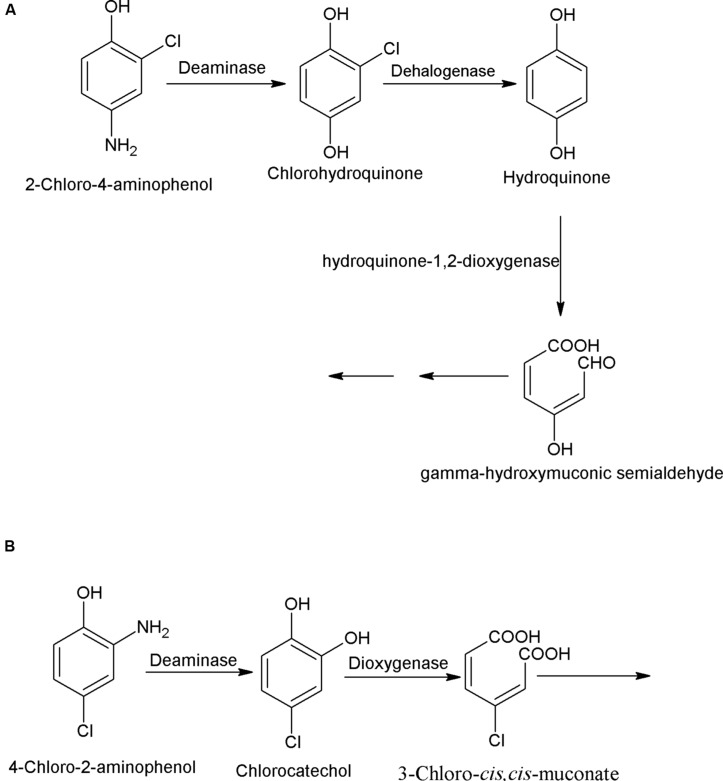
Chloroaminophenols (chlorinated derivatives of aminophenols) are widely used in dye synthesis. In this subsection, pathways for bacterial degradation of 2-chloro-4-aminophenol (2C4AP) and 4-chloro-2-aminophenol (4C2AP) are described. The degradation pathway of 2C4AP was studied in an Arthrobacter sp. SPG that utilized 2C4AP as its sole source of carbon and energy (Arora et al., 2014a). The initial step of the 2C4AP degradation is deaminase-catalyzed hydrolytic deamination of 2C4AP into chlorohydroquinone (CHQ; Arora et al., 2014a). The next step, catalyzed by a CHQ-dehalogenase involves reductive dehalogenation of CHQ to hydroquinone (HQ). Further degradation of HQ proceeds via ring cleavage, catalyzed by HQ- dioxygenase (Figure 3A).
FIGURE 3.
Bacterial degradation pathway of (A) 2-chloro-4-aminophenol in Arthrobacter sp. SPG (Arora et al., 2014a) and (B) 4-chloro-2-aminophenol in Burkholderia sp. RKJ 800 (Arora et al., 2014b).
Another bacterium, Burkholderia sp. RKJ 800, which utilizes 4C2AP as its sole carbon and energy, degrades it via chlorocatechol (Arora et al., 2014b). The 4C2AP degradation pathway is initiated with hydrolytic deamination of 4C2AP to 4-chlorocatechol by a 4C2AP-deaminase (Arora et al., 2014b). The next step, catalyzed by a 4-chlorocatechol-1,2-dioxygenase involves ring cleavage of 4-chlorocatechol into 3-chloro-cis,cis-muconate (Figure 3B).
Bacterial Degradation of 2-Aminobenzoic Acid (Anthranilate)
Anthranilate is a key metabolite of bacterial degradation of several aromatic compounds, including tryptophan (Hayaishi and Stanier, 1951) indole (Fujioka and Wada, 1968), 4-chloroindole (Arora and Bae, 2015), 2-nitrobenzoate (Cain, 1966), quinaldine (Fetzner, 2000), and carbazole (Nojir et al., 2001). Several aerobic degradation pathways have been proposed for bacterial degradation of anthranilate and these pathways are the catechol pathway (Kim et al., 2015), the gentisate pathway (Cain, 1968), the 3-hydroxyanthranilate pathway (Liu et al., 2010) and the 2-aminobenzoyl-CoA pathway (Altenschmidt and Fuchs, 1992).
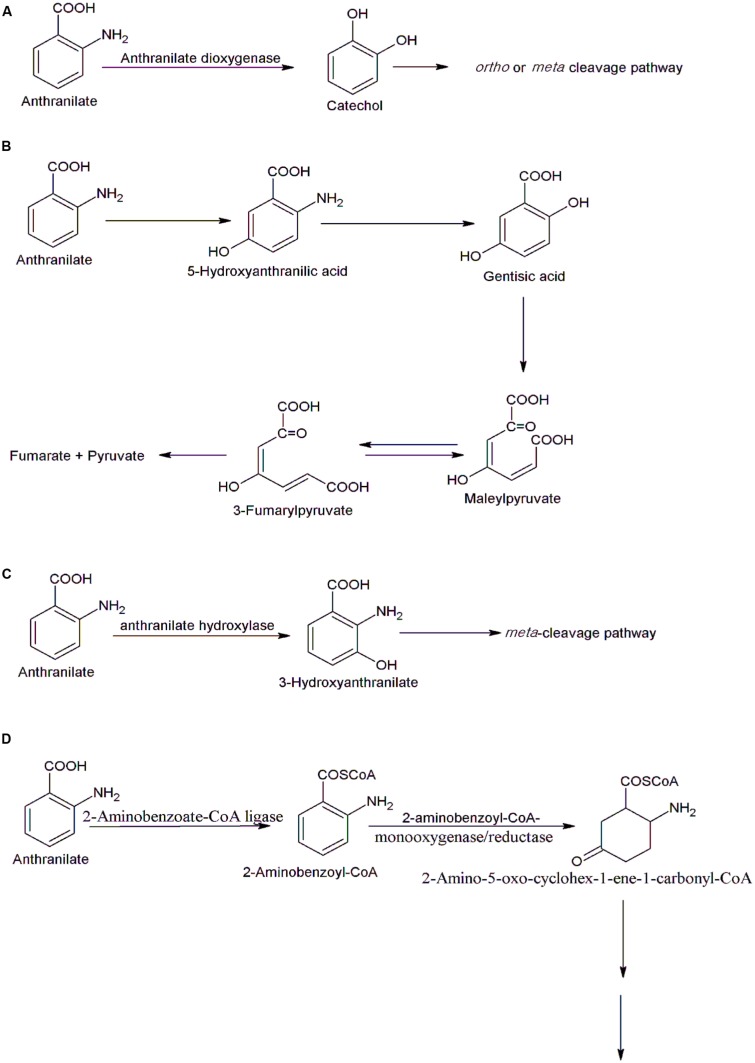
Most of the bacteria degrade anthranilate via the catechol pathway in which anthranilate-1,2-dioxygenase catalyzes conversion of anthranilate to catechol, which is degraded further via the ortho- or meta-cleavage pathway (Chang et al., 2003) (Figure 4A). The enzyme anthranilate-1,2-dioxygenase has been characterized from a number of bacteria (Eby et al., 2001; Chang et al., 2003). In Burkholderia cepacia DBO1, it is a three-component Rieske-type [2Fe-2S] dioxygenase with a reductase, a ferredoxin, and a two-subunit oxygenase (Chang et al., 2003). In Acinetobacter sp. ADP1 (Eby et al., 2001), P. aeruginosa PAO1 (Costaglioli et al., 2012) and P. putida P111, it is a two component complex composed of an oxygenase and a reductase. Kim et al. (2015) cloned and expressed the genes involved in the anthranilate degradation pathway from Pseudomonas sp. PAMC 2593. Two gene clusters have been identified in this strain; the antABC encodes the enzyme anthranilate dioxygenase that converts anthranilate to catechol whereas the catBCA encodes a catechol dioxygenase that cleaves to catechol to cis, cis–muconic acid (Kim et al., 2015).
FIGURE 4.
Various bacterial degradation pathways of anthranilate. (A) The catechol pathway in Burkholderia cepacia DBO1(Chang et al., 2003), Acinetobacter sp. ADP1 (Eby et al., 2001), Pseudomonas aeruginosa PAO1 (Costaglioli et al., 2012), and Pseudomonas sp. PAMC 2593 (Kim et al., 2015), (B) the gentisate pathway in Nocardia opaca (Cain, 1968), (C) the 3-hydroxyanthranilate pathway in Geobacillus thermodenitrificans NG80-2 (Liu et al., 2010), and (D) the 2-aminobenzoyl-CoA pathway in Azoarcus evansii (Schühle et al., 2001).
Nocardia opaca degrades anthranilate via both catechol and gentisate (Cain, 1968). The gentisate pathway is a secondary route of anthranilate degradation in N. opaca. The gentisate pathway proceeds via formation of 5-hydroxyanthranilate, gentisate, maleylpyruvate, and pyruvate (Cain, 1968) (Figure 4B).
The 3-hydroxyanthranilate pathway of anthranilate degradation involves an anthranilate hydroxylase-catalyzed conversion of anthranilate to 3-hydroxyanthranilate that is further degraded via the meta-cleavage (Liu et al., 2010) (Figure 4C). The genes involved in this pathway have been identified and characterized in Geobacillus thermodenitrificans NG80-2 (Liu et al., 2010). The gene encoding anthranilate hydroxylase has been cloned, and expressed in E. coli and the purified protein was FAD-dependent hydroxylase. Two additional enzymes, riboflavin kinase/FMN adenylyltransferase and an FAD reductase, provide FAD for the anthranilate hydroxylase and genes encoding these enzymes were located in the same cluster in which gene encoding hydroxylase was located (Liu et al., 2010).
Another pathway of aerobic degradation of anthranilate was studied in Azoarcus evansii (Altenschmidt and Fuchs, 1992; Schühle et al., 2001). In the initial step, 2-aminobenzoate is activated to 2-aminobenzoyl-CoA by an AMP-forming 2-aminobenzoate-CoA ligase. 2-Aminobenzoyl-CoA is then transformed to a non-aromatic product, 2-amino-5-oxo-cyclohex-1-ene-1-carbonyl-CoA by a flavoenzyme, 2-aminobenzoyl-CoA monooxygenase/reductase (Schühle et al., 2001) (Figure 4D). Further degradation of 2-amino-5-oxo-cyclohex-1-ene-1-carbonyl-CoA occurs by the enzymes of β-oxidation (Schühle et al., 2001). The enzymes involved in the initial steps have been purified and characterized. An enzyme 2-aminobenzoate-CoA ligase is a monomeric protein of 65-kDa whereas 2-aminobenzoyl-CoA monooxygenase/reductase is homodimeric protein of 170 kd. The genes encoding enzymes of anthranilate degradation pathways were located on a small plasmid in Azoarcus evansii (Altenschmidt and Fuchs, 1992). Under anaerobic conditions, cells of Azoarcus evansii converted anthranilate to benzoyl CoA. This is two step reaction catalyzed by a 2-aminobenzoate-CoA ligase and 2-aminobenzoyl-CoA reductase (Schühle et al., 2001). The benzoyl CoA is degraded further via central CoA degradation pathway (Schühle et al., 2001).
Bacterial Degradation of 5-Nitroanthranilate
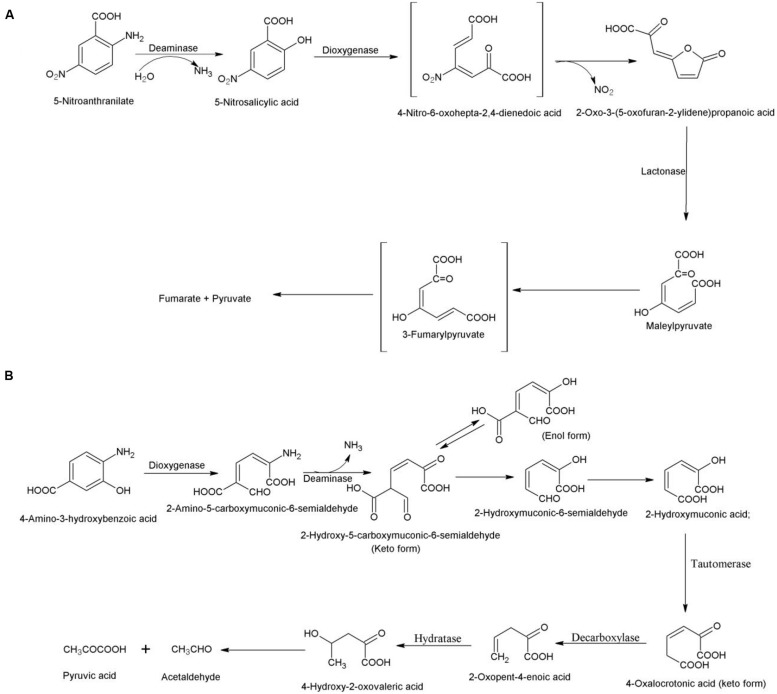
5-Nitroanthranilate is a natural nitroaniline that is produced by the soil bacterium Streptomyces scabiei (the predominant causal agent of common scab of potato in North America; Qu and Spain, 2010). The degradation of this compound was studied in Bradyrhizobium sp. JS329 that utilizes it as its sole source of carbon, nitrogen and energy (Qu and Spain, 2010). The degradation pathway is initiated with hydrolytic deamination of 5-nitroanthranilate to 5-nitrosalicylic acid by 5-nitroanthranilate deaminase. Second step involves 5-nitrosalicylic dioxygenase-catalyzed ring cleavage of 5-nitrosalicylic acid without prior removal of nitro group (Qu and Spain, 2011). The nitro group is eliminated either during the ring fission or immediately following it and the product undergoes spontaneous lactonization. In the next step, lactone is hydrolyzed to maleylpyruvate by a 2-oxo-3-(5-oxofuran-2-ylidene) propanoate lactonase (Figure 5A). The maleylpyruvate is further degraded via 3-fumarylpyruvate (Qu and Spain, 2011).
FIGURE 5.
Bacterial degradation pathway of (A) 5-nitroanthranilate in Bradyrhizobium sp. JS329 (Qu and Spain, 2010) and (B) 4-amino-3-hydroxybenzoate in Bordetella sp. 10d (Orii et al., 2006).
Bacterial Degradation of 4-Amino-3-Hydroxybenzoate
The degradation pathway of 4-amino-3-hydroxybenzoate was studied in 4-amino-3-hydroxybenzoate-assimilating Bordetella sp. 10d (Orii et al., 2006). The degradation pathway is initiated with conversion of 4-amino-3-hydroxybenzoate to 2-amino-5-carboxymuconic-6-semialdehyde by a 2-amino-3-hydroxybenzoate-2,3-dioxygenase (Orii et al., 2006). The next step, catalyzed by 2-amino-5-carboxymuconic-6-semialdehyde deaminase involves deamination of 2-amino-5-carboxymuconic 6-semialdehyde to 5-carboxymuconic-6-semialdehyde that undergoes non-enzymatic carboxylation to form 2-hydroxymuconic-6-semialdehyde (Figure 5B). The hydroxymuconic-6-semialdehyde dehydrogenates to 2-hydroxymuconic acid that is converted to 4-oxalocrotonate by 4-oxalocrotonate tautomerase (Orii et al., 2006). In the next step, 4-oxalocrotonate decarboxylase catalyzes decorboxylation of 4-oxalocrotonate to 2-oxopent-4-enoic acid that is converted to 4-hydroxy-2-oxovaleric acid by a 2-oxopent-4-enoate hydratase. 4-Hydroxy-2-oxovaleric acid is metabolized to pyruvic acid and acetaldehyde (Orii et al., 2006). The enzymes, 2-amino-3-hydroxybenzoate-2,3-dioxygenase and 2-amino-5-carboxymuconic-6-semialdehyde deaminase have been cloned and characterized from Bordetella sp. 10d (Murakami et al., 2004; Takenaka et al., 2009).
Bacterial Degradation of Methylanilines and Their Derivatives
In this subsection, the pathways for the bacterial degradation of methylanilines and their derivatives are summarized. P. testosterone can use 4-methylaniline (p-toluidine) as its sole source of carbon and energy and degraded it via 4-methylcatechol and 2-hydroxy-5-methyl-cis,cis-muconate semialdehyde (Raabe et al., 1984). The initial oxidation of p-toluidine resulted in formation of 4-methylcatechol that ring-cleaved to 2-hydroxy-5-methyl-cis,cis-muconate semialdehyde by a meta-pyrocatechase (Raabe et al., 1984). Anaerobic degradation of 4-methylaniline was studied in anaerobic sulfate-reducing bacterium, Desulfobacula toluolica Tol2 that transformed it into p-aminophenylacetic acid and phenylacetic acid as dead end products (Raber et al., 1998).
Another bacterium, P. cepacia strain CMA1 utilized 3-chloro-4-methylaniline as its sole source of carbon and energy and degraded it via liberation of ammonium and chloride (Stockinger et al., 1992). Authors anticipated that the initial step of degradation of 3-chloro-4-methylaniline in strain CMA1 is an aniline oxygenase-catalyzed reaction with possible formation of chloromethylcatechol that degraded further via an ortho-cleavage pathway (Stockinger et al., 1992). Fuchs et al. (1991) reported co-metabolism of 2-methylaniline, 4-chloro-2-methylaniline, and 3-chloro-2-methylaniline in the presence of ethanol as additional carbon source by two strains of Rhodococcus rhodochrous, wild-type strain CTM and its spontaneous mutant strain CTM-1 (Fuchs et al., 1991). Strain CTM degraded 2-methylaniline via 3-methylcatechol that degraded further via a meta-cleavage pathway (Fuchs et al., 1991). A spontaneous mutant strain CTM-1 lacking the enzyme of meta-cleavage pathway degraded 2-methylcatechol via the ortho-cleavage pathway (Fuchs et al., 1991). Strain CTM degraded 4-chloro-2-methylaniline via 5-chloro-3-methylcatechol that degraded further via the ortho-cleavage pathway (Fuchs et al., 1991). Strain CTM degraded 3-chloro-2-methylaniline via 4-chloro-3-methylcatechol that was further converted to 2-hydroxy-5-chloro-6-oxoheptanoic acid which was accumulated in media (Fuchs et al., 1991).
Bacterial Degradation of Chloroanilines
Chloroanilines including monochloroanilines and dichloroanilines are chloro derivatives of aniline, which are widely used for in the industrial production of dyes, cosmetics, pharmaceutical products, and herbicides. In this subsection, the pathways for the bacterial degradation of monochloroanilines and dichloroanilines are summarized. Many bacteria have been isolated and characterized with their ability to degrade monochloroanilines (4-chloroaniline, 3-chloroaniline, and 2-chloroaniline). Examples are Pseudomonas sp. JL2, Delftia acidovorans CA28, Comamonas testosteroni 12, Acinetobacter baumannii CA2, P. putida CA16, Delftia tsuruhatensis H1, and Acinetobacter baylyi GFJ2 (Latorre et al., 1984; Loidl et al., 1990; Boon et al., 2001; Vangnai and Petchkroh, 2007; Zhang et al., 2010a; Hongsawat and Vangnai, 2011). The initial step of monochloroaniline degradation is an oxidative deamination of monochloroaniline to the corresponding chlorocatechol by aniline dioxygenase. The chlorocatechol is degraded further via either the ortho-cleavage pathway or the meta-cleavage pathway. Most of chloroaniline degrading bacteria degrade aniline via the ortho-cleavage pathway. P. acidovorans CA50 mineralized 2-chloroaniline via the modified ortho-cleavage pathway (Hinteregger et al., 1994). Brevundimonas diminuta INMI KS-7 also degrades 3-chloroaniline and 4-chloroaniline via the ortho-cleavage pathway with formation of 4-chloropyrocatechol, 3-chloromuconic acid, maleylacetic acid and 3-ketoadipic acid (Surovtseva et al., 1986). The aniline degradation via the meta-cleavage pathway has been observed in Diaphorobacter PCA039, which metabolizes 4-chloroaniline via 4-chlorocatechol, 2-hydroxy-5-chloromuconic semialdehyde, 5-chloro-4-oxalocrotonate, and 5-chloro-2-oxo-4-hydroxypentanoate (Zhang et al., 2010b). Similarly, C. testosteroni 12 metabolizes 3-chloroaniline via the meta-cleavage pathway (Boon et al., 2000). Król et al. (2012) showed that chloroaniline dioxygenase of C. testosteroni WDL7 is a multicomponent enzyme consisting of large and small subunits of dioxygenase (encoded by dcaA1, dcaA2), and a reductase (encoded by dcaA3). The large and small subunits of chloroaniline dioxygenase of Comamonas testosteroni WDL7 shows significant amino acid sequence identity with aniline dioxygenase large and small subunits of aniline-utilizing bacteria, Delftia acidovorans 7N, P. putida UCC22, Delftia tsuruhatensis AD9, and Frateuria sp. ANA-18 (Fukumori and Saint, 1997; Murakami et al., 2003; Urata et al., 2004; Liang et al., 2005). Nitisakulkan et al. (2014) reported oxidation of chloroanilines by the P. putida T57 toluene dioxygenase. E. coli expressing the P. putida toluene dioxygenase gene complex (products of the todC1C2BA genes) catalyzes 1,2- and 2,3-dioxygenation of 4-chloroaniline to form 4-chlorocatechol and 2-amino-5-chlorophenol. 5-chloropyrogallol is also formed due to dioxygenation of 4-chlorocatechol.
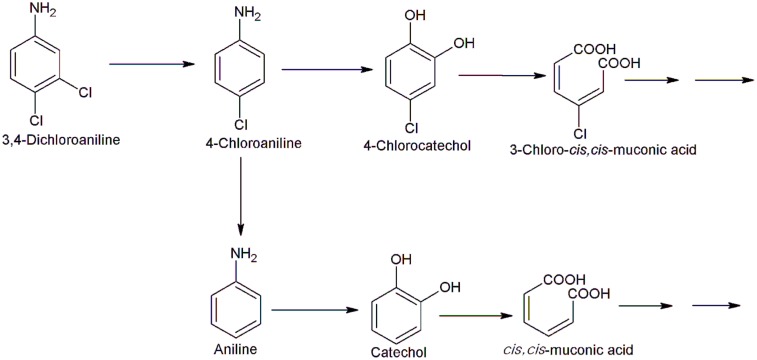
Bacterial degradation of dichloroanilines has also been investigated. Bacillus megaterium IMT21 and Rhodococcus sp. T1-1 utilized five isomers of dichloroaniline including 3,4-dichloroaniline, 2,3-dichloroaniline, 2,4-dichloroaniline, 2,5-dichloroaniline, and 3,5-dichloroanilines as their sole source of carbon and energy (Lee et al., 2008; Yao et al., 2011). Strain IMT21 degrades 2,3-dichloroaniline, 2,4-dichloroaniline and 2,5-dichloroaniline via dichloroaminophenol, and 3,4- and 3,5-dichloroaniline via dichloroacetanilide (Yao et al., 2011). However, no metabolite was detected in degradation of any of the dichloroaniline isomers by strain T1-1 (Lee et al., 2008). Another bacterium, Alcaligenes faecalis H1 mineralized 3,4-dichloroaniline via an initial oxidative deamination with formation of 4,5-dichloropyrocatechol (Surovtseva et al., 1993) whereas P. fluorescens 26-K mineralized 3,4-dichloroaniline via 4-amino-2-chlorophenol through initial dehalogenation and subsequent hydroxylation (Travkin et al., 2003). Brevundimonas diminuta INMI KS-7 degrades 3,4-dichloroaniline via 4,5-dichloropyrocatechol and dichloromuconic acid (Surovtseva et al., 1986). A strain of Pseudomonas sp. degrades 3.4-dichloroaniline in the presence of aniline via 4,5-dichlorocatechol, 3,4-dichloromuconate, 3-chlorobutenolide, 3-chloromaleylacetate, and 3-chloro-4-ketoadipate (You and Bartha, 1982). Branching of degradation pathway of 3,4-dichloroaniline was observed in Acinetobacter baylyi strain GFJ2 that utilized it as its sole source of carbon energy (Hongsawat and Vangnai, 2011). The initial step of degradation involves dehalogenation of 3,4-dichloroaniline to 4-chloroaniline which is further degraded via two different routes (Figure 6). In first route, 4-chloroaniline undergoes dehalogenation to produce aniline, which is further degraded via catechol and the ortho-cleavage pathway. In second route, 4-chloroaniline undergoes dioxygenation to 4-chlorocatechol and ammonia. Further degradation of 4-chlorocatechol proceeds via the ortho-ring cleavage pathway (Hongsawat and Vangnai, 2011).
FIGURE 6.
Bacterial degradation pathway of 3,4-dichloroaniline in Acinetobacter baylyi GFJ2 (Hongsawat and Vangnai, 2011).
Anaerobic degradation of 3,4-dichloroaniline was observed in a Rhodococcus sp. 2 that degraded it through reductive deamination to form 1,2-dichlorobenzene (Travkin et al., 2002). Other metabolites (3,4-dichloroacetanilide and 3,4-dichloro-N-(3,4-dichlorophenyl) benzamide) were also detected as transformation products of this anaerobic reaction (Travkin et al., 2002).
Conclusion
Bacterial degradation of aniline and its chloro and methyl derivatives generally occurs via formation of corresponding catechols that degrade further via either the ortho-cleavage or the meta-cleavage pathway. The mechanism of catechol formation in aniline degradation has recently been postulated and the related genes and enzymes have been well-characterized. Future works on proteomics may increase our understanding towards bacterial degradation of aniline.
Bacterial degradation pathways for aminophenols and chloroaminophenols have also been studied. The degradation of these compounds generally initiated via either the ring cleavage or the hydrolytic deamination. The genes and enzymes involved in the aminophenol degradation have also been characterized whereas the genomics of the degradation pathways of chloroaminophenols have yet not studied.
Diverse mechanisms of the anthranilate degradation have been reported and four aerobic metabolic pathways including the catechol pathway, the gentisate pathway, the 3-hydroxyanthranilate pathway, and the 2-aminobenzoyl-CoA pathway have been proposed. Amongst, the catechol pathway is the most common route for anthranilate degradation.
Little is known about bacterial degradation of other monocyclic aromatic amines. More bacteria should be isolated by the enrichment method using monocyclic aromatic amines as substrates and the biochemical and molecular characterization of biodegradation of monocyclic aromatic amines should be carried out in these bacteria.
Author Contributions
PA collected all the relevant publications, arranged the general structure of the review, drafted the text, and produced figures.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Altenschmidt U., Fuchs G. (1992). Novel aerobic 2-aminobenzoate metabolism. Purification and characterization of 2-aminobenzoate-CoA ligase, localisation of the gene on a 8-kbp plasmid, and cloning and sequencing of the gene from a denitrifying Pseudomonas sp. Eur. J. Biochem. 205 721–727. 10.1111/j.1432-1033.1992.tb16835.x [DOI] [PubMed] [Google Scholar]
- Arora P. K., Bae H. (2015). Biodegradation of 4-chloroindole by Exiguobacterium sp. PMA. J. Hazard. Mater. 284 261–268. 10.1016/j.jhazmat.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Arora P. K., Mohanta T., Srivastava A., Bae H., Singh V. (2014a). Metabolic pathway for degradation of 2-chloro-4-aminophenol by Arthrobacter sp. SPG. Microb. Cell Fact. 13:164 10.1186/s12934-014-0164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P. K., Srivastava A., Singh V. P. (2014b). Novel degradation pathway of 4-chloro-2-aminophenol via 4-chlorocatechol in Burkholderia sp. RKJ 800. Environ. Sci. Pollut. Res. Int. 21 2298–2304. 10.1007/s11356-013-2167-y [DOI] [PubMed] [Google Scholar]
- Besaratinia A., Tommasi S. (2013). Genotoxicity of tobacco smoke-derived aromatic amines and bladder cancer: current state of knowledge and future research directions. FASEB J. 27 2090–2100. 10.1096/fj.12-227074 [DOI] [PubMed] [Google Scholar]
- Boon N., Goris J., De Vos P., Verstraete W., Top E. M. (2000). Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl. Environ. Microbiol. 66 2906–2913. 10.1128/AEM.66.7.2906-2913.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon N., Goris J., De Vos P., Verstraete W., Top E. M. (2001). Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67 1107–1115. 10.1128/AEM.67.3.1107-1115.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B. (1966). Induction of an anthranilate oxidation system during the metabolism of ortho-nitrobenzoate by certain bacteria. J. Gen. Microbiol. 42 197–217. 10.1099/00221287-42-2-197 [DOI] [PubMed] [Google Scholar]
- Cain R. B. (1968). Anthranilic acid metabolism by microorganisms. Formation of 5-hydroxyanthranilate as an intermediate in anthranilate metabolism by Nocardia opaca. Antonie van Leeuwenhoek 34 417–432. 10.1007/BF02046464 [DOI] [PubMed] [Google Scholar]
- Chang H. K., Mohseni P., Zylstra G. J. (2003). Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185 5871–5881. 10.1128/JB.185.19.5871-5881.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chengbin X., Jun N., Hai Y., Xudong S., Jiye H. U. (2009). Biodegradation of aniline by a newly isolated Delftia sp. XYJ6. Chin. J. Chem. Eng. 17 500–505. 10.1016/S1004-9541(08)60237-2 [DOI] [Google Scholar]
- Chirino B., Strahsburger E., Agulló L., González M., Seeger M. (2013). Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS ONE 8:e75746 10.1371/journal.pone.0075746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. T. (2000). Mutagenicity and carcinogenicity of aromatic amines metabolically produced from azo dyes. J. Environ. Sci. Health C 18 51–74. 10.1080/10590500009373515 [DOI] [Google Scholar]
- Chung K. T., Kirkovsky L., Kirkovsky A., Purcell W. P. (1997). Review of mutagenicity of monocyclic aromatic amines: quantitative structure-activity relationships. Mutat. Res. 387 1–16. 10.1016/S1383-5742(97)00019-7 [DOI] [PubMed] [Google Scholar]
- Costaglioli P., Barthe C., Claverol S., Brözel V. S., Perrot M., Crouzet M., et al. (2012). Evidence for the involvement of the anthranilate degradation pathway in Pseudomonas aeruginosa biofilm formation. Microbiologyopen 3 326–339. 10.1002/mbo3.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruin L., Josephy P. (2002). Perspives on the chemical etiology of breast cancer. Environ. Health Perspect. 110 119–128. 10.1289/ehp.02110s1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruin L., Pawliszyn J., Josephy P. (2002). Detection of monocyclic aromatic amines, possible mammary carcinogens, in human milk. Chem. Res. Toxciol. 12 78–82. 10.1021/tx980168m [DOI] [PubMed] [Google Scholar]
- Dupret J. M., Dairou J., Busi F., Silar P., Martins M., Mougin C., et al. (2011). “Pesticide-derived aromatic amines and their biotransformation,” in Pesticides in the Modern World – Pests Control and Pesticides Exposure and Toxicity Assessment ed. Stoytcheva Dr. M. (Rijeka: InTech; ) 601–614. 10.5772/18279 [DOI] [Google Scholar]
- Eby D. M., Beharry Z. M., Coulter E. D., Kurtz D. M., Neidle E. L. (2001). Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1. J. Bacteriol. 183 109–118. 10.1128/JB.183-1.109-118.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzner S. (2000). Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2,4-dioxygenases. Naturwissenschaften 87 59–69. 10.1007/s001140050011 [DOI] [PubMed] [Google Scholar]
- Fuchs K., Schreiner A., Lingens F. (1991). Degradation of 2-methylaniline and chlorinated isomers of 2-methylaniline by Rhodococcus rhodochrous strain CTM. J. Gen. Microbiol. 137 2033–2039. 10.1099/00221287-137-8-2033 [DOI] [PubMed] [Google Scholar]
- Fujii T., Takeo M., Maeda Y. (1997). Plasmid-encoded genes specifying aniline oxidation from Acinetobacter sp. strain YAA. Microbiology 143(Pt 1), 93–99. 10.1099/00221287-143-1-93 [DOI] [PubMed] [Google Scholar]
- Fujioka M., Wada H. (1968). The bacterial oxidation of indole. Biochim. Biophys. Acta 158 70–78. 10.1016/0304-4165(68)90073-1 [DOI] [PubMed] [Google Scholar]
- Fukumori F., Saint C. P. (1997). Nucleotide sequences and regulational analysis of genes involved in conversion of aniline to catechol in Pseudomonas putida UCC22 (pTDN1). J. Bacteriol. 179 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard M., Vallaeys T., Vorhölter F. J., Minoia M., Werlen C., Sentchilo V., et al. (2006). The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188 1999–2013. 10.1128/JB.188.5.1999-2013.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayaishi O., Stanier R. Y. (1951). The bacterial oxidation of tryptophan. III. Enzymatic activity of cell-free extracts from bacteria employing the aromatic pathway. J. Bacteriol. 62 691–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinteregger C., Loidl M., Streichsbier F. (1994). Pseudomonas acidovorans: a bacterium capable of mineralizing 2-chloroaniline. J. Basic Microbiol. 34 77–85. 10.1002/jobm.3620340203 [DOI] [PubMed] [Google Scholar]
- Hongsawat P., Vangnai A. S. (2011). Biodegradation pathways of chloroanilines by Acinetobacter baylyi strain GFJ2. J. Hazard. Mater. 186 1300–1307. 10.1016/j.jhazmat.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Kahng H. Y., Kukor J. J., Oh K. H. (2000). Characterization of strain HY99, a novel microorganism capable of aerobic and anaerobic degradation of aniline. FEMS Microbiol. Lett. 190 215–221. 10.1111/j.1574-6968.2000.tb09289.x [DOI] [PubMed] [Google Scholar]
- Kim D., Yoo M., Kim E., Hong S. G. (2015). Anthranilate degradation by a cold-adapted Pseudomonas sp. J. Basic Microbiol. 55 354–362. 10.1002/jobm.201300079 [DOI] [PubMed] [Google Scholar]
- Król J. E., Penrod J. T., McCaslin H., Rogers L. M., Yano H., Stancik A. D., et al. (2012). Role of IncP-1β plasmids pWDL7::rfp and pNB8c in chloroaniline catabolism as determined by genomic and functional analyses. Appl. Environ. Microbiol. 78 828–838. 10.1128/AEM.07480-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre J., Reineke W., Knackmuss H. J. (1984). Microbial metabolism of chloroanilines: enhanced evolution by natural genetic exchange. Arch. Microbiol. 140 159–165. 10.1007/BF00454919 [DOI] [PubMed] [Google Scholar]
- Layton D. W., Bogen K., Knize M. G., Hatch F. T., Johnson V. M., Felton J. S. (1995). Cancer risk of heterocyclic amines in cooked foods: an analysis and implications for research. Carcinogenesis 16 39–52. 10.1093/carcin/16.1.39 [DOI] [PubMed] [Google Scholar]
- Lee J. B., Sohn H. Y., Shin K. S., Kim J. S., Jo M. S., Jeon C. P., et al. (2008). Microbial biodegradation, and toxicity of vinclozolin, and its toxic metabolite 3,5-dichloroaniline. J. Microbiol. Biotechnol. 18 343–349. [PubMed] [Google Scholar]
- Liang Q., Takeo M., Chen M., Zhang W., Xu Y., Lin M. (2005). Chromosome-encoded gene cluster for the metabolic pathway that converts aniline to TCA-cycle intermediates in Delftia tsuruhatensis AD9. Microbiology 151(Pt 10) 3435–3446. 10.1099/mic.0.28137-0 [DOI] [PubMed] [Google Scholar]
- Liu X., Dong Y., Li X., Ren Y., Li Y., Wang W., et al. (2010). Characterization of the anthranilate degradation pathway in Geobacillus thermodenitrificans NG80-2. Microbiology 156 589–595. 10.1099/mic.0.031880-0 [DOI] [PubMed] [Google Scholar]
- Loidl M., Hinteregger C., Ditzelmüller G., Ferschl A., Streichsbier F. (1990). Degradation of aniline and monochlorinated anilines by soil-born Pseudomonas acidovorans strains. Arch. Microbiol. 155 56–61. 10.1007/BF00291275 [DOI] [Google Scholar]
- Murakami S., Hayashi T., Maeda T., Takenaka S., Aoki K. (2003). Cloning and functional analysis of aniline dioxygenase gene cluster, from Frateuria species ANA-18, that metabolizes aniline via an ortho-cleavage pathway of catechol. Biosci. Biotechnol. Biochem. 67 2351–2358. 10.1271/bbb.67.2351 [DOI] [PubMed] [Google Scholar]
- Murakami S., Nakanishi Y., Kodama N., Takenaka S., Shinke R., Aoki K. (1998). Purification, characterization, and gene analysis of catechol 2,3-dioxygenase from the aniline-assimilating bacterium Pseudomonas species AW-2. Biosci. Biotechnol. Biochem. 62 747–752. 10.1271/bbb.62.747 [DOI] [PubMed] [Google Scholar]
- Murakami S., Sawami Y., Takenaka S., Aoki K. (2004). Cloning of a gene encoding 4-amino-3-hydroxybenzoate 2,3-dioxygenase from Bordetella sp. 10d. Biochem. Biophys. Res. Commun. 314 489–494. 10.1016/j.bbrc.2003.12.111 [DOI] [PubMed] [Google Scholar]
- Nishino S. F., Spain J. C. (1993). Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol. 59 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitisakulkan T., Oku S., Kudo D., Nakashimada Y., Tajima T., Vangnai A. S., et al. (2014). Degradation of chloroanilines by toluene dioxygenase from Pseudomonas putida T57. J. Bios. Bioeng. 117 292–297. 10.1016/j.jbiosc.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Nojir H., Sekiguchi H., Maeda K., Urata M., Nakai S., Yoshida T., et al. (2001). Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J. Bacteriol. 183 3663–3679. 10.1128/JB.183.12.3663-3679.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Shimizu S., Suzuki S. (1983). A screening method for detection of aromatic amines in environmental samples based on their methemoglobin formation activitiesin vitro. Bull. Environ. Contam. Toxicol. 1983 31 278–284. 10.1007/BF01608699 [DOI] [PubMed] [Google Scholar]
- Orii C., Takenaka S., Murakami S., Aoki K. (2006). Metabolism of 4-amino-3-hydroxybenzoic acid by Bordetella sp. strain 10d: a different modified meta-cleavage pathway for 2-aminophenols. Biosci. Biotechnol. Biochem. 70 2653–2661. 10.1271/bbb.60264 [DOI] [PubMed] [Google Scholar]
- Park H. S., Kim H. S. (2000). Identification and characterization of the nitrobenzene catabolic plasmids pNB1 and pNB2 in Pseudomonas putida HS12. J. Bacteriol. 182 573–580. 10.1128/JB.182.3.573-580.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pira E., Piolatto G., Negri E., Romano C., Boffetta P., Lipworth L., et al. (2010). Bladder cancer mortality of workers exposed to aromatic amines: a 58-year follow-up. J. Natl. Cancer Inst. 102 1096–1099. 10.1093/jnci/djq214 [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Denissenko M. F., Olivier M., Tretyakova N., Hecht S. S., Hainaut P. (2002). Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21 7435–7451. 10.1038/sj.onc.1205803 [DOI] [PubMed] [Google Scholar]
- Qu Y., Spain J. C. (2010). Biodegradation of 5-nitroanthranilic acid by Bradyrhizobium sp. strain JS329. Appl. Environ. Microbiol. 76 1417–1422. 10.1128/AEM.02816-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Spain J. C. (2011). Molecular and biochemical characterization of the 5-nitroanthranilic acid degradation pathway in Bradyrhizobium sp. strain JS329. J. Bacteriol. 193 3057–3063. 10.1128/JB.01188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe T., Appel M., Lingens F. (1984). Degradation of p-toluidine by Pseudomonas testosteroni. FEMS Microbiol. Lett. 25 61–64. 10.1111/j.1574-6968.1984.tb01376.x [DOI] [Google Scholar]
- Raber T., Gorontzy T., Kleinschmidt M., Steinbach K., Blotevogel K. H. (1998). Anaerobic degradation and transformation of p-toluidine by the sulfate-reducing bacterium Desulfobacula toluolica. Curr. Microbiol. 37 172–176. 10.1007/s002849900359 [DOI] [PubMed] [Google Scholar]
- Schnell S., Schink B. (1991). Anaerobic aniline degradation via reductive deamination of 4-aminobenzoyl-CoA in Desulfobacterium anilini. Arch. Microbiol. 155 183–190. 10.1007/BF00248615 [DOI] [Google Scholar]
- Schühle K., Jahn M., Ghisla S., Fuchs G. (2001). Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 183 5268–5278. 10.1128/JB.183.18.5268-5278.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper P. L., Kim M. Y., Sun H. L. P., Wogan G. N., Tannenbaum S. R. (2010). Monocyclic aromatic amines as potential human carcinogens: old is new again. Carcinogenesis 31 50–58. 10.1093/carcin/bgp267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabbert R., Schäfer K. H., Biefel C. (2003). Rustemeier K: analysis of aromatic amines in cigarette smoke. Rapid Commun. Mass Spectrom. 17 2125–2132. 10.1002/rcm.1161 [DOI] [PubMed] [Google Scholar]
- Steck S. E., Gaudet M. M., Eng S. M., Britton J. A., Teitelbaum S. L., Neugut A. I., et al. (2007). Cooked meat and risk of breast cancer: lifetime versus recent dietary intake. Epidemiology 18 373–382. 10.1097/01.ede.0000259968.11151.06 [DOI] [PubMed] [Google Scholar]
- Stellman J. (ed.) (1998). “Aromatic Amino Compounds,” in Encyclopaedia of Occupational Health and Safety 4th Edn Vol. IV (Genewa: International Labour Office; ) 104.94–104.118. [Google Scholar]
- Stockinger J., Hinteregger C., Loidl M., Ferschl A., Streichsbier F. (1992). Mineralization of 3-chloro-4-methylaniline via an ortho-cleavage pathway by Pseudomonas cepacia strain CMA1. Appl. Microbial. Biotechol. 38 421–428. 10.1007/bf00170098 [DOI] [Google Scholar]
- Surovtseva E. G., Ivoylov V. S., Karasevich Y. (1986). Metabolism of chlorinated anilines in Pseudomonas diminuta. Mikrobiologiya 55 591–595. [Google Scholar]
- Surovtseva E. G., Sukhikh A. P., Ivoilov V. S. (1993). Isozymes of the pathway for aniline and 4-chloroaniline preparatory metabolism in Alcaligenes sp. Mikrobiologyia 61 99–106. [Google Scholar]
- Suter W., Ahiabor R., Blanco B., Locher F., Mantovani F., Robinson M., et al. (1996). Evaluation of the in vivo genotoxic potential of three carcinogenic aromatic amines using the Big Blue transgenic mouse mutation assay. Environ. Mol. Mutagen. 28 354–362. [DOI] [PubMed] [Google Scholar]
- Takenaka S., Koshiya J., Okugawa S., Takata A., Murakami S., Aoki K. (2011). Fe-superoxide dismutase and 2-hydroxy-1,4-benzoquinone reductase preclude the auto-oxidation step in 4-aminophenol metabolism by Burkholderia sp. strain AK-5. Biodegradation 22 1–11. 10.1007/s10532-010-9369-5 [DOI] [PubMed] [Google Scholar]
- Takenaka S., Murakami S., Shinke R., Hatakeyama K., Yukawa H., Aoki K. (1997). Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and catalytic properties of the purified enzyme. J. Biol. Chem. 272 14727–14732. 10.1074/jbc.272.23.14727 [DOI] [PubMed] [Google Scholar]
- Takenaka S., Okugawa S., Kadowaki M., Murakami S., Aoki K. (2003). The metabolic pathway of 4-aminophenol in Burkholderia sp. strain AK-5 differs from that of aniline and aniline with C-4 substituents. Appl. Environ. Microbiol. 69 5410–5413. 10.1128/AEM.69.9.5410-5413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka S., Sato T., Koshiya J., Murakami S., Aoki K. (2009). Gene cloning and characterization of a deaminase from the 4-amino-3-hydroxybenzoate-assimilating Bordetella sp. strain 10d. FEMS Microbiol. Lett. 298 93–98. 10.1111/j.1574-6968.2009.01699.x [DOI] [PubMed] [Google Scholar]
- Takenaka S., Setyorini E., Kim Y. J., Murakami S., Aoki K. (2005). Constitutive synthesis of enzymes involved in 2-aminophenol metabolism and inducible synthesis of enzymes involved in benzoate, p-hydroxybenzoate, and protocatechuate metabolism in Pseudomonas sp. strain AP-3. Biosci. Biotechnol. Biochem. 69 1033–1035. 10.1271/bbb.69.1033 [DOI] [PubMed] [Google Scholar]
- Takeo M., Ohara A., Sakae S., Okamoto Y., Kitamura C., Kato D., et al. (2013). Function of a glutamine synthetase-like protein in bacterial aniline oxidation via γ-glutamylanilide. J. Bacteriol. 195 4406–4414. 10.1128/JB.00397-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travkin V., Baskunov B. P., Golovlev E. L., Boersma M. G., Boeren S., Vervoort J., et al. (2002). Reductive deamination as a new step in the anaerobic microbial degradation of halogenated anilines. FEMS Microbiol. Lett. 209 307–312. 10.1111/j.1574-6968.2002.tb11149.x [DOI] [PubMed] [Google Scholar]
- Travkin V. M., Solyanikova I. P., Rietjens I. M., Vervoort J., van Berkel W. J., Golovleva L. A. (2003). Degradation of 3, 4-dichloro-and 3, 4-difluoroaniline by Pseudomonas fluorescens 26-K. J. Environ. Sci. Health B 38 121–132. 10.1081/PFC-120018443 [DOI] [PubMed] [Google Scholar]
- Turesky R. (2007). Formation and biochemistry of carcinogenic heterocyclic aromatic amines in cooked meats. Toxicol. Lett. 168 219–227. 10.1016/j.toxlet.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Urata M., Uchida E., Nojiri H., Omori T., Obo R., Miyaura N., et al. (2004). Genes involved in aniline degradation by Delftia acidovorans strain 7N and its distribution in the natural environment. Biosci. Biotechnol. Biochem. 68 2457–2465. 10.1271/bbb.68.2457 [DOI] [PubMed] [Google Scholar]
- Vangnai A. S., Petchkroh W. (2007). Biodegradation of 4-chloroaniline by bacteria enriched from soil. FEMS Microbiol. Lett. 268 209–216. 10.1111/j.1574-6968.2006.00579.x [DOI] [PubMed] [Google Scholar]
- Yao X. F., Khan F., Pandey R., Pandey J., Mourant R. G., Jain R. K., et al. (2011). Degradation of dichloroaniline isomers by a newly isolated strain, Bacillus megaterium IMT21. Microbiology 157(Pt 3) 721–726. 10.1099/mic.0.045393-0 [DOI] [PubMed] [Google Scholar]
- You I. S., Bartha R. (1982). Stimulation of 3,4-dichloroaniline mineralization by aniline. Appl. Environ. Microbiol. 44 678–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. L., He D., Chen J. M., Liu Y. (2010a). Biodegradation of 2-chloroaniline, 3-chloroaniline, and 4-chloroaniline by a novel strain Delftia tsuruhatensis H1. J. Hazard. Mater. 179 875–882. 10.1016/j.jhazmat.2010.03.086 [DOI] [PubMed] [Google Scholar]
- Zhang T., Ren H. F., Liu Y., Zhu B. L., Liu Z. P. (2010b). A novel degradation pathway of chloroaniline in Diaphorobacter sp. PCA039 entails initial hydroxylation. World J. Microbiol. Biotechnol. 26 665–673. 10.1007/s11274-009-0221-1 [DOI] [Google Scholar]
- Zhang T., Zhang J., Liu S., Liu Z. (2008). A novel and complete gene cluster involved in the degradation of aniline by Delftia sp. AN3. J. Environ. Sci. (China: ) 20 717–724. 10.1016/S1001-0742(08)62118-X [DOI] [PubMed] [Google Scholar]